Abstract
Background and aims: The mucosa associated flora of the large intestine is important in determining mucosal function although what controls its composition is unknown. This study has determined the effect of the prebiotic carbohydrates oligofructose and inulin on the mucosal flora.
Methods: An in vitro chemostat model of both planktonic and surface associated bacteria was used followed by an intervention study in 29 subjects undergoing colonoscopy.
Subjects: Fourteen subjects, recruited from colonoscopy waiting lists, supplemented their diet for two weeks with a mix of 7.5 g of oligofructose and 7.5 g inulin. Fifteen subjects were recruited at the time of colonoscopy and given no supplement. Multiple endoscopic biopsies were taken from the caecum, transverse and descending colon, and rectum. The mucosal flora was characterised by culture and to species level by cellular fatty acid profiles. Cell proliferation was assessed by immunohistochemical staining for minichromosome maintenance protein 2, Ki67, and proliferating cell nuclear antigen.
Results: In vitro prebiotics increased surface counts of bifidobacteria from 6.6 to 7.3 log10 colony forming units (CFU) per slide (p<0.0006) with no significant changes in planktonic bacteria. In the feeding study, prebiotics increased mucosal bifidobacteria (log CFU/g mucosa (SEM)) in both the proximal (control 5.3 (0.4) v prebiotic 6.3 (0.3)) (p = 0.059) and distal (control 5.2 (0.3) v prebiotic 6.4 (0.3)) colon (p = 0.01). Lactobacilli were also increased (3.0 (0.1) v 3.7 (0.2) (p = 0.02) in the proximal and 3.1 (0.1) v 3.6 (0.2) (p = 0.04) in the distal colon, respectively). There were significantly more eubacteria in fed subjects but no changes in total anaerobes clostridia, bacteroides, or coliforms, nor in proliferation indices.
Conclusion: Prebiotic carbohydrates can change the composition of the mucosa associated flora significantly.
Keywords: mucosa associated floral, prebiotic, bifidobacteria, biofilm, large intestine
Bacteria that inhabit the large bowel can be divided into planktonic, namely those that could be considered to be free living in the lumen of the bowel, and the mucosa associated flora (MAF). There is a large body of literature describing the planktonic flora of the colon and its functions but much less is known about mucosal populations. While the planktonic flora clearly play an important role in digestion through fermentation,1,2 vitamin synthesis,3 metabolism of xenobiotics,4 and provide a barrier to invading pathogens,5 the mucosal flora, because of their location, are increasingly thought to be critical in determining epithelial health through their ability to exclude pathogens, stimulate development of the immune system, and moderate inflammatory responses.6–9
The growth and metabolism of established planktonic flora is controlled largely by dietary substrates that escape digestion in the small bowel, and probably also by endogenous secretions of the gut.1,10,11 What influences the MAF is largely unknown although there is clearly cross talk between epithelial cells, bacteria, and the gut immune system.12–15 The role of diet has not been studied, but carbohydrates such as oligofructose, inulin,16,17 and galacto-oligosaccharides18 are known to stimulate selectively the growth of bifidobacteria and lactobacilli in the planktonic flora of the colon (prebiotic effect) and thereby contribute to barrier function.6,19,20 If the composition of the MAF was also to be moderated by diet, then the potential exists for contributing to epithelial cell health.
In the present study, we have used an in vitro model of the large bowel that allows both planktonic and surface associated bacteria to be studied and have used it to show that the prebiotic carbohydrates oligofructose and inulin selectively stimulate the growth of bifidobacteria in the surface associated flora. We went on to feed these carbohydrates to healthy patients scheduled to have a colonoscopy and examined their effect on the MAF in all regions of the large bowel.
PATIENTS AND METHODS
Fresh faecal specimens were obtained from six healthy volunteers (aged 24–55 years) who had no previous history of gastrointestinal disease and had not taken antibiotics for at least three months prior to the study. Faeces were collected separately from urine in a plastic bag suspended in a metal frame over a toilet and processed immediately.
Prebiotic feeding study
Twenty nine subjects were recruited from the colonoscopy waiting list at Addenbrookes Hospital. Exclusion criteria included: known or suspected inflammatory bowel disease, diarrhoea, colorectal carcinoma, and exposure to antibiotics in the preceding three months. Written consent was obtained from each patient, and the study approved by both the Medical Research Council Dunn Nutrition Ethics Committee and the Local Research Ethics Committee.
Prebiotics
Carbohydrates used were inulin (Raftiline HP) and oligofructose (Raftilose P95). Both were supplied by Orafti (Tienen, Belgium). Raftiline HP contains 99.5% inulin extracted from chicory roots. It comprises molecules of glucose-fructose (GF)N type, with N ranging from 5 to 60, and an average degree of polymerisation of 10. Raftilose P95 contains oligofructose 93.2% (GFN and FN, β(2, 1) in a ratio of 2:1) and glucose + fructose + sucrose 6.8%.
All chemicals, unless otherwise specified, were supplied by Sigma Ltd (Poole, Dorset, UK).
Batch culture experiments
Faecal slurries were prepared by homogenising fresh faeces in anaerobic culture media and sieving using a 250 μm aperture. The culture media contained (g/l): soluble starch 5.0; pectin 1.0; xylan 1.0; guar gum 1.0; arabinogalactan 1.0; casein 5.0; peptone 2.5 (Oxoid, Basingstoke, UK); tryptone 2.5 (Oxoid); yeast extract 4.5 (Oxoid); porcine gastric mucin type II 2.5; NaCl 4.5; K2HPO4 0.4; MgCl2 0.15; NH4Cl 0.4; KCl 0.25; cysteine 0.8; vitamin B12 0.005; haemin 0.01; and Tween 80 1.0. A vitamin solution (1 ml/l) was added which consisted of (mg/l): menadione 1.0; biotin 2.0; pantothenate 10.0; nicotinamide 5.0; p-aminobenzoic acid 5.0; and thiamine 4.0.
Duplicate batch culture fermenters (Soham Scientific, Ely, UK) were fitted with a mucin bait (that is, a series of glass slides suspended in a glass rack within the culture). These slides were precoated with an agar/mucin mix (1% agar (Oxoid), 3% porcine gastric mucin type I). The fermenters (working volume 300 ml) were inoculated with 10% (w/v) faecal slurry and 7.5 g/l oligofructose, and 7.5 g/l inulin was added to one vessel. The vessels were continually stirred, sparged with oxygen free nitrogen, and maintained at a pH of 5.8 and temperature of 37°C. Samples of liquid culture (1 ml) and surface associated cultures (one coated glass slide) were removed from each fermenter after 0, 3, 6, 12, and 24 hours and serially diluted in an anaerobic cabinet (10:10:80; H2:CO2:N2 atmosphere) with pre-reduced half strength peptone water. This model system for studying biofilm populations is described in detail elsewhere.21 Triplicate plates were then inoculated with 0.1 ml samples and incubated aerobically or anaerobically, as appropriate, at 37°C. Bacteria were counted on Wilkins-Chalgren agar (Oxoid; total anaerobes), nutrient agar (Oxoid; total aerobes), Wilkins-Chalgren agar plus gram negative supplements (Oxoid; bacteroides), Beerens agar (bifidobacteria), Rogosa agar (Oxoid; lactobacilli), and perfringens agar plus supplements (Oxoid; clostridia). Subsequently, bacteria were characterised to the genus level on the basis of morphology, Gram reaction, spore production, and metabolism using API 20A (Bio-Merieux, Lyon, France).22
Feeding study
Twenty nine volunteers were recruited. Fourteen subjects (eight men and six women; mean age 59 years (range 35–72)) supplemented their usual diet with a mixture of 2.5 g inulin and 2.5 g oligofructose three times a day for two weeks prior to their booked colonoscopy appointment. All kept a daily diary of supplements taken and recorded any side effects. Indications for colonoscopy were: previous colorectal polyps, a family history of colorectal carcinoma, iron deficiency anaemia, and rectal bleeding, Fifteen control subjects were recruited at the time of their colonoscopy appointment (six men and nine women; mean age 59 years (range 31–81)). Indications for colonoscopy were previous colorectal polyps, family history of colorectal carcinoma, iron deficiency anaemia, and rectal bleeding. All 29 subjects followed a low fibre diet the day prior to the procedure and took Picolax (sodium picosulphate and magnesium citrate; Ferring Pharmaceuticals, Feltham, Middlesex, UK), one sachet 24 hours and one 16 hours before the colonoscopy, which was performed under midazolam (Roche Products Ltd, Welwyn Garden City, UK) and pethidine sedation. Endoscopic biopsy samples were taken from macroscopically clean mucosa (no visible faecal material) using sterilised forceps from the caecum, transverse colon, descending colon, and rectum.
Bacteriological analysis of biopsies
Biopsy samples from the caecum and descending colon were transferred immediately into sterile pre-weighed bijoux bottles containing half strength peptone water. These were weighed and immediately removed to an anaerobic cabinet where samples were homogenised by hand and serially diluted and plated out as described above with the addition of MacConkey agar No 2 (Oxoid).
Bacterial isolates were characterised to species level on the basis of their cellular fatty acid profiles. Cell pellets from 30 ml of overnight culture in peptone yeast extract broth were saponified in 3.75 M NaOH in aqueous methanol (50% vol/vol). Fatty acids were then methylated by addition of two volumes of 3.25 N HCl in aqueous methanol prior to extraction. The Microbial Identification System (MIDI Inc. Newark, Delaware, USA) described previously23 was used to generate a cellular fatty acid profile for each isolate and identification obtained by comparison with the Moore Standard Library for anaerobic bacteria (http://www.midi-inc.com/pages/databases.html).
Short chain fatty acids were measured as previously described.16
Proliferation markers
Other biopsies from the caecum, transverse colon, descending colon, and rectum were fixed in formalin and processed to paraffin. Proliferation in colorectal epithelial cells was assessed by immunohistochemical staining for the cell cycle markers minichromosome maintenance protein 2 (Mcm-2), Ki67, and proliferating cell nuclear antigen (PCNA), as described previously.24,25 Stained sections were examined by a single observer (SJL) using a Nikon Eclipse E600 microscope. For every sample, the frequency of expression of each marker was determined in 2–4 well orientated complete crypts by calculating a labelling index (LI), representing the percentage of epithelial cell nuclei that stained positively in 2 to “n” well oriented complete crypts. (LI values were compared using the Wilcoxon signed rank test for paired data.)
Data and statistical analysis
For each experiment, data were analysed using the Excel statistical package. For the bacteriological and cell proliferation analysis, estimation of probability of difference between groups was undertaken by the Student t test.
RESULTS
Batch cultures
Surface associated populations were established on mucin coated slides after three hours of incubation. However, the results shown in table 1 ▶ are at 12 hours when the bacterial growth rate in the system was high and, as can be seen from the short chain fatty acid concentrations (table 2 ▶), a stable system existed. Total anaerobe to aerobe ratio was similar in both surface and planktonic populations at 100:1. The effect of the added prebiotic mix was to increase numbers of bifidobacteria. This prebiotic action was significant only for surface counts which increased by fivefold in the supplemented vessel, and by 2.5-fold in the planktonic phase (NS). Surface lactobacilli increased twofold and clostridia decreased twofold but these changes were not statistically significant.
Table 1.
Mean viable bacterial counts (mean (SEM)) from six batch cultures at 12 hours incubation with and without 1.5% w/v added prebiotic
| Bacteria | Planktonic counts (log10 CFU/ml culture) | Surface counts (log10 CFU/slide) | ||
| Control | +Prebiotic | Control | +Prebiotic | |
| Total aerobes | 6.9 (0.3) | 6.8 (0.2) | 4.9 (0.5) | 5.0 (0.4) |
| Total anaerobes | 9.7 (0.2) | 9.6 (0.3) | 7.8 (0.3) | 7.6 (0.3) |
| Bacteroides | 9.5 (0.2) | 9.4 (0.3) | 7.5 (0.3) | 7.2 (0.2) |
| Bifidobacteria | 8.4 (0.1) | 8.8 (0.2) | 6.6 (0.2) | 7.3 (0.2)* |
| Lactobacilli | 3.5 (0.6) | 4.1 (0.3) | 2.2 (0.3) | 2.5 (0.4) |
| Clostridia | 6.5 (0.3) | 6.2 (0.4) | 4.8 (0.6) | 4.4 (0.5) |
Significant difference: control versus prebiotic, *p<0.006
Table 2.
Short chain fatty acids, lactate, and succinate concentrations (mean (SEM) mM) in batch culture media with and without 1.5% w/v added prebiotic over 24 hours
| Time (h) | Acetate | Propionate | Butyrate | Valerate | Caproate | Lactate | Succinate | |||||||
| Con | Pre | Con | Pre | Con | Pre | Con | Pre | Con | Pre | Con | Pre | Con | Pre | |
| 0 | 13 (1.2) | 13 (1.2) | 5 (0.7) | 5 (0.7) | 3 (0.7) | 3 (0.7) | 0.7 (0.2) | 0.7 (0.2) | 0.7 (0.2) | 0.7 (0.2) | 1.7 (04) | 1.7 (0.4) | 1.2 (0.5) | 1.2 (0.5) |
| 3 | 43 (4.1) | 76 (13.5) | 17 (3.3) | 22 (4.9) | 14 (2.9) | 17 (3.3) | 2.3 (0.7) | 2.2 (0.6) | 1.3 (0.5) | 1.4 (0.4) | 4.8 (1.9) | 12 (3.0) | 3.3 (2.4) | 5.3 (2.7) |
| 6 | 69 (4.9) | 160 (20.4) | 28 (3.3) | 48 (16.7) | 29 (4.5) | 43 (8.2) | 7 (2.4) | 4.2 (1.4) | 3.6 (1.8) | 2.1 (1.1) | 3.1 (1.9) | 14 (8.0)* | 3.0 (2.4) | 5.4 (3.4) |
| 12 | 90 (4.1) | 200 (13.9)* | 40 (2.4) | 65 (18.4) | 42 (2.0) | 89 (5.3)* | 18 (4.9) | 20 (4.9) | 7.4 (2.3) | 11 (6.5) | 3.5 (2.9) | 26 (9.4)* | 4.0 (3.4) | 7.4 (6.8) |
| 24 | 99 (5.3) | 208 (13.5)* | 45 (3.7) | 57 (8.9) | 54 (4.1) | 96 (2.4)* | 23 (5.3) | 17 (4.5) | 10 (2.9) | 23 (10.2) | ||||
Con, control; Pre, prebiotic.
Significantly different from control, *p<0.001.
Short chain fatty acid concentrations increased progressively over the first 12 hours of the batch culture and then showed very similar levels at 24 hours. Concentrations of acetate and butyrate were significantly higher with prebiotic at 12 hours and 24 hours, as might be expected with the extra substrate provided. The molar ratio of acetate:propionate:butyrate was 52:23:24 at 12 hours in control vessels and 56:18:25 with prebiotic, which were not significantly different. Lactate concentrations also increased with prebiotic, as did caproate, but no such changes were seen with valerate or succinate.
FEEDING STUDY
Compliance and tolerability
The prebiotic supplement was well tolerated by subjects although all reported an increase in flatulence (mild-moderate in 10, severe in four). Other side effects observed were bloating, (mild-moderate in seven, severe in two) and a laxative effect (mild-moderate in 10, severe in one), as noted in previous studies.18,26 Scores were similar for both weeks of the supplementation period. Compliance with the supplement was excellent, with subjects taking on average 97% of the supplements (range 81–100%).
Effect on mucosal flora
Overall, total anaerobe counts were unaffected by the addition of the prebiotic to the diet (table 3 ▶). A prebiotic effect was seen in both the right and left colon. Mucosa associated bifidobacteria were detected in 12 of 14 of the supplemented group compared with seven of 15 of the control group. The effect of the prebiotic was to give a difference in mean counts of 1 order of magnitude (1 log10) greater. Analysis of bacterial populations at the species level revealed Bifidobacterium angulatum as the dominant bifidobacteria in both groups, comprising 71% of the isolates from the supplemented and 50% of those from the control group. Mucosal lactobacilli were detected in eight of 14 of the fed subjects compared with only one subject in the control group. Overall numbers were 0.5–0.7 log10 greater with the prebiotic. Eubacteria, the second most numerous faecal isolates,27 were detected in 10 of 14 supplemented subjects and five of 15 control subjects. Eubacteria showed large differences between the groups with 1.5–1.6 log10 greater numbers with the prebiotic. All of these changes were statistically significant. In contrast, numbers of mucosal coliforms and thus total aerobes were lower by 0.6 and 0.5 logs with the prebiotic although the range of counts was very wide (4.2–8.8 in the control group, 3–8.2 in the supplemented group) and the result was not statistically significant. Prebiotic feeding did not change total bacteroides numbers and bacteroides species distribution. Overall, numbers of clostridia were unaffected by the prebiotic supplementation although marked species differences were noted, with Clostridium innocuum dominating the supplemented group (66% of all clostridia identified) but representing only 10% of control clostridia, 50% of which were identified as Clostridium cocleatum, a species not detected in the supplemented group. Total anaerobe numbers remained steady and the anaerobe:aerobe ratio was approximately 100:1 in both groups.
Table 3.
Effect of supplementing diet with 15 g/day prebiotic mix on mucosa associated flora of the right and left colon
| Bacteria | Proximal colon | Distal colon | ||
| Control | +Prebiotic | Control | +Prebiotic | |
| Total aerobes | 6.4 (0.4) | 5.9 (0.4) | 6.4 (0.3) | 5.9 (0.4) |
| Coliforms | 6.2 (0.4) | 5.6 (0.4) | 6.2 (0.3) | 5.7 (0.4) |
| Total anaerobes | 8.5 (0.2) | 8.6 (0.2) | 8.7 (0.1) | 8.6 (0.1) |
| Bacteroides | 8.1 (0.3) | 8.3 (0.2) | 8.3 (0.2) | 8.5 (0.2) |
| Bifidobacteria | 5.3 (0.4) | 6.3 (0.3)* | 5.2 (0.3) | 6.4 (0.3)† |
| Eubacteria | 4.5 (0.3) | 6.0 (0.4)‡ | 4.6 (0.3) | 6.1 (0.3)§ |
| Lactobacilli | 3.0 (0.1) | 3.7 (0.2)¶ | 3.1 (0.1) | 3.6 (0.2)** |
| Clostridia | 5.1 (0.3) | 4.9 (0.3) | 5.0 (0.3) | 4.9 (0.3) |
Values are expressed as mean (SEM) in log CFU/g mucosa.
Significance of difference, control versus prebiotic: *p = 0.059, †p = 0.01, ‡p = 0.003, §p = 0.002, ¶p = 0.017,**p = 0.04.
Proliferation indices
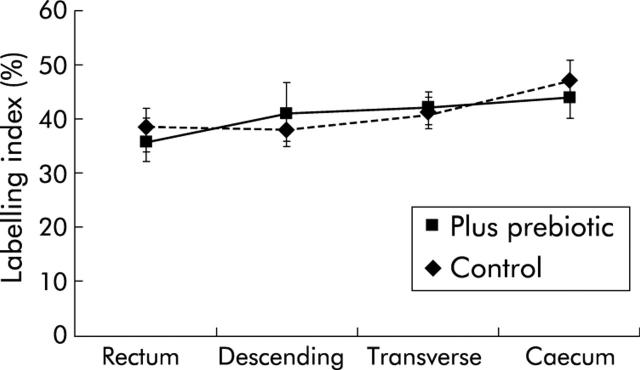
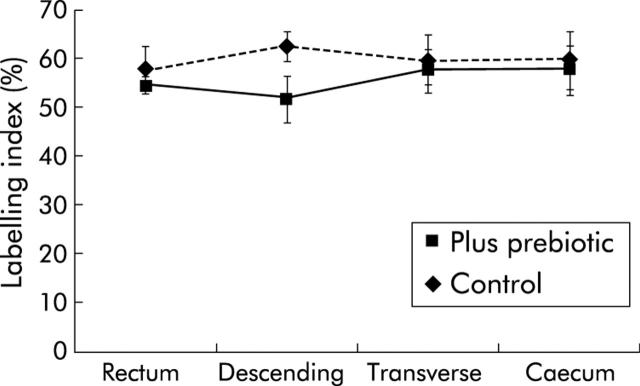
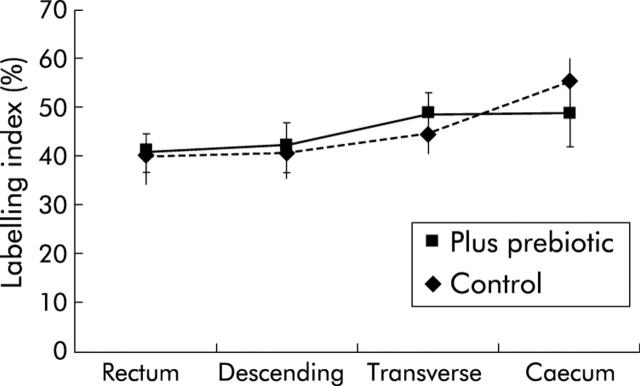
The mucosa was macroscopically normal in all subjects and this appearance did not change with the prebiotic supplement. For all biopsies, more cells were stained with the antibody to MCM-2 protein than with those to PCNA and Ki67. As shown in figs 1 ▶–3 ▶, the prebiotic supplementation had no effect on labelling indices for any of the markers of mucosal proliferation at the four sites of the colon studied.
Figure 1.
K167 labelling index from four sites of the colon in control subjects and those fed prebiotic. Values are mean (SEM).
Figure 3.
Minichromosome maintenance protein 2 labelling index from four sites of the colon in control subjects and those fed prebiotic. Values are mean (SEM).
DISCUSSION
These studies have shown clearly, both by the use of an in vitro model and in vivo, that the composition of surface associated flora can be influenced by substrate and hence by diet. Moreover, the dietary changes required are very small; only a few grams of sugar-like carbohydrates were added to subjects’ food each day. Equally important is the nature of the change in the flora. An increase in bifidobacteria and lactobacilli in the gut, either by the use of pro or prebiotics, is currently considered a healthy objective. There are no previously reported studies in humans of any dietary factor that can be shown to alter the composition of the MAF. In a recent paper, Kleessen and colleagues28 showed in human flora associated rats that feeding an oligofructose/inulin mixture increased counts of mucosa associated bifidobacteria in the colon and improved mucosal architecture and mucus secretion.
In the in vitro model, the overall composition of the flora was the same whether in the planktonic or surface associated phase (table 1 ▶), although bacterial densities were lower at 12 hours on the agar-mucin surface. A clear bifidogenic effect was seen on the mucus baits with no notable changes in other genera or in the planktonic bacteria. This absence of a selective effect on the planktonic bifidobacteria may reflect the fact that truly free living (that is, not surface associated) bacteria do not exist in the luminal phase of the large bowel (see below). Alternatively, the use of multiple carbohydrate substrates in the batch culture medium, in addition to oligofructose and inulin, may have reduced the importance of the prebiotics in determining the pattern of overall growth in the mixed culture.
Short chain fatty acid concentrations increased substantially with the additional prebiotic substrate but no change in molar ratios was seen and thus these carbohydrates cannot be seen as providing selective increases in butyrate over other short chain fatty acids, although overall they are a good source of butyrate. These findings are similar to those observed in human studies of prebiotic feeding and faecal composition.16 The increase in lactate with added prebiotic is probably a consequence of the presence of rapidly metabolisable substrate in excess.
In the in vivo study, MAF reflected very much the bacterial composition that has been reported for planktonic and faecal populations. This is perhaps not surprising because the notion that a true planktonic flora exists in the large intestine is probably incorrect. Most, if not all, of the flora in the hind gut that are not mucosa associated are attached to particulate matter.29,30 Macfarlane and colleagues (in 1995 and 1997) showed that the composition of the flora attached to particles in the large bowel is very similar to that which is “free living” but that metabolically they are different. Increased activities of polysaccharidases, glycosidases, proteases, and arylamidases, and mucus degrading enzymes are seen in the particle associated species, probably reflecting the nature of the underlying surface material which will largely comprise plant cell walls, entrapped in a viscoelastic gel of the many mucins that are secreted into the gut. End products of fermentation also differ, with surface associated communities producing more acetate than planktonic flora.31
Addition of only 15 g/day of inulin and oligofructose produced significant changes in the MAF in both the proximal and distal colon. Most importantly, counts of bifidobacteria increased by 1 log10 or more, the principal species being B angulatum. The increase in this particular genus is the classic prebiotic effect demonstrated previously in vitro32–34 and in vivo in faecal flora.16–18 The essence of prebiosis, however, is the selectivity of the effect. In the present study, the absence of significant increases in bacteroides, clostridia, coliforms, and total aerobes and anaerobes is reassuring. Numbers of lactobacilli also increased significantly, but this genus, like bifidobacteria, is thought to be part of the beneficial or protective flora of the gut. We also observed significant changes in eubacteria, an observation not previously noted in studies of prebiosis. It is not clear why this should occur in MAF and not planktonic or faecal flora. However, it is known that bacteria living in biofilms on surfaces show altered substrate use and efficiency of metabolism as well as resistance to host defence mechanisms and antibiotics.35–37 Metabolic consortia build up and substrate use becomes efficient. It is possible therefore that eubacteria benefited from cross feeding of carbohydrate residues from the initial actions of bifidobacteria and lactobacilli on prebiotics. Many eubacteria are saccharolytic.
Inulin and oligofructose are fructans that are not hydrolysed by pancreatic enzymes and escape digestion in the small bowel.38–40 Bifidobacteria have relatively high amounts of β-fructosidase that is selective for β1–2 glycosidic bonds in fructans.41 Subsequent transport mechanisms and sites of hydrolysis may also be faster. After oligosaccharide hydrolysis, monomers then serve as an efficient growth substrate for the bifidus pathway of hexose fermentation.42 In addition, the inhibitory effects of bifidobacterial growth on other colonic organisms6 are likely to assist in the competitive influence that occurred with oligofructose and inulin. Lactobacilli can also ferment FOS,43 and human feeding studies have shown that faecal populations of these bacteria can be increased in some subjects when fed prebiotics.18,44 Data presented in this study show that lactobacilli appear to compete equally effectively for prebiotic substrates at the mucosal surface.
In any study of the gut microflora, methodology is always an important issue. Culture based methods are known to introduce bias towards organisms that are easier to culture and recently, studies using molecular techniques have improved our knowledge of bacteria that are more difficult to culture and which form a significant portion of the microbiota. Culture based methodologies will never account for the entire microbial population, are slow, require immediate sample handling and identification by a variety of morphological, metabolic, and biochemical techniques. Phenotypic methods of identification are unlikely to match the discriminatory power of some genotypic tools. However, culture based methods are well established, cost effective, sensitive, and allow direct comparison with the many similar studies reported in the literature. They give valuable information of the likely metabolic potential of the overall population. Molecular analyses are important in contributing to the gaps in our knowledge but no method is infallible and other factors need to be considered with these newer methods. They are also subject to bias—for example, bacteria that are more susceptible to lysis, probe permeability, and amplification will be more readily detected. There are still a very limited number of probes available at species level. FISH methods are almost impossible to quantify and molecular techniques in general have lower sensitivities. Techniques such as DGGE (molecular fingerprinting) are not quantitative, and the metabolic contribution of “molecular species” cannot be determined. Both methodologies continue to contribute to our understanding of the large bowel ecosystem but it is important to consider the limitations of each technique, particularly when comparing populations of absolute cell numbers to those given as a proportion of the total bacteria present. Similar population sizes have been found by both in situ hybridisations and bacterial cell culture for organisms such as bifidobacteria and eubacteria, but the size of the total microflora is underestimated by the latter method, which would alter the proportional value.45,46 However, it currently remains prudent to consider evidence using both methodologies and this study clearly shows a significant change in the culturable portion of the MAF.
Widely differing results for the composition of the MAF have been reported. This is likely to be partly because of the different, and changing, methods referred to above, now used in bacteriology. Swidsinski and colleagues9 found low total counts, by conventional culture techniques, of MAF in 40 control subjects although these bacteria were similar in composition to faecal (planktonic) flora. However, biopsy specimens were vigorously washed before culturing. Much higher MAF counts were found in patients with active ulcerative colitis in this study. Schultsz and colleagues7 also observed very low counts of MAF in nine controls undergoing colonoscopy. Specimens were not washed but were fixed in formalin embedded in paraffin and bacteria detected by non-radioactive detection of rRNA by in situ hybridisation. Again, high counts were found in ulcerative colitis patients. Poor staining in the controls was ascribed to the MAF being inactive or dead, poor antibody penetration, and the barrier effect of mucus. Others have shown a numerous and diverse MAF in colonic or rectal mucosal from either sudden death victims47 or from colonoscopic and rectal biopsies of healthy subjects.8,48–51 In these reports, however, standard culturing techniques were used and biopsies were either not washed or washed gently to remove visible adherent faecal material. Similarly, Zoetendal and colleagues52 took biopsies at colonoscopy from the colons of 10 individuals and used 16S RNA probes to analyse bacterial diversity on paraformaldehyde fixed samples. Bacterial counts up to 106 were found in seven of 10 subjects but showed fewer species diversity than samples of faecal flora. Whether these bacteria are largely in the mucus layer, rather than adherent to epithelial cells, is a likely possibility but their proximity to the mucosal and thus interaction with epithelial and immune cells is an important interface. In situations where mucus secretion is thought to be abnormal, epithelial adherent populations exist.7,9
Alterations in the MAF had no effect on cell cycle entry by mucosal epithelial cells at four different sites in the large bowel. Our observation that Mcm-2 was present in significantly greater numbers of epithelial cells than the traditionally used proliferation markers Ki67 and PCNA is consistent with data from other studies.25,53 MCM proteins are the most sensitive markers available of cell cycle entry, being abundant in nuclei throughout the cell cycle and lost rapidly following differentiation, as occurs in large bowel crypts.25 It should be noted that Mcm-23, Ki67, and PCNA all essentially provide information concerning the cell cycle state of the stained tissue and more subtle differences in rates of cycling would not be identified using our approach.
Does the composition of the MAF matter? Almost certainly yes. Gut flora clearly interact with the mucosal immune system11–15 which is important in the process of developing tolerance to commensal flora. In mucosal disorders such as ulcerative colitis, this tolerance may have broken down.54–56 Changing the composition towards a flora with increased bifidobacteria and lactobacilli is thought to be beneficial to the host.57 These genera contain few if any pathogenic species and are a major part of the large bowel defence against invading pathogenic organisms through their capacity to secrete peptides that inhibit pathogen growth.6,58,59 When given as probiotics, lactobacilli adhere to the colonic and rectal mucosa46,60 although probably do not permanently colonise the gut epithelium.61 Probiotic bacteria are clearly established as being a benefit in the prevention of antibiotic associated diarrhoea62,63 and reducing the duration of infectious diarrhoea in children.64 Animal models indicate that they may have a protective effect against the development of colorectal cancer65,66 an effect that can be strengthened by combination with a prebiotic.67
The health benefits of prebiotic carbohydrates have yet to be established. Apart from some benefits to calcium absorption, no clear value has been shown.68 However, research in this area is relatively recent while the benefits of probiotics have been known for 100 years.
Moving the composition of the MAF towards one with increased numbers and more diverse species of bifidobacteria and lactobacilli should be beneficial to colonic health. If this can be achieved by a small change in the carbohydrate composition of the diet, then new possibilities emerge for public health and for preventive strategies by the gastroenterologist.
Figure 2.
Proliferating cell nuclear antigen labelling index from four sites of the colon in control subjects and those fed prebiotic. Values are mean (SEM).
Acknowledgments
SJL was supported by a grant from the Addenbrookes Hospital Trust Fund
Abbreviations
MAF, mucosa associated flora
G, glucose
F, fructose
Mcm-2, minichromosome maintenance protein 2
PCNA, proliferating cell nuclear antigen
LI, labelling index
REFERENCES
- 1.Macfarlane GT, Cummings JC. Diet and the metabolism of intestinal bacteria. In: Brostoff J, Challacombe SJ, eds. Food allergy and intolerance. Amsterdam: Elsevier Science, 2002:321–41.
- 2.Williams BA, Verstegen MWA, Tamminga S. Fermentation in the large intestine of single-stomached animals and its relationship to animal health. Nutr Res Rev 2001;14:207–27. [DOI] [PubMed] [Google Scholar]
- 3.Houghton LA, Green TJ, Donovan UM, et al. Association between dietary fiber intake and the folate status of a group of female adolescents. Am J Clin Nutr 1997;66:1414–21. [DOI] [PubMed] [Google Scholar]
- 4.Rowland IR. Toxicology of the colon: role of the intestinal microflora. In: Gibson GR, Macfarlane GT, eds. Human colonic bacteria. Role in nutrition and, physiology, and pathology. Boca Raton: CRC Press, 1995:155–74.
- 5.Gorbach SL, Barza M, Guilano M, et al. Colonization resistance of the human intestinal microflora: testing the hypothesis in normal volunteers. Eur J Clin Microbiol Infect Dis 1988;7:98–102. [DOI] [PubMed] [Google Scholar]
- 6.Gibson GR, Wang X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol 1994;77:412–20. [DOI] [PubMed] [Google Scholar]
- 7.Schultsz C , Berg FVD, Fiebo W, et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 1999;117:1089–97. [DOI] [PubMed] [Google Scholar]
- 8.Pathmakanthan S , Thornley JP, Hawkey CJ. Mucosally associated bacterial flora of the human colon: quantitative and species specific differences between normal and inflamed colonic biopsies. Microb Ecol Health Dis 1999;11:169–74. [Google Scholar]
- 9.Swidsinski A , Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002;122:44–54. [DOI] [PubMed] [Google Scholar]
- 10.Stephen AM, Cummings JH. Mechanism of action of dietary fibre in the human colon. Nature 1980;284:283–4. [DOI] [PubMed] [Google Scholar]
- 11.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 2002;22:283–307. [DOI] [PubMed] [Google Scholar]
- 12.Sartor RB. Induction of mucosal immune responses by bacteria and bacterial components. Curr Opin Gastroenterol 2001;17:555–61. [DOI] [PubMed] [Google Scholar]
- 13.Hooper LV, Wong MH, Thelin A, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 2001;291:881–4. [DOI] [PubMed] [Google Scholar]
- 14.Macpherson AJ, Gatto D, Sainsbury E, et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000;288:2222–6. [DOI] [PubMed] [Google Scholar]
- 15.Lu L , Walker A. Pathologic and physiologic interactions of bacteria with the gastrointestinal epithelium. Am J Clin Nutr 2001;73:11224–30S. [DOI] [PubMed] [Google Scholar]
- 16.Gibson GR, Beatty ER, Wang X, et al. Selective stimulation of Bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 1995;108:975–82. [DOI] [PubMed] [Google Scholar]
- 17.Kleessen B , Sykura B, Zunft H-J, et al. Effects of inulin and lactose on faecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am J Clin Nutr 1997;65:1397–492. [DOI] [PubMed] [Google Scholar]
- 18.Ito M , Deguchi Y, Miyamori A, et al. Effects of administration of galactooligosaccharides on the human faecal microflora, stool weight and abdominal sensation. Microb Ecol Health Dis 1990;3:285–92. [Google Scholar]
- 19.Schley PD, Field CJ. The immune-enhancing effects of dietary fibres and prebiotics. Br J Nutr 2002;87:S221–30. [DOI] [PubMed] [Google Scholar]
- 20.Zopf D , Roth S. Oligosaccharide anti-infective agents. Lancet 1996;347:1017–21. [DOI] [PubMed] [Google Scholar]
- 21.Macfarlane GT, Macfarlane S. Activities of human colonic mucin degrading bacteria on surfaces and in biofilms. In: Corfield A, ed. Mucin, methods and protocols. Totowa: Humana Press, 2000:439–52.
- 22.Macfarlane GT, Cummings JH, Allison C. Protein degradation by human intestinal bacteria. J Gen Microbiol 1986;132:1647–56. [DOI] [PubMed] [Google Scholar]
- 23.Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 2001;48:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams GH, Romanowski P, Morris L, et al. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci U S A 1998;95:14932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman A , Morris LS, Mills AD, et al. The minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res 1999;5:2121–32. [PubMed] [Google Scholar]
- 26.Stone-Dorshow T , Levitt MD. Gaseous response to ingestion of a poorly absorbed fructo-oligosaccharide sweetener. Am J Clin Nutr 1987;46:61–5. [DOI] [PubMed] [Google Scholar]
- 27.Finegold SM, Sutter VL, Mathisen GE. Normal indigenous intestinal flora. In: Hentges DJ, ed. Human intestinal microflora in health and disease. New York: Academic Press, 1983:3–31.
- 28.Kleessen B , Hartmann L, Blaut M. Fructans in the diet cause alterations of intestinal mucosal architecture, released mucins and mucosa-associated bifidobacteria in gnotobiotic rats. Br J Nutr 2003;89:597–606. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane GT, Macfarlance S. Human intestinal ‘biofilm’ communities. In: Lappin-Scott H, ed. The life and death of biofilm. Cardiff: BioLine, 1995:83–7.
- 30.Macfarlane S , McBain AJ, Macfarlane GT. Consequences of biofilm and sessile growth in the large intestine. Adv Dental Res 1997;11:59–68. [DOI] [PubMed] [Google Scholar]
- 31.Macfarlane S , Cummings JH, Macfarlane GT. Bacterial colonisation of surfaces in the large intestine. In: Roberfroid MB, ed. Colonic microbiota, nutrition and health. Netherlands: Kluwer Academic Publishers, 1999:71–87.
- 32.Yazawa K , Tamura Z. Search for sugar sources for selective increase of bifidobacteria. Bifidobacteria Microflora 1982;1:39–44. [Google Scholar]
- 33.Tamura Z . Nutriology of bifidobacteria. Bifidobacteria Microflora 1983;2:3–16. [Google Scholar]
- 34.Wang X , Gibson GR. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J Appl Bacteriol 1993;75:373–80. [DOI] [PubMed] [Google Scholar]
- 35.Costerton JW, Cheng K-J, Geesey GG, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol 1987;41:435–64. [DOI] [PubMed] [Google Scholar]
- 36.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999;284:1318–22. [DOI] [PubMed] [Google Scholar]
- 37.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet 2001;358:135–8. [DOI] [PubMed] [Google Scholar]
- 38.Bach-Knudsen KE, Hessov I. Recovery of inulin from Jerusalem artichoke (Helianthus tuberosus L.) in the small intestine of man. Br J Nutr 1995;74:101–3. [DOI] [PubMed] [Google Scholar]
- 39.Ellegard L , Andersson H, Bosaeus I. Inulin and oligofructose do not influence the absorption of cholesterol, or the excretion of cholesterol, Ca, Mg, Zn, Fe, or bile acids but increases energy excretion in ileostomy subjects. Eur J Clin Nutr 1997;51:1–5. [DOI] [PubMed] [Google Scholar]
- 40.Molis C , Flourie B, Ouarne F, et al. Digestion, excretion and energy value of fructooligosaccharides in healthy humans. Am J Clin Nutr 1996;64:324–8. [DOI] [PubMed] [Google Scholar]
- 41.De Vries W , Stouthamer AH. Carbohydrate metabolism in Bifidobacterium bifidum var pennsylvanicus. Biochim Biophys ACTA 1967;136:415–25. [DOI] [PubMed] [Google Scholar]
- 42.Scardovi V . The fructose-6-phosphate shunt as a peculiar pattern of hexose degradation in the genus Bifidobacterium. Biochim Biophys ACTA 1965;15:19–24. [Google Scholar]
- 43.Kaplan H , Hutkins RW. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl Environ Microbiol 2000;66:2682–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams CH, Witherly SA, Buddington RK. Influence of dietary neosugar on selected bacterial groups of the human faecal microbiota. Microb Ecol Health Dis 1994;7:91–7. [Google Scholar]
- 45.Langendijk PS, Schut F, Jansen GJ, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microb 1995;61:3069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwiertz A , le Bay G, Blaut M. Quantification of different Eubacterium spp. in human fecal samples with species-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 2000;66:375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croucher SC, Houston AP, Bayliss CE, et al. Bacterial populations associated with different regions of the human colon wall. Appl Environ Microbiol 1983;45:1025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson M-L, Molin G, Jeppsson B, et al. Administration of different Lactobacillus strains in fermented oatmeal soup: In vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol 1993;59:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poxton IR, Brown R, Sawyerr A, et al. Mucosa-associated bacterial flora of the human colon. J Med Microbiol 1997;46:85–91. [DOI] [PubMed] [Google Scholar]
- 50.Poxton IR, Brown R, Sawyerr AF. The mucosal anaerobic gram-negative bacteria of the human colon. Clin Infect Dis 1997;25:S111–13. [DOI] [PubMed] [Google Scholar]
- 51.Hartley CL, Neumann CS, Richmond MH. Adhesion of commensal bacteria to the large intestine wall in humans. Infect Immun 1979;23:128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from faeces. Appl Environ Microbiol 2002;68:3401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott IS, Morris LS, Bird K, et al. A novel immunohistochemical method to estimate cell cycle state and phase in archival tissue: implications for estimation of outcome in colorectal cancer. J Pathol 2003;201:187–97. [DOI] [PubMed] [Google Scholar]
- 54.Macpherson A , Khoo UY, Forgacs I, et al. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut 1996;38:365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology 2002;123:689–99. [DOI] [PubMed] [Google Scholar]
- 56.Sartor RB. Intestinal microflora in human and experimental inflammatory bowel disease. Curr Opin Gastroenterol 2001;17:324–30. [DOI] [PubMed] [Google Scholar]
- 57.Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Report of a Joint FAO/WHO Expert Consultation. Cordoba, Argentina: FAO/WHO, 2001.
- 58.Lievin V , Peiffer I, Hudault S, et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 2000;47:646–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hopkins MJ, Macfarlane GT. Non-digestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl Environ Microbiol 2003;69:1920–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alander M , Korpela R, Saxelin M, et al. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett Appl Microbiol 1997;24:361–4. [DOI] [PubMed] [Google Scholar]
- 61.Bezkorovainy A . Probiotics: determinants of survival and growth in the gut. Am J Clin Nutr 2001;73:399S–405. [DOI] [PubMed] [Google Scholar]
- 62.D’Souza AL, Rajkumar C, Cooke J, et al. Probiotics in prevention of antibiotic associated diarrhoea: meta-analysis. BMJ 2002;324:1361–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cremonini F , Di Caro S, Nista EC, et al. Meta-analysis: the effect of probiotic administration on antibiotic-associated diarrhoea. Aliment Pharmacol Ther 2002;16:1461–7. [DOI] [PubMed] [Google Scholar]
- 64.Huang JS, Bousvaros A, Lee JW, et al. Efficacy of probiotic use in acute diarrhea in children: A meta-analysis. Dig Dis Sci 2002;47:2625–34. [DOI] [PubMed] [Google Scholar]
- 65.Wollowski I , Rechkemmer G, Pool-Zobel BL. Protective role of probiotics and prebiotics in colon cancer. Am J Clin Nutr 2001;73:451–5S. [DOI] [PubMed] [Google Scholar]
- 66.Cummings JH, Macfarlane GT. Bacteria in the pathogenesis of colorectal cancer. In: Scheurlen M, ed. Exogenous factors in colonic carcinogenesis. Amsterdam: Kluwer Academic Publishers, 2002:180–91.
- 67.Bolognani F , Rumney CJ, Pool-Zobel BL, et al. Effect of lactobacilli, bifidobacteria and inulin on the formation of aberrant crypt foci in rats. Eur J Nutr 2001;40:293–300. [DOI] [PubMed] [Google Scholar]
- 68.Roberfroid M , Gibson G. Nutritional health benefits of inulin and oligofructose. Br J Nutr 2002;87:S1–311.11895145 [Google Scholar]