Abstract
Background/Aims: Hepatitis C virus (HCV) infection results in a high frequency of chronic disease. The aim of this study was to identify early prognostic markers of disease resolution by performing a comprehensive analysis of viral and host factors during the natural course of acute HCV infection.
Methods: The clinical course of acute hepatitis C was determined in 34 consecutive patients. Epidemiological and virological parameters, as well as cell mediated immunity (CMI) and distribution of human leukocyte antigens (HLA) alleles were analysed.
Results: Ten out of 34 patients experienced self-limiting infection, with most resolving patients showing fast kinetics of viral clearance: at least one negative HCV RNA test during this phase predicted a favourable outcome. Among other clinical epidemiological parameters measured, the self-limiting course was significantly associated with higher median peak bilirubin levels at the onset of disease, and with the female sex, but only the latter parameter was independently associated after multivariate analysis. No significant differences between self-limiting or chronic course were observed for the distribution of DRB1 and DQB1 alleles. HCV specific T cell response was more frequently detected during acute HCV infection, than in patients with chronic HCV disease. A significantly broader T cell response was found in patients with self-limiting infection than in those with chronic evolving acute hepatitis C.
Conclusion: The results suggest that host related factors, in particular sex and CMI, play a crucial role in the spontaneous clearance of this virus. Most importantly, a negative HCV RNA test and broad CMI within the first month after onset of the symptoms represent very efficacious predictors of viral clearance and could thus be used as criteria in selecting candidates for early antiviral treatment.
Keywords: hepatitis C, acute infection, chronicity rate, immune response, HLA
Acute hepatitis C virus (HCV) infection is usually a mild and asymptomatic disease,1 yet it is characterised by a high rate of chronic evolution, approaching 90% according to some studies.2–7 Nevertheless, complete recovery from acute HCV infection has been reported to occur in up to 30–50% of cases with community acquired symptomatic acute hepatitis C.8–15 It is still not clear whether this high rate of chronicity reflects differences in the viral or host characteristics. 3,6–9,11,16–22 The host immunogenetic background9,17,18,23–26 and, particularly, the characteristics of the host immune response in the initial phase of the infection are emerging as key factors in determining disease evolution. Data from several groups have shown that virus specific T helper cell and cytotoxic T lymphocyte (CTL) responses are associated with control of acute HCV viremia and clearance of the virus in the chimpanzee model of infection27–32 as well as in humans.8,10–12,33–37 However, T cell responses have been studied using a limited set of previously mapped peptides in a small number of patients.
Understanding the virus host interactions, which enable a proportion of patients with acute infection to clear HCV, is probably a key to the development of more effective treatment and prevention strategies. To this end, we enrolled a large cohort of HCV acutely infected individuals and evaluated clinical epidemiological parameters, distribution of human leukocyte antigens (HLA) class II alleles, breadth and magnitude of CD4+ and CD8+ T cell responses in patients with acute hepatitis C in both those who resolve the infection and those who develop chronic infection. Subjects with already established chronic HCV infection and healthy bone marrow donors were also included in the analysis to better define the role of cellular mediated immunity (CMI) and of the HLA class II polymorphism.
PATIENTS AND METHODS
Study population
The study cohort included 34 consecutive patients with acute hepatitis C (seven women, 27 men, median age 30 years, range 20–61 years), referred to infectious disease clinics or attending drug dependency units, between March 1999 and August 2001. Diagnosis of acute HCV infection was based on (1) high levels of serum alanine aminotransferase (ALT) at least 10 times above the upper limit of normal; (2) seroconversion assessed by third generation enzyme linked immunosorbent assay, or anti-HCV positivity at the time of the diagnosis with an anti-HCV negative test in the previous 12 months; (3) presence of HCV RNA in at least the first serum sample, and (4) sudden onset of liver disease symptoms. Twelve subjects with an anti-HCV positive test at the time of the diagnosis, but without an anti-HCV negative test in the previous 12 months were also diagnosed as having acute hepatitis C on the basis of their clinical state, anamnestic characteristics, and a retrospective diagnostic confirmation consisting in a change in the serological profile, with an increasing number of reactivities against further HCV antigens15,38,39 by recombinant immunoblot assay (RIBA). Only one patient, who showed anti-HCV and HCV RNA positive tests, had normal ALT values (20 U/L) at enrolment in the study. Slightly altered ALT values were observed corresponding to new positive HCV RNA detections during the follow up. This patient was diagnosed as having acute HCV infection because he was symptomatic (but not jaundiced), had an anti-HCV negative test 10 months before the disease onset, and reported a recognised risk factor for the acquisition of the infection in the two months before the disease onset. Furthermore in the RIBA test performed on follow up sera, a progressive, increasing number of reactivities against the HCV antigens were detected in this patient. Alternative causes of acute hepatitis, such as other viral infection, autoimmunity, alcohol, drugs, and toxins were excluded. Patients with concomitant immunological disorders or with HIV coinfection were also excluded from the study.
Patients were scheduled to undergo a medical visit and to take a blood sample for biochemical, virological, and immunological assessment at the onset of the disease (month 0: start of the observation) and after 1, 3, 6, 12 months, and at four month intervals for a total of 24 months or until the patients eventually underwent antiviral therapy because of progression to chronic hepatitis. All observations reported in this study refer to the natural history of the infection (that is, in the absence of therapy) unless otherwise specified.
Self-limiting HCV infection was defined as normalisation of serum ALT levels in association with clearance of serum HCV RNA within six months after the onset of the disease. At least two successive negative HCV RNA measurements 6–12 months apart from the first negative result was used as further criterion for the diagnosis of self-limiting HCV infection. Patients who showed detectable HCV RNA in serum for more than six months with or without persistent increased ALT levels were considered to have developed a chronic infection.
A cohort of 56 patients with histological confirmed chronic HCV related liver disease was used to compare the profile and the strength of the T cell immune response in late chronic versus acute infection. Blood samples from 40 healthy unrelated bone marrow donors (anti-HCV negative and without evidence of liver disease) were used as controls to represent HLA distribution in the uninfected population.
All patients gave informed consent before entering the study. The protocol and all the procedures of the study were conducted in conformity with the ethical guidelines of the Declaration of Helsinki.
Anti-HCV antibody testing
Anti-HCV antibodies were measured by Microparticle Enzyme Immunoassay (MEIA) HCV version 3 (AXSYM System, Abbott Diagnostics, Wiesbaden, Germany). Anti-HCV positivity was confirmed by second generation RIBA (Deciscan, Diagnostic Sanofi Pasteur, Marnes la Coquette, France).
HCV RNA determination
Levels of serum HCV RNA were determined by using Amplicor HCV Monitor Kit version 3 (Roche Diagnostic Systems, Branchburg, NJ, USA; detection limit: 600 UI/ml).
When viral load was lower than 600 UI/ml, HCV RNA was evaluated by a qualitative PCR with a detection limit of 100 copies/ml. HCV RNA was isolated from serum by Qiamp viral RNA kit (Qiagen, Hilden, Germany). Five μl of purified RNA was used to generate cDNA in a 20 μl reaction mixture containing 10 U of RNase inhibitor (Promega Inc, Madison, WI, USA), 50 ng random examer, and 200 μM (each) deoxynucleoside triphosphate. After incubation at 70°C for 10 minutes, reverse transcription was carried out at 37°C for three hours with 50 U of Moloney murine leukaemia virus reverse transcriptase (Life Technologies, Gaithersburg, MD, USA). cDNA was amplified with primers for 5′UTR by nested PCR. Primer sequences were as follows: B1 (outer sense) 5′ AAC TAC TGT CTT CAC GCA GAA 3′; A1 (outer reverse) 5′ GAT GCA CGG TCT ACG AGA CCT C 3′; B2 (inner sense) 5′ ATG GCG TTA GTA GTG 3′; A2 (inner reverse) 5′ GCG ACC CAA CAC TAC TCG GCT 3′. PCR mixture contained 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 200 μM (each) deoxynucleoside triphoshate, 0.5 μM primers, and 1.75U of Taq Gold Polymerase (Roche Molecular Systems Inc, Branchburg, NJ, USA) in a total volume of 50 μl. One tenth of the product from the first round of amplification was used for the second PCR round. Cycling parameters consisted of 35 cycles of 1 minute 95°C (plus 15 minutes in the first cycle); 30 seconds at 45°C and 1 minute at 72°C for the first round and 30 cycles at 95°C (plus 15 minutes in the first cycle); 30 seconds at 50°C and 1 minute at 72°C for the second round.
HCV genotype in PCR positive sample was assessed using a reverse hybridisation line probe assay (Inno-LiPA HCV Kit Innogenetics, Gent, Belgium).
HLA typing
Genomic DNA was isolated from peripheral blood monuclear cells (PBMC) according to salting out method.40 HLA typing of DRB1 and DQB1 class II alleles was performed by molecular tests in low resolution. HLA class II DRB1*11, DRB1*07, DQB1*03, and DQB1*05 alleles were typed also in high resolution. Low resolution molecular typing of HLA class II alleles was performed by reverse hybridisation with sequence specific oligonucleotide probes following amplification of the second exon of the DRB1 and DQB1 genes (PPCR-SSO) with biotinylated primers (Inno Lipa, Innogenetics, Ghent, Belgium).41 High resolution molecular typing of HLA-DRB1 and HLA-DQB1 alleles was performed by PCR with sequence specific primers (PCR-SSP)42 using a commercial kit (Dynal Biotech Ltd, Bromborough, UK) according to the manufacturer’s instruction.
Interferon (IFN) γ enzyme linked immunospot (ELIspot)
Synthetic peptides (20 amino acids in length and overlapping by 10 residues) were derived from the sequence of HCV BK strain.43 All peptides were synthesised with free N-terminal amine and free C-terminal carboxylate, using standard Fmoc solid phase methods44 and were purified by preparative high pressure liquid chromatography. The peptides were reconstituted in 100% DMSO at 20 mg/ml and used in the assay at 5 μg/ml. To facilitate the analysis, the 216 peptides were combined in seven pools covering Core, NS3 protease (NS3p), NS3 helicase (NS3h), NS4, NS5A, and NS5B (split in two pools NS5B-I and NS5B-II).
Polyvinyl difluoride membrane plates (96 well; Millipore MAIPS 4510) were coated with 10 μg/ml of antihuman IFN-γ monoclonal antibody (MAbTech clone# 1-D1K) in sterile phosphate buffered saline (PBS). Cryopreserved PBMC were thawed, recovered overnight at 37°C, 5% CO2 and added to the wells at two different cell concentrations (200 000 and 100 000 cells/well) in 100 μl of R10 medium (RPMI medium 1640 (GibcoBRL, Ghatersburg, MD, USA), 10% fetal bovine serum (HyClone, Logan, UT, USA), supplemented with 10 mM HEPES buffer, 2 mM L-glutamine, 50 U/ml of penicillin, and 50 μg/ml of streptomycin (all GibcoBRL). Five μg/ml peptide solutions were added, while concanavalin A and DMSO served as positive and negative controls, respectively. After incubation for 18–20 hours at 37°C in 5% CO2, plates were washed with PBS/0.05% Tween 20, and 100 μl/well of 1 μg/ml biotinylated mouse antihuman IFN-γ mAb (MAbTech clone 7-B6-1) in 0.5% BSA/PBS was added per well and incubated for three hours at RT. The assay was developed by incubation with alkaline phosphatase conjugated antibiotin mAb (Vector Laboratories, Burlingame, CA, USA) diluted to 1:750 in 0.5% BSA/PBS followed by incubation with BCIP/NBT substrate solution (Pierce, Rockford, IL, USA). Plates were washed with deionised water and dried before automated counting by the ELIspot reader system (ELR03 AID, Elispot Scientific, Strassberg, Germany).
To differentiate between antigen specific and background production of IFN-γ, a cut off value was defined based on fold increase over background and overall spot count. To this end, PBMC from 20 HCV seronegative subjects were stimulated using individual HCV peptide pools and compared with cells incubated in the absence of peptides (“mock”). Values measured in the presence of peptides were divided by the mock value to determine fold increase over background. In this analysis, the background ranged between 0 and approximately 55 spots per million cells. Therefore, spot counts within the range of 0–55 spots/million cells (SFC/106 cells) may not be clearly antigen specific. The mock ratios were similar regardless of antigens and we estimated the maximum mock ratio as the sample average plus twice the sample standard deviation.
Based on these analyses, we have defined threefold over mock plus at least 55 specific spots/million cells as a stringent cut off for the definition of a significant positive response.
Statistical analysis
The Mann-Whitney test was used for continuous variables to assess differences between distributions. The χ2 test and Fisher’s exact test were used for comparisons of proportions. A p value of <0.05 was considered significant. A multiple regression model was constructed to explore the independent association of the clinical-epidemiological variables with the outcome of disease. The adjusting variables were selected because of their association (p<0.10) in the previous analyses.
Comparison of HLA alleles frequencies in different groups (resolving, chronically evolving, and bone marrow donors) was carried out using the Fisher’s exact test. The observed probability (p) was corrected (pc) according to Bonferroni.
The Bio Medical Data Processing statistical package (University of California, Los Angeles, CA, USA) was used for all calculations above cited. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) with Fleiss’ quadratic confidence intervals (CI) were calculated by Epi-info statistical package.
RESULTS
Patients’ characteristics
A total of 34 subjects with acute hepatitis C (seven women, 27 men; median age 30 years, range 20–61 years) were included in the study (table 1 ▶). The most frequent at risk exposures reported within six months before disease onset were: intravenous drug use (19 subjects), surgical or medical invasive procedures (nine), accidental needle injuries (two), and sexual intercourse with a HCV positive individual (one). No recognised risk factor could be identified for three subjects. HCV genotype was determined in 31 patients, showing a clear prevalence of genotypes 3a and 1b (12 subjects each), followed by genotype 1a (three), 2a/2c (two), and 1a/1b or 2b (one). All but one subject (97%) were symptomatic at the onset of the disease and 23 of them (68%) had jaundice.
Table 1.
Characteristics of patients with self-limiting and chronically evolving acute HCV infection
| Parameter | Number of patients | Self-limiting | Chronic course |
| Number | 34 | 10 (29%) | 24 (71%) |
| M\F | 27\7 | 5\5*‡ | 22\2* |
| Age (years); median (range) | 30 (20–61) | 40, 5 (20–61) | 29 (20–56) |
| Risk factor | |||
| IVDU | 19 | 4 | 15 |
| Invasive procedure | 9 | 4 | 5 |
| Needle | 2 | 1 | 1 |
| HCV+ve sexual partner | 1 | 0 | 1 |
| No data | 3 | 1 | 2 |
| ALT (U/L); median (range) | 1058 (20–3276) | 1201.5 (20–1829) | 861.5 (107–3276) |
| AST (U/L); median (range) | 519 (29–1962) | 618.5 (29–1962) | 496.5 (56–1751) |
| Peak total bilirubin (mg/dl); median (range) | 5.78 (0.43–31.8) | 7.6 (0.9–31.8)† | 3.4 (0.4–14.0)† |
| RNA titer (UI/ml); median (range) | 29950 (590–2.38×106) | 32250 (590–2.38×106) | 29950 (590–1.94×106) |
| Genotype | |||
| 1a | 3 | 1 | 2 |
| 1b | 12 | 4 | 8 |
| 1a/1b | 1 | 0 | 1 |
| 2a/2c | 2 | 0 | 2 |
| 2b | 1 | 1 | |
| 3a | 12 | 3 | 9 |
| No data | 3 | 2 | 1 |
| Symptoms at the onset (n. pts) | 33 (97%) | 10 (100%) | 23 (96%) |
| Jaundice at the onset (n. pts) | 23 (68%) | 9 (90%) | 14 (58%) |
| Follow up (month); median (range) | 13 (6–29) | 12 (6–29) | 13 (6–26) |
*p<0.01 self-limiting v chronic course (univariate analysis).
†p<0.05 self-limiting v chronic course (univariate analysis).
‡OR = 19.4 (CI 1.65 to 227).
Median peak ALT and bilirubin levels at the onset of the disease were 1058 U/L (range 20–3276 U/L) and 5.78 mg/dL (range 0.43–31.80), respectively. Median HCV RNA titre at the entry into the study was 29.950 UI/mL (range 590–2.38×106). All samples with viral load lower than 600 UI/mL resulted HCV RNA negative when tested by a qualitative PCR.
Clinical outcome
Clinical outcome was determined after a follow up of at least six months (median 13; range 6–29). Ten out of 34 subjects (29%) normalised serum ALT levels and displayed undetectable HCV RNA within the first six months after the onset of the disease, with half of them achieving normalisation of both virological and biochemical parameters within the first four weeks from the onset. In these subjects, ALT remained normal and HCV RNA undetectable until the end of the follow up (median period 12 months; range 6–29 months) (fig 1 ▶). Thus, they were considered to have a self-limiting acute infection.
Figure 1.
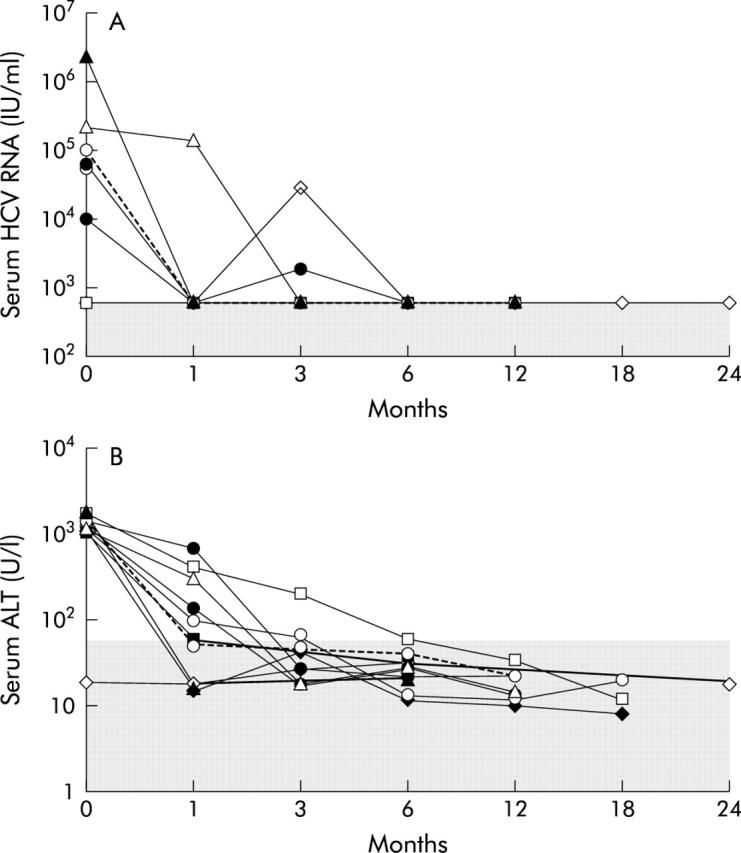
Follow up of HCV RNA and ALT level in patients with self-limiting HCV acute infection. (A) Viral load. Different symbols represent data points for each individual subject. Viral loads are indicated on the vertical axis. The shaded area represents the range of values below the threshold of sensitivity of the employed HCV RNA determination method. The time scale is shown on the horizontal axis. (B) ALT levels. Different symbols represent data points for each individual subject. ALT values are indicated on the vertical axis. The shaded area represents the normal range of values. The time scale is shown on the horizontal axis.
Twenty four subjects who showed increased serum ALT levels and/or detectable serum HCV RNA for more than six months after disease onset were considered to have developed a persistent infection. Their median follow up period was 13 months (range 6–26 months). Eighteen of these patients (75%) had detectable HCV RNA and persistently high ALT values throughout the entire follow up period, while four patients tended to normalise serum ALT values, but had persistently detectable HCV RNA in their serum. Two patients tested negative for HCV RNA at six and 12 months after disease onset, respectively, but could not be further evaluated to assess an eventual delayed resolution of their infection.
All subjects with self-limiting infection showed at least a negative HCV RNA test within three months of follow up, while eight and six of them had a normal ALT determination or a combination of negative HCV RNA and normal ALT, respectively (fig 1 ▶). When such an analysis was carried out among patients with chronic evolution, only five of them displayed a negative HCV RNA test within three months from the onset of the infection, whereas seven and three patients had a normal ALT determination or tested negative for both viremia and increased ALT levels, respectively. The differences between subjects with self-limiting infection and those with chronic evolution in the detection frequency of at least one HCV RNA negative test or a normal ALT value determination or both within the third month from the disease onset were statistically significant (p<0.01, p<0.05, and p<0.01, respectively). A similar figure was obtained by analysing the same parameters already within the first month from the onset of the symptoms (9/10 v 4/24 for HCV RNA, p<0.01; 6/10 v 4/24 for ALT, p<0.05; 6/10 v 2/24 for both parameters at the same time, p<0.01). Focusing on the detection of HCV RNA within the first month from the disease onset, 90% (95% CI 54.1 to 99.5) of the resolving patients had at least a negative test (sensitivity), whereas 83.3% (95% CI 61.8 to 94.5) of the patient with chronic evolution had not at least a negative HCV RNA determination (specificity). The post-test probability of recovery (or PPV) following a positive result (that is, at least a negative HCV RNA test) and the post-test probability of chronic evolution (or NPV) following a negative result (all determinations positive for HCV RNA) of such a test were 69.2% (95% CI 38.9 to 89.6) and 95.2% (95% CI 74.1 to 99.8), respectively.
With regard to the other clinical-epidemiological features (table 1 ▶), females experienced a self-limiting course of disease in a significantly higher proportion than males (5/7 v 5/27; p<0.01) and patients with a resolving hepatitis had higher median peak bilirubin level at the onset of disease (7.66 mg/dL v 3.39 mg/dL; p<0.05). After multivariate analysis including age, sex, ALT, and bilirubin levels at the onset, the only independent factor associated to a self-limiting course of acute hepatitis C was female sex (OR = 19.4; 95% CI 1.65 to 227).
Multispecific T cell response during the early phase of acute HCV infection is associated to disease resolution
To define the role of adaptive T cell immunity in determining the course of disease, we measured the breadth and magnitude of anti-HCV CD4+ and CD8+ T cell responses ex vivo in PBMC of 30 acutely infected subjects by the quantitative and functional IFN-γ ELIspot assay. As resolution of HCV infection occurs in most cases within the first three months from the onset of the infection,8,11,13–15 this analysis was performed for each patient on PBMC collected at the time of the enrolment (T = 0) and after one month (T = 1). Furthermore, to avoid bias towards preselected epitopes and HLA alleles, we used as a source of antigen a panel of 216 peptides covering about 72% of the HCV polyprotein and spanning the Core and NS3-NS5B region of a 1b HCV isolate.
The results of this analysis showed that HCV infection induced CMI in 18 out of 30 acutely infected patients. This high frequency of T cell response is twice as much the rate of disease resolution we observed in the same cohort (60% v 27%), implying that T cell response per se is not sufficient to achieve spontaneous clearance of the virus.
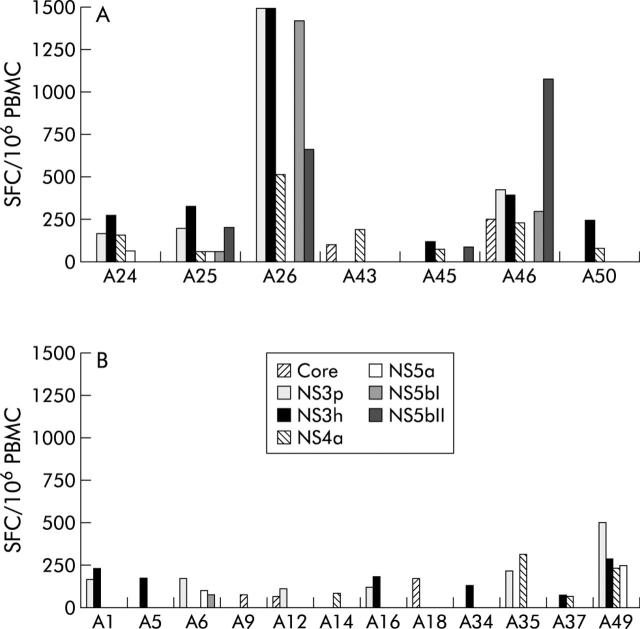
Eight of the 30 HCV acutely infected subjects screened by IFNγ-ELIspot assay had a self-limiting infection. All but one of these individuals developed an HCV specific CMI (fig 2A ▶), whereas 12 of 22 patients who developed a persistent infection (fig 2B ▶) displayed anti-HCV T cell responses, but the difference between the two groups was not statistically significant. All T cell responding subjects with self-limiting acute hepatitis C displayed a multispecific response: A43, A45, and A50 responded to two or three antigens, A24 to 4, A26 to 5, and the last two subjects (A25 and A46) to six out of the seven antigens tested (fig 3 ▶). In contrast, individuals with a chronic evolution targeted fewer antigens: 10 responded to one or two antigens and just two subjects (A6 and A49) showed a response targeting three or four antigens. The difference in the breadth of the T cell response between the two groups was statistically significant (p<0.01). On the contrary, no significant statistical difference was found between individuals with a self-limiting and those with chronic evolution, as regards the overall median strength of CMI response evaluated by adding up the reactivities against individual antigens (784 SFC/106 PBMC, range 412–6872 v 448 SFC/106, range 240–1510; p = 0.063).
Figure 2.
Magnitude and breadth of the T cell response during the early phase of HCV infection in subjects with (A) acute self-limiting infection (n = 7) and with (B) a chronic evolution (n = 12). PBMC samples collected at the time of enrollment (T = 0) and one month after (T = 1), were tested by IFN-γ ELIspot assay against seven peptide pools corresponding to Core, NS3 protease (NS3p), NS3 helicase (NS3h), NS4, NS5a, and NS5b (split in two pools NS5b-I and NS5b-II). For each patient, results from the time point showing the highest frequency of HCV specific CMI were reported. It was T = 0 for patients A26, A45, A50, A1, A9, A12, A16, A18, A34, and A35 and T = 1 for patients A24, A25, A43, A46, A5, A6, A14, A37, and A49. To simplify, only responses above the threshold defined using seronegative subjects (see Materials and Methods) are shown. For all the remaining subjects ELIspot responses were negative at both time points tested. Numbers represent spot forming cells (SFC)/106 PBMCs. In subjects A26 the response against NS3p and NS3h exceed the upper value of the scale.
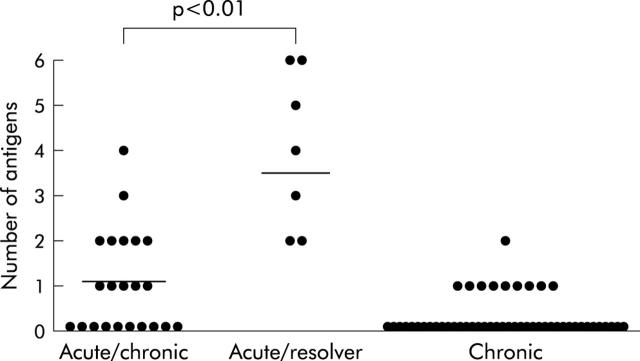
Figure 3.
Breadth of anti-HCV T cell response during acute and chronic infection: the number of HCV antigens recognised by each individual is shown as a single dot. Breadth of the response was compared between individuals with self-limited acute hepatitis, or acute hepatitis C and chronic evolution and the patients with late chronic infection. For each patient with acute infection we have plotted the higher response measured at either T = 0 or T = 1. Horizontal lines indicate median numbers of recognised peptide pools for each group. Statistically significant difference between groups was calculated by Mann Whitney test.
When we evaluated the ability of a multispecific CMI response (⩾3 antigens) to predict a favourable outcome in these 30 acutely infected patients, sensitivity and specificity were 62.5% (95% CI 25.9 to 89.8) and 90.9% (95% CI 69.4 to 98.4), respectively. The post-test probability of recovery following a positive response and the post-test probability of chronic evolution following a negative response were 71.4% (95% CI 30.3 to 94.9) and 87.0% (95% CI 65.3 to 96.6), respectively. By combining this test with the detection of at least a negative HCV RNA test within one month from disease onset, sensitivity, specificity, post-test probability of recovery following a positive result and post-test probability of chronic evolution following a negative result of the two combined test were 50.0%, (95% CI 17.4 to 82.6) 100% (95% CI 81.5 to 100), 100% (95% CI 39.6 to 100), and 84.6% (95% CI 64.3 to 95.0), respectively, demanding that both tests gave a positive response (for HCV RNA it means at least a negative determination in the first month) to consider the test result as positive. Demanding that either or both tests gave a positive response to consider the test result as positive sensitivity, specificity, post-test probability of recovery following a positive result, and post-test probability of chronic evolution following a negative result of the two combined test were 100% (95% CI 17.4 to 82.6), 77.3% (95% CI 54.2 to 91.3), 61.5% (95% CI 32.3 to 84.9), and 100% (95% CI 77.1 to 100), respectively.
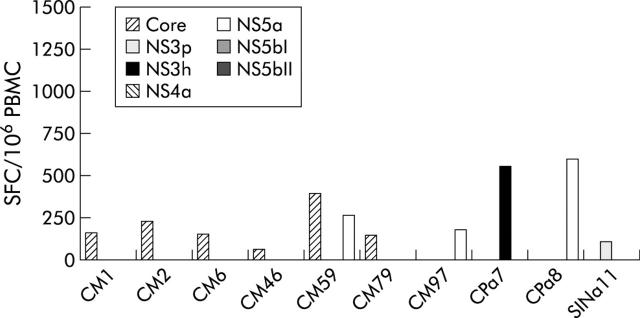
To further extend these findings, we measured HCV specific CMI in a cohort of 56 chronically infected patients by IFN-γ ELIspot assay. Patients with chronic hepatitis C were older than acutely infected patients (median 59 years, range 31–75 years), but no statistically significant difference was found between them with regard to sex and genotype distribution. In this cross sectional study, only 10 patients (18%) were found to be positive by ELIspot assay (fig 4 ▶). The magnitude of the overall responses in patients with chronic hepatitis C was similar to that observed in acutely infected subjects presenting a chronic evolution and clearly lower than that observed in acutely infected subjects showing a self-limiting course (range 201–1464 SFC/106 PBMC, with a median value of 346; fig 4 ▶). Most importantly, CMI was directed against a single antigen in nine of the 10 chronic patients with a positive T cell response, whereas in the remaining patient it targeted two peptide pools (fig 3 ▶). The difference in the frequency of IFNγ positive responses observed between these patients and subjects with self-limiting acute hepatitis C was statistically significant (p<0.01).
Figure 4.
Magnitude and breadth of the T cell response in chronically infected patients. PBMC samples were tested by IFN-γ ELIspot assay against the seven HCV peptide pools. Only responses above the threshold defined using seronegative subjects (see Materials and Methods) are shown.
Overall, these data indicate that a strong T cell response targeting multiple HCV antigens in the early phase of infection is a distinctive feature of individuals who clear the virus.
HLA class II typing, HCV-specific CMI, and outcome of acute hepatitis C
The frequency of DRB1 (table 2 ▶) and HLA DQB1 (table 3 ▶) alleles was determined in both resolving and chronically evolving acutely HCV infected subjects and compared to that of a control population of 40 healthy bone marrow donors. No statistically significant difference was observed in the distribution of these alleles among these groups of subjects after Bonferroni correction. Interestingly, allele DRB1*1101 (that was more frequent in resolving patients than in those with chronic evolution and less frequent in patients with chronic evolution than in controls; tables 2 ▶ and 3 ▶) was detected in two female subjects (A26 and A46) who showed the strongest and broadest T cell response as measured by IFN-γ ELIspot assay (fig 2A ▶). Both individuals showed a rapid resolution of their infection with HCV clearance and ALT normalisation occurring within the first (A26) and second (A46) month from disease onset.
Table 2.
DRB1 alleles distribution in patients with self-limiting HCV acute infection (SLI), with persistent HCV infection (PI) and in controls
| Allele | SLI | PI | Controls | SLI v PI | SLI v controls | PI v controls | SLI+PI v controls | ||||
| DRB1 | (n = 10) | (n = 24) | (n = 40) | p | pc | p | pc | p | pc | p | pc |
| *01 | 1 (10%) | 2 (8.3 %) | 3 (7.5%) | NS | NS | NS | NS | ||||
| *03 | 3 (30%) | 3 (12.5%) | 3 (7.5%) | NS | NS | NS | NS | ||||
| *04 | 2 (20%) | 4 (16.7%) | 8 (20%) | NS | NS | NS | NS | ||||
| *0701 | 3 (30%) | 10 (41.7%) | 7 (17.5%) | NS | NS | 0.026 | NS | 0.029 | NS | ||
| *11 | 4 (40%) | 13 (54.2%) | 25 (62.5%) | NS | NS | NS | NS | ||||
| *1101 | 2 (20%) | 2 (8.3%) | 11 (27.5%) | NS | NS | 0.048 | NS | NS | |||
| *1104 | 2 (20%) | 11 (45.8%) | 15 (37.5%) | NS | NS | NS | NS | ||||
| *12 | 0 | 1 (4.2%) | 2 (5%) | NS | NS | NS | NS | ||||
| *13 | 1 (10%) | 4 (16.7%) | 9 (22.5%) | NS | NS | NS | NS | ||||
| *14 | 2 (20%) | 3 (12.5%) | 1 (2.5%) | NS | NS | NS | NS | ||||
| *15 | 1 (10%) | 4 (16.7%) | 4 (10%) | NS | NS | NS | NS | ||||
| *16 | 2 (20%) | 2 (8.3%) | 5 (12.5) | NS | NS | NS | NS | ||||
NS, not significant.
Table 3.
DQB1 alleles distribution in patients with self-limiting HCV acute infection (SLI), with persistent HCV infection (PI) and in controls
| Allele | SLI | PI | Controls | SLI v PI | SLI v controls | PI v controls | SLI+PI v controls | ||||
| DQB1 | (n = 10) | (n = 24) | (n = 40) | p | pc | p | pc | p | pc | p | pc |
| *02 | 4 (40%) | 9 (37.5%) | 8 (20%) | NS | NS | NS | NS | ||||
| *0301 | 4 (40%) | 15 (62.5%) | 27 (67.5%) | NS | NS | NS | NS | ||||
| *0302 | 2 (20%) | 4 (16.7%) | 3 (7.5%) | NS | NS | NS | NS | ||||
| *0303 | 2 (20%) | 2 (8.3%) | 2 (5%) | NS | NS | NS | NS | ||||
| *05 | 6 (60%) | 8 (33.3 %) | 12 (30%) | NS | NS | NS | NS | ||||
| *0501 | 2 (20%) | 2 (8.3%) | 6 (15%) | NS | NS | NS | NS | ||||
| *0502 | 2 (20%) | 3 (12.5%) | 5 (12.5%) | NS | NS | NS | NS | ||||
| *0503 | 2 (20%) | 3 (12.5%) | 1 (2.5%) | NS | NS | NS | NS | ||||
| *06 | 1 (10%) | 5 (20.8%) | 12 (30%) | NS | NS | NS | NS | ||||
NS, not significant.
DISCUSSION
A number of recent studies have shown that spontaneous elimination of HCV most often occurs in the first few months of infection, and that once chronic infection is established spontaneous viral clearance is rare.8,11,13–15,45 Thus, the initial months after infection seem to be of crucial importance for the subsequent course of disease. However, a prospective study on a statistically significant number of acutely infected subjects, which measured clinical, virological, and, most importantly, cell mediated immunity without bias for the patient haplotype was still missing.
On the basis of the clinical criteria adopted to define the outcome of acute HCV infection, 29% (10/34) of the patients in our study cohort experienced a self-limiting infection. This figure is consistent with previous reports on patients with symptomatic acute HCV infection, for which significant rates of resolution were measured.8,10–12,14,34
A significant proportion of females experienced a self-limiting disease with respect to males and patients with a resolving hepatitis had higher median peak bilirubin level at the onset of the infection in respect to patients with a chronic evolution. However, after multivariate analysis only sex resulted to be independently associated to the disease outcome. The higher peak bilirubin levels in subjects with a resolving course of infection, as well as the tendency to have higher peak ALT concentrations, probably reflects a strong antiviral immune response in these individuals.11 The association of the female sex with a self-limiting course of HCV infection and with a better response to interferon α therapy, has already been reported by other authors.8,9,11,17,18,46–48 One possible explanation for this finding is that host immune response might be positively influenced by female sex hormones, thus favouring HCV clearance.17,46
Among the 10 subjects with self-limiting infection, seven achieved virus clearance within one month and eight within three months after the onset. Similarly, normalisation of ALT values occurred within one or three months in six and eight subjects, respectively. In addition, we have found that detection of at least one negative HCV RNA test, with or without a normal ALT value, within one or three months from the onset of symptoms, was significantly associated with resolution of acute HCV infection. These findings suggest that repeated evaluation of HCV RNA status until the third month from the onset of disease can serve as a good predictor of the outcome of the infection and can be used as useful criterion to decide the beginning of antiviral treatment. Previous studies have shown that HCV specific CMI can be detected in individuals who resolved infection, but very rarely in chronic patients, supporting the view that the cellular arm of anti-HCV immunity plays a major role in control of viral infection.8,10,27,33–37 However, the majority of the existing information was gathered employing a limited number of epitopes, mostly restricted by the HLA A2.1 haplotype,34–37 an approach that clearly misses many responses.49,50 The cohort of acutely infected patients described in this study provided for a unique opportunity to evaluate the influence of adaptive T cell immunity in the outcome of HCV infection without these biases. A first question we wished to answer was whether HCV induces a T cell response in most of the infected individuals or if the virus can overcome T cell surveillance by providing only weak immunological stimuli. By ex vivo IFNγ ELIspot assay using a complete set of overlapping peptides covering most of the HCV polyprotein, we found a significantly higher frequency of responses in acutely infected individuals than in a group of control patients with a long history of persistent infection (60% v 18%). Furthermore, among those individuals of the latter group where chronic infection persisted in the face of measurable T cell response, fewer antigens were targeted, suggesting that over time T cell reactivity becomes focused on a lower number of epitopes than those observed during the early phase of infection. Preliminary data from our cohort of acutely infected individuals also indicate that in patients with chronic evolution T cell immunity becomes undetectable after a few months from the onset of the infection. These data support previous observations indicating that HCV induces significant CMI during the early phase of infection, but that this response is not sustained.8,31,33–35
Because of the difficulty in using peptides matching the infecting genotypes in each patient, we have used a panel of peptides derived from a 1b HCV isolate corresponding to regions highly conserved among different strains. T cell response was measured in subjects bearing different HLA alleles and infected by different viral genotypes. All but one subjects infected by 3a genotype were scored positive in our assay, consistent with previous findings of cross genotype T cell reactivity for epitopes located within the NS3-NS5 region.51,52 Therefore, even if there is some underestimation of the strength and the frequency of CMI responses in the study cohort, we think our approach remains a viable option to address ex vivo T cell responses in large cohorts of acutely and chronically infected subjects
The high frequency of T cell responses measured within the first month from disease onset suggested that a significant proportion of chronically evolving patients would also develop anti-HCV CMI. In fact, about half (12/22) of the acutely infected subjects who subsequently developed a chronic infection displayed a T cell response during the first four weeks from the onset of the symptoms. Although the difference in the frequency of T cell responses between resolving and chronically evolving subjects was evident, it did not reach statistical significance, possibly also because of the limited number of individuals analysed. These data imply that, if present, the immunological signature of a self-limiting course of disease is not simply the presence of specific T cells, but rather consists of some qualitative feature of this response, which is missing in patients progressing toward chronicity. Consistently with this hypothesis, a T cell response targeting multiple HCV antigens in the early phase of the infection emerged as a statistically significant distinctive feature of individuals who cleared the virus. This finding is in agreement with the notion that a response against multiple antigens may reduce the likelihood of escaping immune response through antigenic variation, as recently shown in the chimpanzee model.26,54,55
When applied in our cohort to predict a favourable course of infection, the detection of a multispecific T cell response (⩾3 antigens) by IFNγ ELIspot assay showed low sensitivity and post-test probability of recovery following a positive test. This test’s characteristics imply that we might consider as having an unfavourable course—and eventually subject to early treatment—a non-negligible proportion of subjects who indeed will resolve the infection.
What is the mechanism for the lower spectrum (or total lack) of anti-HCV T cell responses in chronically evolving subjects? In some models of infection, prolonged exposure to high amounts of viral antigens could lower immune responses both in terms of number as well as function of antigen specific cells.3,56–58 In line with this hypothesis, we observed that CMI induced in chronically evolving patients was on average of lower intensity than that measured in subjects with self-limiting acute infection. Our observation of similar viral loads at the onset of the symptoms in subjects with self-limiting acute infection and those with chronic evolution does not rule out this hypothesis because hepatic measurements of viremia could not be performed for ethical reasons. Finally, other mechanisms like deficient induction of T helper cell response, incomplete T cell maturation, or liver induced immunotolerance may be responsible for the observed attenuation of the immune response in chronically evolving patients.
Recent studies have described important associations between some HLA alleles, particularly DRB1*1101 and/or DQB1*0301, and clearance of HCV.9,17,18,23–26 We found no statistically significant difference in the distribution of HLA DRB1 and DQB1 alleles between patients with resolving infection and those with chronic evolution probably because of the insufficient number of individuals analysed. It is worthy of note that the strongest and broadest HCV specific T cell response with a rapid clearance of the virus, was found in patients carrying haplotype DRB1*1101. Further prospective studies on larger cohorts of HCV acutely infected individuals are needed to exhaustively evaluate the role of specific HLA class II alleles in the outcome of the infection and to characterise the mechanisms leading to T cell failure during primary infection. Notwithstanding this, our results suggest that induction of a broad CMI already within one month from the onset of the symptoms, and negative detections of HCV RNA during this period are predictive of a favourable outcome of HCV acute infection. Indeed, when these tests were used in combination in the first month from disease onset, to predict a favourable outcome, demanding that either or both of them were positive, we were able to identify all patients with a self-limiting infection, even though about 20% of patients with a predicted favourable courses (false positive) finally showed a chronic evolution. Nonetheless, it is necessary to underline that these estimates were based on a small sample of patients, and so they need to be further confirmed by analysing prospectively a larger group of acutely infected individuals.
These findings could provide for a convenient alternative to the immediate treatment of all patients with acute hepatitis C as suggested by Jaeckel et al, who recently reported sustained response of 95% with starting treatment of patients within 4–16 weeks after infection.58 The very high rate of response found by these authors could have been due to the adjuvant effect of anti-HCV CMI which is often attenuated (if not absent) in the chronic phase of infection. Thus, by implementing a T cell assay together with RNA detection tests within the first month from the onset of symptoms, one could select with high diagnostic sensitivity a group of patients with high probability to resolve the infection, preventing them having to undergo an early antiviral treatment. If such a combined test failed in predicting a favourable course (false positive results), treatment could still achieve high frequency of sustained virological response if started within the first six months from disease onset.14,15,58
Acknowledgments
This study was supported in part by the Viral Hepatitis Project, Istituto Superiore di Sanità (D. Leg.vo. 30/12/1992 n. 502).
*OTHER MEMBERS OF THE ACUTE HEPATITIS C ITALIAN STUDY GROUP ARE AS FOLLOWS: G Taliani (Institute of Infectious Diseases, University of Florence, Florence); S Buonocore, G Lettieri, P Pierri (Cotugno Hospital, Infectious Diseases Unit, Naples, Italy); F Meneghetti (Padova Hospital, Infectious and Tropical Diseases Unit, Padova, Italy); T Stroffolini (San Giacomo Hospital, Liver Unit, Rome, Italy); G Maio (A. Rummo Hospital, Infectious Diseases Unit, Benevento, Italy); R Francavilla (Bisceglie Hospital, Infectious Diseases Unit, Bisceglie, Italy); P Chiriacò (A. Perrino Hospital, Infectious Diseases Unit, Brindisi, Italy); U Baldi (Umberto I Hospital, Infectious Diseases Unit, Nocera Inferiore, Italy); P Bellissima (Gravina Hospital, Infection Diseases Unit, Caltagirone, Italy); L Cosco, T Ferraro (Pugliese Ciaccio, Hospital, Infectious Diseases Unit, Catanzaro, Italy); N Petrosillo, P Scognamiglio, M R Capobianchi (National Institute of Infectious Diseases, Lazzaro Spallanzani, Rome, Italy); V Mellace, F Montesano (Soverato, Drug Dependency Unit, ASL 7, Soverato, Italy); G Audino (Drug Dependency Unit ASL 7, Catanzaro, Italy); C De Stefano, M Giustra (Department of the Dependencies ASL 11, Reggio Calabria, Italy); A Caterini (Viterbo Hospital, Infectious Diseases Unit, Viterbo, Italy); V Guadagnino (Institute of Infectious Diseases, University of Catanzaro, Catanzaro, Italy); M Cuccia (Epidemiology and Prevention Service, AUSL 3, Catania, Italy); O Zuccaro(Institute of Hygiene, University of Rome Tor Vergata, Rome, Italy); L Laurenti (Department of Cell Biotechnology and Hematology, University “La Sapienza”, Rome, Italy); C Scottà (Department of Cellular and Development Biology, University “La Sapienza”, Rome, Italy).
Abbreviations
ALT, alanine aminotransferase
CI, confidence interval
CMI, cell mediated immunity
CTL, cytotoxic T lymphocyte
DMSO, dimethyl sulfoxide
ELIspot, enzyme linked immunospot
HCV, hepatitis C virus
HLA, human leukocyte antigen
IFN, interferon
IVDU, intravenous drug use
MEIA, microparticle enzyme immunoassay
NPV, negative predictive value
PBMC, pheripheral blood mononuclear cells
PBS, phosphate buffered saline
PPV, positive predictive value
RIBA, recombinant immunoblot assay
SFC, spot forming cells
REFERENCES
- 1.Orland JR, Wright TL, Cooper S. Acute hepatitis C. Hepatology 2001;33:321–7. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Hepatitis C. the clinical spectrum of disease. Hepatology. 1997;26(Suppl. 1):15S–20. [DOI] [PubMed]
- 3.Alberti A , Chemello L, Bevegnu L. Natural history of hepatitis C. J Hepatol 1999;31 (Suppl 1) :17–24. [DOI] [PubMed] [Google Scholar]
- 4.Alter MJ, Margolis HS, Krawczynski K, et al. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med 1992;327:1899–905. [DOI] [PubMed] [Google Scholar]
- 5.Mattson L , Sönnerborg A, Weiland O. Outcome of acute symptomatic non-A, non-B hepatitis: A 13-year follow-up study of hepatitis C virus markers. Liver 1993;13:274–8. [DOI] [PubMed] [Google Scholar]
- 6.Barrera JM, Bruguera M, Erilla MG, et al. Persistent hepatitis C viremia after acute self-limiting post-transfusion hepatitis. Hepatology 1995;21:639–44. [PubMed] [Google Scholar]
- 7.Villano SA, Vlahov D, Nelson KE, et al. Persistence of viremia and the importance of long-term follow-up after Acute hepatitis C infection. Hepatology 1999;29:908–14. [DOI] [PubMed] [Google Scholar]
- 8.Gerlach JT, Diepolder HM, Jung MC, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology 1999;117:933–41. [DOI] [PubMed] [Google Scholar]
- 9.Arlic L , Fort M, Izopet J, et al. Genes of the Major Histocopatibility Complex class II influence the outcome of hepatitis C virus infection. Gastroenterology 1997;113:1675–81. [DOI] [PubMed] [Google Scholar]
- 10.Missale G , Bertoni R, Lamonaca V, et al. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigour of the anti-viral cell-mediated immune response. J Clin Invest 1996;98:706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamal SM, Rasenak JW, Bianchi L, et al. Acute hepatitis C without and with schistosomiasis: correlation with hepatitis C-specific CD4+ T-cell and cytokine response. Gastroenterology 2001;121:646–56. [DOI] [PubMed] [Google Scholar]
- 12.Tsay SL, Liaw YF, Chen MH, et al. Detection of type 2-like T-helper cells in hepatitis C virus infection: implications for hepatitis C virus chronicity. Hepatology 1997;25:449–58. [DOI] [PubMed] [Google Scholar]
- 13.Hofer H , Watkins-Riedel T, Janata O, et al. Spontaneous viral clearance in patients with acute hepatitis C can be predicted by repeated measurements of serum viral load. Hepatology 2003;37:60–4. [DOI] [PubMed] [Google Scholar]
- 14.Santantonio T , Sinisi E, Guastadisegni A, et al. Natural course of acute hepatitis C: a long-term prospective study. Dig Liver Dis 2003;35:104–13. [DOI] [PubMed] [Google Scholar]
- 15.Gerlach JT, Diepolder HM, Zachoval R, et al. Acute Hepatitis C: High rate of both spontaneous and treatment-induced viral clearance. Gastroenterology 2003;125:80–8. [DOI] [PubMed] [Google Scholar]
- 16.Tong MJ, El-Farra NS, Reikes AR, et al. Clinical outcomes after transfusion-associated hepatitis C. N Engl J Med 1995;332:1463–6. [DOI] [PubMed] [Google Scholar]
- 17.Alric L , Fort M, Izopet J, et al. Study of host- and virus-related factors associated with spontaneous hepatitis C virus clearance. Tissue Antigens 2000;56:154–8. [DOI] [PubMed] [Google Scholar]
- 18.Thursz M , Yallop R, Goldin R, et al. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Research. Lancet 1999;354:2119–24. [DOI] [PubMed] [Google Scholar]
- 19.Chu CW, Hwang SJ, Luo JC, et al. Comparison of clinical, virologic and pathologic features in patients with acute hepatitis B and C. J Gastroenterol Hepatol 2001;16:209–14. [DOI] [PubMed] [Google Scholar]
- 20.Amoroso P , Rapicetta M, Tosti ME, et al. Correlation between virus genotype and chronicity rate in acute hepatitis C. J Hepatol 1998;28:939–44. [DOI] [PubMed] [Google Scholar]
- 21.Dusheiko G , Weiss HS, Brown D, et al. Hepatitis C virus genotypes: an investigation of type-specific differences in geographic origin and disease. Hepatology 1994;19:13–18. [PubMed] [Google Scholar]
- 22.Farci P , Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 2000;288:339–44. [DOI] [PubMed] [Google Scholar]
- 23.Cramp MI, Carucci P, Underhill J, et al. Association between HLA class II genotype and spontaneous clearance of hepatitis C viremia. J Hepatol 1998;29:207–13. [DOI] [PubMed] [Google Scholar]
- 24.Minton EJ, Smillie D, Neal KR, et al. Association between MHC class II alleles and clearance of circulating hepatitis C virus. Members of the Trent Hepatitis C Virus Study Group. J Infect Dis 1998;178:39–44. [DOI] [PubMed] [Google Scholar]
- 25.Mangia A , Gentile R, Cascavilla I, et al. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J Hepatol 1999;30:984–9. [DOI] [PubMed] [Google Scholar]
- 26.Harcourt G , Hellier S, Bunce M, et al. Effect of HLA class II genotype on T helper lymphocyte response and viral control in hepatitis C virus infection. J Viral Hepatitis 2001;8:174–9. [DOI] [PubMed] [Google Scholar]
- 27.Thimme R , Bukh J, Spangenberg HC, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A 2002;99:15661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper S , Erickson AL, Adams EJ, et al. Analysis of a successful immune response against hepatitis C virus. Immunity 1999;10:439–49. [DOI] [PubMed] [Google Scholar]
- 29.Shoukry NA, Grakoui A, Houghton M, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med 2003;197:1645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grakoui A , Shoukry NA, Woolard DJ, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science 2003;302:659–62. [DOI] [PubMed] [Google Scholar]
- 31.Bassett SE, Guerra B, Brasky K, et al. Protective immune response to Hepatitis C Virus in chimpanzees rechallenged following clearance of primary infection. Hepatology 2001;33:1479–87. [DOI] [PubMed] [Google Scholar]
- 32.Weiner AJ, Paliard X, Selby M, et al. Intrahepatic genetic inoculation of Hepatitis C virus RNA confers cross-protective immunity. J Virol 2001;75:7142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grüner NH, Gerlach TJ, Jung MC, et al. Association of hepatitis C virus-specific CD8+ T cells with viral clearance in acute hepatitis C. J Infect Dis 2000;181:1528–36. [DOI] [PubMed] [Google Scholar]
- 34.Takaki A , Wiese M, Maertens G, et al. Cellular immune response persists and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. NatMed 2000;6:578–82. [DOI] [PubMed]
- 35.Lechner F , Wong D, Dunbar R, et al. Analysis of successful immune response in persons infected with HCV. J Exp Med 2000;9:1499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grüner N , Lechner F, Jung M, et al. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J Virol 2001;75:5550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thimme R , Oldach D, Chang K-M, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med 2001;194:1395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cacopardo B , Fatuzzo F, Benanti F, et al. Long-term study of anti-HCV reactivity using 3d generation recombinant immunoblot assay in acute post-transfusion hepatitis. Recenti Prog Med 1995;86:378–81. [PubMed] [Google Scholar]
- 39.Lelie PN, Cuypers TM, Reesink HW, et al. Pattern of serological Markers in transfusion-transmitted hepatittis C virus infection using second-generation HCV assay. J Med Virol 1992;37:203–9. [DOI] [PubMed] [Google Scholar]
- 40.Miller SA, Dykes DD, Polesky HF, et al. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Research 1988;16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura A , Sasazuki T. Reference protocol for the HLA DNA typing technique. In: Aizawa M, Sasazuki T, eds. 1991. Proceedings of the 11th International Histocompatibility Workshop and Conference. HLA: Oxford University Press, 1991:397–419.
- 42.Olerup O , Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours; an aletrnative to serological Typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 1992;39:225. [DOI] [PubMed] [Google Scholar]
- 43.Takamizawa A , Mori C, Fuke I, et al. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol 1991;65:1105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atherton E , Sheppard RC. Solid phase peptide synthesis, a practical approach. Oxford: IRL Press, 1989.
- 45.Larghi A , Zuin M, Crosignani A, et al. Outcome of an outbreak of acute hepatitis C among healthy volunteers participating in pharmacokinetics studies. Hepatology 2002;36:993–1000. [DOI] [PubMed] [Google Scholar]
- 46.Kenny-Walsh E . Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N Engl J Med 1999;340:1228–33. [DOI] [PubMed] [Google Scholar]
- 47.Izopet J , Payen JL, Alric L, et al. Baseline level and early suppression of serum HCV RNA for predicting sustained complete response to alpha-interferon therapy. J Med Virol 1998;54:86–91. [DOI] [PubMed] [Google Scholar]
- 48.Alric L , Izopet J, Fort M, et al. Study of the association between major histocompatibility complex class II genes and the response to interferon alpha in patients with chronic hepatitis C infection. Hum Immunol 1999;60:516–23. [DOI] [PubMed] [Google Scholar]
- 49.Wong DK, Dudley DD, Afdhal NH, et al. Liver-derived CTL in hepatitis C virus infection, breadth and specificity of responses in a cohort of persons with chronic infection. J Immunol 1998;160:1479–88. [PubMed] [Google Scholar]
- 50.Lauer GM, Ouchi K, Chung RT, et al. Comprehensive analysis of CD8+ T cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol 2002;76:6104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koziel MJ, Dudley D, Afdhal N, et al. HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokine release. J Clin Invest 1995;96:2311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang KM, Rehermann B, McHutchison JG, et al. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest 1997;100:2376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosen HR, Hinrichs DJ, Gretch DR, et al. Association of multispecific CD4(+) response to hepatitis C and severity of recurrence after liver transplantation. Gastroenterology 1999;117:926–32. [DOI] [PubMed] [Google Scholar]
- 54.Erickson AL, Kimura Y, Igarashi S, et al. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 2001;15:883–95. [DOI] [PubMed] [Google Scholar]
- 55.Moskophidis D , Laine E, Zinkernagel RM. Peripheral clonal deletion of antiviral memory CD8 T cells. Eur J Immunol 1993;23:3306–11. [DOI] [PubMed] [Google Scholar]
- 56.Vogel TU, Allen TM, Altman JD, et al. Functional impairment of simian immunodeficiency virus-specific CD8 T cells during the chronic phase of infection. J Virol 2001;75:2458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lechner F , Gruener NH, Urbani S, et al. CD8 T lymphocytes responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol 2000;30:2479–87. [DOI] [PubMed] [Google Scholar]
- 58.Jaeckel E , Cornberg M, Wedemeyer H, et al. Treatment of acute hepatitis C with interferon alfa-2b. N Engl J Med 2001;345:1452–7. [DOI] [PubMed] [Google Scholar]