Abstract
Background: In coeliac disease (CD) patients, the dominant DQ2-Α-I-gliadin peptide recognised by CD4 T cells is contained within peptide sequence 57–73 (p57-73) of Α-gliadin. This peptide sequence is also located within a 33-mer protease resistant gliadin fragment and therefore is likely to play an important role in the pathogenesis of CD.
Aims: Our aim was to determine whether a B cell epitope was present within the immunodominant T cell epitope of Α-gliadin and, if so, to elucidate its sequence and determine the importance of deamidation and/or modification of the amino acid at position 65 for IgA binding.
Patients and methods: A cohort of CD patients, disease controls, and healthy individuals were examined. Serum IgA antibodies to the native and modified p57-73 fragment of Α-gliadin were analysed using enzyme linked immunosorbent assays. Peptide scanning experiments were further used to elucidate the B cell epitope.
Results and conclusion: IgA antibodies to p57-73 were found in 29/72 (40.2%) endomysial antibody positive patients, all of whom had CD. The peptide antibody appeared to be present when patients were on a diet containing gluten and declined on a gluten free diet. The p57-73 antibody was very specific for CD (98%) and had a sensitivity of 56%. The amino acid at position 65 was not important for IgA binding but was crucial for T cell recognition of p57-73. Pentapeptide PXPQP emerges as a potentially strong candidate for the IgA binding motif in this region of Α-gliadin. This study shows that a significant proportion of newly diagnosed CD patients have an antibody response to the immunodominant T cell epitope.
Keywords: coeliac disease, antigliadin IgA, T cell epitope, B cell epitope
Coeliac disease (CD) is a common inflammatory disorder of the small intestine that is triggered by gluten in wheat and prolamines in rye and barley.1–3 This condition is strongly associated with the HLA class II molecules DQ2 or DQ8 and is believed to be a T cell mediated autoimmune disorder.1 At least 50 stimulatory peptides in gluten have been isolated from patients with CD.4–8 A unique 33-mer peptide fragment within Α-2-gliadin is thought to be particularly important in initiating the immune response to gluten as it contains a number of T cell stimulatory epitopes that are resistant to intestinal proteases.8 An immunodominant T cell epitope (peptide sequence 57-73) within the 33-mer Α-2-gliadin fragments has been identified in peripheral blood and small intestinal T cells of patients with CD.2,5 Deamidation of glutamine at position 65 in this T cell epitope by tissue transglutaminase is crucial for CD4 T cell recognition and activation.2,5
Patients with untreated CD have elevated levels of antigliadin and antiendomysium antibodies. Tissue transglutaminase is the autoantigen of endomysium antibodies and plays an important role in the pathogenesis of CD, as described previously.9 The role, if any, of antigliadin in the development of CD is less well understood and in general antigliadin antibodies are believed to be markers of small intestinal damage in this disorder but are relatively non-specific.9 In addition, there is significantly less information on the presence and role of B cell epitopes within gliadin in comparison with studies investigating the presence of T cell stimulatory epitopes.2,10–13
The aims of this study were firstly to determine whether anti-p57-73 IgA gliadin antibodies were present in patients in CD, and secondly to establish if peptide length, deamidation, or amino acid substitutions influenced antibody binding to the T cell immunodominant epitope.
METHODS
Patient samples
A total of 192 serum samples were investigated for the presence of anti-p57-73 gliadin antibodies. Subjects studied were divided as follows: 72 CD (41 untreated CD, 25 patients on a successful gluten free diet (GFD), and six CD of unknown dietary status), disease controls (n = 70), assay controls (10 patients with rheumatoid factor, eight patients with IgA paraproteins, and eight patients with a raised polyclonal IgA greater than 5 g/l), and 24 healthy laboratory volunteers. Ethics approval for use of stored serum samples was obtained from the Central Oxfordshire Ethics Committee. All patients had previously been typed for HLA DQ antigens and were either HLA DQ2 or DQ8 positive.
Assay of EMA and gliadin antibodies
IgA endomysium antibodies (EMA) were determined after incubation of serum with primate oesophagus tissue (The Binding Site, UK) by indirect immunofluorescence. Serum IgA was measured using rate nephelometry (Beckman Coulter, UK). Samples with an IgA value of less than 0.1 g/l were excluded from the study. Total IgG and IgA gliadin antibodies were measured by enzyme linked immunosorbent assay (ELISA; Aesku Lab, Germany).
IgA anti-p57-73 Α-gliadin ELISA
Maxisorp microtitre plates (Life Technologies, UK) were coated with 100 μl of peptide 57-73 per well at 5 μg/ml (diluted in Tris buffered saline (TBS), pH 7.5). The 57-73 peptide, used in this study, is a common sequence found in various alpha-gliadins and is identical to the Α-gliadin protein sequence (DNA accession No Q41545) described by Kasarda and colleagues14 and differing from gene derived sequences by substitution of S73 for Leu or Pro. The coated plates were incubated at 37°C for two hours before being left overnight for 16 hours at 4°C. The plates were washed four times with TBS (pH 8.0) with 0.01 mol/l EDTA plus 0.1% Tween 20 (TTBS) using a Dynex plate washer (Thermo Life Sciences, Basingstoke, UK) before being blocked with 100 μl/well of 10% fetal calf serum in TTBS for one hour at room temperature. After four washes, serum samples were diluted 1:100 in TTBS and 100 μl added to the plates in duplicate for one hour at room temperature. After a further wash step, antihuman IgA peroxidase conjugate (Sigma A0295) was diluted 1/2000 in TTBS and 100 μl/well incubated at room temperature for one hour. Substrate (100 μl of 1 mg/ml O-phenylenediamine (Sigma P8287) plus 0.06% H2O2 in 0.1 mol/l sodium citrate buffer) was then added to every well and developed in the dark at room temperature for 15 minutes. Plates were stopped with 50 μl/well of 0.18 M sulphuric acid before being read at 490 nm on a Dynex plate reader (Thermo Life Sciences, Basingstoke, UK). Buffer blanks and positive and negative controls were included on every assay. As the data were not normally distributed, the 95th percentile of the control sample optical densities (ODs) was used as a cut off15 for the p57-73 ELISA (see fig 1 ▶).
Figure 1.
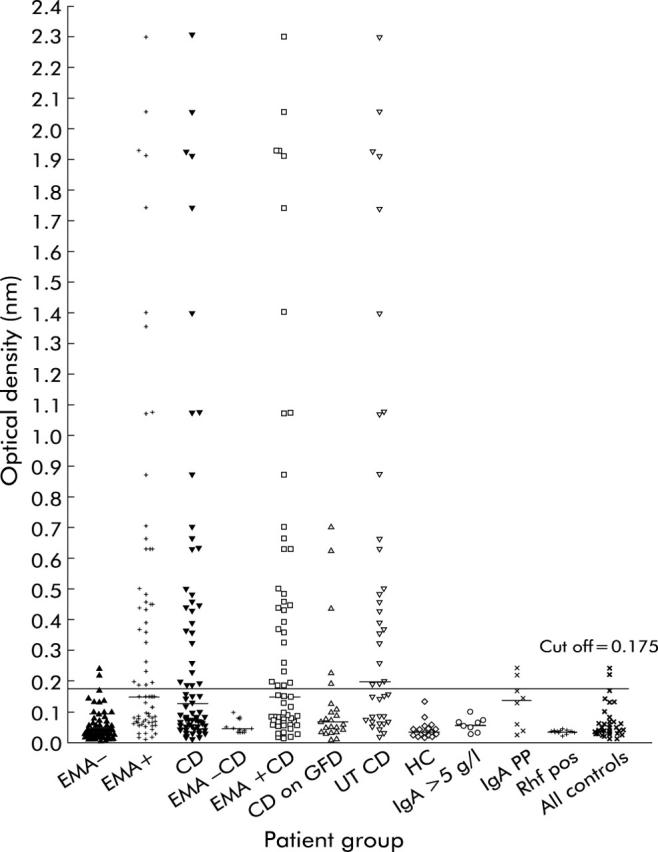
Optical densities (ODs) of samples to peptide 57-73 arranged in groups. Α-gliadin peptide 57-73 antibody levels of coeliac patients, disease controls, and healthy controls. Some samples appear in more than one column if they meet the criteria. Bars in each group represent the median for that group. The 95th percentile of the control sample ODs was used as a cut off (0.175 nm) for the p57-73 ELISA and is indicated by the black line. CD, coeliac disease; GFD, gluten free diet; EMA, endomysial antibody; UT CD, untreated CD; HC, healthy control; Rhf, rheumatoid factor positive; PP, paraprotein.
Amino acid substitution at position 65 of p57-73 ELISA
The assay was carried out using the same protocol as p57-73 ELISA but plates were coated with p57-73 and with lysine, arginine, proline, glutamine, or glutamic acid at position 65 of Α-gliadin at a concentration of 5 μg/ml in TBS. A mixture of peptide positive and negative samples was tested in duplicate on these plates. Each plate was repeated twice and mean results used.
Scanning peptides along the p57-73 region
To elucidate the B cell epitope, nine biotinylated octapeptides were made by frame shifting one amino acid at a time to span the whole p57-73 sequence. Column A of the Streptawell plate (streptavidin coated plates; Roche Diagnostics, UK) was coated with biotinylated p57-73 and the rest of the plate was coated with one of nine short biotinylated peptides (see fig 3 ▶). Peptides were coated in TBS coating buffer at 5 μg/ml and incubated at 4°C for 16 hours. The assay was then carried out as described previously, except the antihuman IgA peroxidase conjugate could be used at a lower concentration of 1:8000 as the biotinylated peptide bound tightly to the plates. A mixture of peptide positive and negative samples were tested in duplicate on these plates. Each plate was repeated twice and mean results used.
Figure 3.
Amino acid sequence of peptide 57-73 and peptides 1–9. Amino acid sequence of peptide 57-73 and biotinylated peptides 1–9 that were used to elicit a potential B cell epitope for IgA binding. PXPQP, the proposed B cell epitope or smaller breakdown products exposed in peptides 5–7, is underlined.
Statistics
One way measures ANOVA and Spearman correlation were performed using GraphPad Prism version 3.00 for Windows (GraphPad Software, San Diego California, USA). Positive and negative predictive values of this assay for CD were calculated using 41 untreated newly diagnosed CD patients that were EMA positive and 103 EMA negative non-CD samples.
RESULTS
Proportion of IgA anti-p57-73 Α-gliadin peptide antibodies from EMA positive and negative subjects
Four different sera (two anti-p57-73 positive and two p57-73 negative) examined over 10 different ELISA assays were used to determine inter- and intra-assay coefficients of variation (CV) for the ELISA. Interassay CV varied from 9.3% to 15.9%, and intra-assay CV varied from 3.2% to 14.7%. To validate p57-73 specificity in this ELISA, direct inhibition studies using Α-gliadin were carried out. Overnight incubation of p57-73 positive sera with three different concentrations of Α-gliadin resulted in maximal 82% inhibition of p57-73 antibody binding in the ELISA (data not shown) suggesting good specificity for gliadin in this assay.
IgA anti-p57-73 gliadin peptide antibodies were found in 29 of 72 EMA positive samples and in two EMA negative samples (fig 1 ▶). The latter samples had an IgA paraprotein. The proportion of patients with untreated CD who had IgA anti-p57-73 gliadin peptides antibodies was 57% whereas only 14% of patients on a GFD had anti-p57-72 gliadin peptide antibodies. All patients on a GFD diet who were gliadin peptide positive had endomysium antibodies. IgA peptide 57-73 antibody levels and IgA gliadin antibody titres were found to correlate with a high degree of significance (r = 0.70, p<0.0001, n = 71, Spearman correlation, data not shown).
The sensitivity and specificity of the anti-p57-73 gliadin peptide antibody for the diagnosis of untreated CD were 56% and 98%, respectively. The positive predictive value was 92% and the negative predictive value was 86%.
Time course of the disappearance of anti-p57-73 antibodies on a GFD
For one patient, sera were available over a 26 month period from diagnosis through to the start and maintenance on a GFD. Five samples spanning this period were tested for EMA and anti-p57-73 IgA antibodies. Patient A was strongly peptide and EMA positive at the point of diagnosis (table 1 ▶). Two months later, the patient had biopsy proven coeliac disease and was on a GFD. Successive samples monitoring the progress of the GFD showed that the anti-p57-73 antibody waned markedly from as early as two months post GFD and became almost negative. EMA took longer to become negative.
Table 1.
Anti-p57-73 antibody values of patient A over time from diagnosis through to a successful gluten free diet
| Date of sample | EMA result | Peptide antibody OD |
| 07.1999 | Positive | 1.742 |
| 09.1999 | Positive | 0.524 |
| 10.1999 | Positive | 0.278 |
| 03.2000 | Positive | 0.125* negative |
| 09.2001 | Negative | 0.078* negative |
The optical density (OD) of the cut off level was 0.175 nm.
EMA, endomysial antibody.
Effect of a truncated peptide on anti-p57-73 gliadin antibody binding
A shorter peptide (62-68), from the centre of peptide 57-73, was used in the ELISA to see whether this shortened sequence bound IgA antibodies. IgA antibodies (n = 47) from the same subject binding to sequences p57-73 and p62-68 of Α-gliadin gave equivalent OD values suggesting that the binding motif of 57-73 incorporates the peptide 62-68 (data not shown)
Binding in the p57-73 Α-gliadin ELISA with alternative amino acids at position 65
The sequence of peptide 57-73 of Α-gliadin recognised by CD4 T cells is deamidated at position 65, with a glutamine altered to glutamic acid (3,6). To ascertain whether the amino acid at position 65 is also important for IgA antibody binding, experiments with four different amino acids (proline, lysine, arginine, and glutamic acid) at position 65 (p65) of peptide 57-73 were carried out. Figure 2 ▶ shows ODs of 18 sera samples, all anti-p57-73 antibody positive, against the five peptides with different p65 amino acids. No significant difference between the p65 amino acid substituted peptides could be demonstrated (p = 0.266, n = 18, one way ANOVA).
Figure 2.
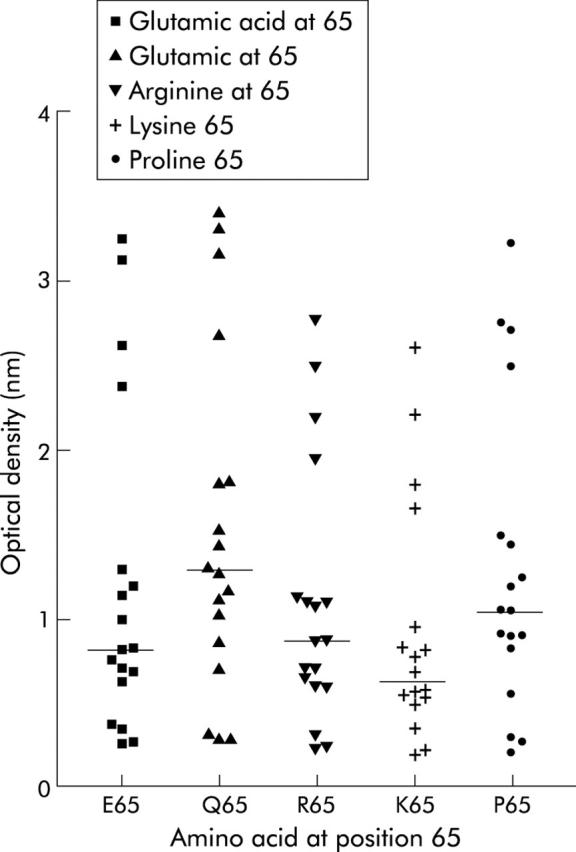
Optical densities of peptide positive and negative samples against peptides with different amino acids at position 65. Binding of 18 different sera representing p57-73 peptide positive and negative samples were compared in five different ELISAs where the amino acid at position 65 of p57-73 was altered. Bars indicate median values.
Scanning the p57-73 region of Α-gliadin to determine specific B cell epitopes
To identify the antibody binding region of the peptide 57-73, nine octapeptides, spanning the whole 57-73 sequence, were examined in the ELISA (fig 3 ▶). Any significant changes in binding should elucidate specific binding sequences and thus the B cell epitopes. Peptides were biotinylated to eliminate ELISA plate coating variability. An initial experiment ascertained that biotinylating peptides along 57-73 did not alter binding of anti-p57-73 IgA antibodies. The distribution of sample binding and the cut off values remained the same for both unbiotinylated and biotinylated peptides (data not shown). Thirty nine samples (29 EMA positives and 10 EMA negatives) with a range of anti-peptide 57-73 antibody levels were tested against these nine peptides in ELISAs. Figure 4 ▶ shows binding to the nine biotinylated peptides for a range of representative samples with ODs less than 2. Figure 4 ▶ demonstrates that, in general, binding to different peptides appears uniform across the whole sequence, inferring a short common motif or a polyclonal response to the region.
Figure 4.
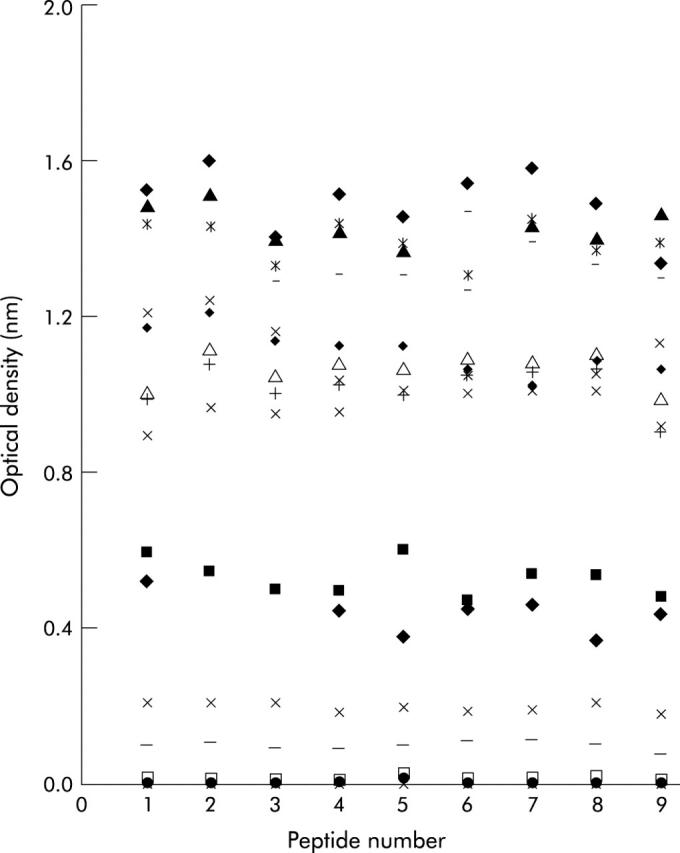
Comparing optical densities of the same sera against nine octapeptides of gliadin 57-73. Binding of 18 different sera (a range of representative sera with optical densities less than 2) to nine biotinylated octapeptides peptides spanning peptide 57-73 is shown.
DISCUSSION
In this study, we found that 57% of untreated patients with CD had IgA gliadin peptide antibodies against the immunodominant T cell epitope 57-73 (DQ2-Α-I-gliadin). IgA antibodies to this epitope are highly specific for untreated CD, unlike IgA antibodies to whole gliadin and to different epitopes of Α-gliadin.2,10 The ELISA showed good positive (92%) and negative (86%) predictive values and was highly specific for CD (98%). The limited sensitivity of the anti-p57-73 gliadin peptide antibodies implies that it will not be useful as a diagnostic screening tool for CD. Serial serum samples obtained from a single patient showed that the p57-73 IgA antibody disappeared on a GFD. Further analysis is required to establish whether this assay has a role in monitoring compliance with a GFD.
Although T cells require deamidation of glutamine at position 65 to recognise the Α-2-gliadin 57-73 peptide sequence, it is unclear whether deamidation plays an important role in antibody recognition. Our data showing that deamidation of glutamine at position 65 does not influence IgA binding is in agreement with the findings of Aleanzi and colleagues13 who however showed that deamidation of glutamine within this immunodominant epitope did enhance IgG binding to this peptide in some patients. A number of groups have reported that deamidation of glutamine residues in γ-gliadin promotes IgG and IgA gliadin peptide binding,2,13 suggesting that the role of this process in antibody recognition is complex and may vary for different gliadins and different antibody isotypes.
Substitutions of glutamine at position 65 by either positively or negatively charged amino acids did not have a statistically significant effect on IgA binding to the 57-73 gliadin peptide sequence, supporting the view that IgA binding is not markedly influenced by the presence of glutamine deamidation at position 65.
Peptide scanning experiments identified the hexapeptide QXQPFP (X being P, Q, or L) shown previously2,13 to be a significant IgA and IgG epitope, as one potential epitope for IgA binding within p57-73 (fig 3 ▶). QXQPFP occurs only at the beginning of p57-73 (fig 3 ▶) and the uniform binding observed across the whole p57-73 sequence infers another common motif or a polyclonal antibody response (fig 4 ▶) to this region of Α-gliadin.
Both the 33-mer peptide8 and the shorter p57-73 peptide studied here exhibit highly repetitive sequences. The longer 33-mer has three repetitive amino acid motifs PXPQP (with X being either phenylalanine or tyrosine) repeated four times, LPYPQP repeated three times, and PQPQL repeated three times along its length. Of these, PXPQP is repeated twice within p57-73 whereas LPYPQP is present only once and PQPQL is not present (as the deamidated form of p57-73 was used in the scanning experiments). Figure 3 ▶ shows that 9/9 peptides had at least three of the five amino acids of PXPQP, 8/9 had four, and 6/9 had the complete motif, making PXPQP a potentially strong candidate as the IgA binding motif in this region of Α-gliadin. It is possible that due to its repetitive nature within p57-73, this sequence was not elucidated by the scanning experiment as the motif fragments in peptides 5–7 (fig 3 ▶) may have been enough to sustain antibody binding at the maximal level.
Identification of T cell epitopes and the primary initiator peptide8 opens up the possibility of peptide immunotherapy.16,17 Whole Α-gliadin has been administered intranasally to successfully inhibit gliadin specific T cell responses in a murine model of CD.18 The finding that a peptide, which contains an immunodominant T cell epitope, can elicit an antibody response raises the possibility that peptide immunotherapy could give rise to inappropriate immune responses, such as those described in recent trials involving therapeutic vaccination in patients with Alzheimer’s disease.17,19 Our data show for the first time that antibody responses in patients with untreated CD are directed against the gliadin peptide fragment that contains the immunodominant T cell epitope.
Finally, this work adds to the evidence that antibody responses against immunodominant T cell epitopes do occur in human autoimmune disease.20,21 The finding that this gliadin peptide fragment contains both B and T cell epitopes suggests that it plays an important role in the pathogenesis of coeliac disease as B and T cell interactions play a vital role in induction of immune responses.
Acknowledgments
We would like to thank Jackie Wozniak, Stoke Mandeville Hospital, for performing the IgA and IgG gliadin assays, and the staff at the Department of Immunology, Churchill Hospital, Oxford, for performing the EMA and IgA assays.
Abbreviations
CD, coeliac disease
p57-73, Α-gliadin peptide sequence 57-73
GFD, gluten free diet
EMA, endomysial antibody
Rhf, rheumatoid factor positive
PP, paraprotein
CV, coefficient of variation
ELISA, enzyme linked immunosorbent assay
TBS, Tris buffered saline
OD, optical density
REFERENCES
- 1.Fasano A , Catassi C. Current approaches to diagnosis and treatment of celiac disease: an evolving spectrum. Gastroenterology 2001;120:636–51. [DOI] [PubMed] [Google Scholar]
- 2.Osman AA, Gunnel T, Dietl A, et al. B cell epitopes of gliadin. Clin Exp Immunol 2000;121:248–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson RP, Degano P, Godkin AJ, et al. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med 2000;6:337–42. [DOI] [PubMed] [Google Scholar]
- 4.Sjöström H , Lundin KEA, Molberg Ø, et al. Identification of a gliadin T cell epitope in coeliac disease: general importance of gliadin deamidation for intestinal T cell recognition. Scand J Immunol 1998;48:111–15. [DOI] [PubMed] [Google Scholar]
- 5.Arentz-Hansen H , Körner R, Molberg Ø, et al. The intestinal T cell response to a-gliadin in adult celiac disease is focused on a single deamidates glutamine targeted by tissue transglutaminase. J Exp Med 2000;191:603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van de Wal Y , Kooy Y, Van Veelen P, et al. Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc Natl Acad Sci U S A 1998;95:10050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van de Wal Y , Kooy Y, Van Veelen P, et al. Glutenin is involved in the gluten-driven mucosal T cell response. Eur J Immunol 1999;29:3133–9. [DOI] [PubMed] [Google Scholar]
- 8.Shan L , Molberg Ø, Parrot I, et al. Structural basis for gluten intolerance in celiac sprue. Science 2002;297:2275–9. [DOI] [PubMed] [Google Scholar]
- 9.Murray JA. The widening spectrum of celiac disease. Am J Clin Nutr 1999;69:354–65. [DOI] [PubMed] [Google Scholar]
- 10.Lähdeaho M - L, Vainio E, Lehtinen M, et al. Activation of celiac disease immune system by specific A-gliadin peptides. Cereal Chem 1995;72:475–9. [Google Scholar]
- 11.Ten Dam M , Van de Wal Y, Mearin ML, et al. Anti-A-gliadin antibodies (AGA) in the serum of coeliac children and controls recognize an identical collection of linear epitopes of A-gliadin. Clin Exp Immunol 1998;114:189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krupic̆kova S , Tuc̆ková L, Flegelová Z, et al. Identification of common epitopes on gliadin, enterocytes and calreticulin recognized by anti-gliadin antibodies of patients with coeliac disease. Gut 1999;44:168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aleanzi M , Demonte AM, Esper C, et al. Celiac disease: Antibody recognition against native and selectively deamidated gliadin peptides. Clin Chem 2001;47:2023–8. [PubMed] [Google Scholar]
- 14.Altman DG. Practical statistics for medical research. London, UK: Chapman and Hall, 1991.
- 15.Schuppan D , Hahn EG. Gluten and the gut—lessons for immune regulation. Science 2002;297:2218–20. [DOI] [PubMed] [Google Scholar]
- 16.Mowat AM. Coeliac disease—a future for peptide therapy? Lancet 2000;356:270–1. [DOI] [PubMed] [Google Scholar]
- 17.Maurano F , Siciliano RA, De Giulio B, et al. Intranasal administration of one alpha gliadin can downregulate the immune response to whole gliadin in mice. Scand J Immunol 2001;53:290–5. [DOI] [PubMed] [Google Scholar]
- 18.Nicoll JAR, Wilkinson D, Holmes C, et al. Neuropathology of human Alzheimer disease after immunization with amyloid-β peptide: a case report. Nat Med 2003;9:448–52. [DOI] [PubMed] [Google Scholar]
- 19.Shimoda A , Nakamura M, Ishibashi H, et al. Molecular mimicry of mitochondrial and nuclear autoantigens in primary biliary cirrhosis. Gastroenterology 2003;124:1915–25. [DOI] [PubMed] [Google Scholar]
- 20.Wucherpfenning KW, Catz I, Hausmann S, et al. Recognition of the immunodominant myelin basic protein peptide by autoantibodies and HLA-DR2-restricted T cell clones from multiple sclerosis patients. J Clin Invest 1997;100:1114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasarda DD, Okita TW, Bernardin JE, et al. Nucleic acid (cDNA) and amino acid sequences of alpha-type gliadins from wheat (Triticum aestivum). Proc Natl Acal Sci USA81:4712–16. [DOI] [PMC free article] [PubMed]



