Abstract
Ubiquitin-dependent degradation of regulatory proteins controls many cellular processes, including cell cycle progression, morphogenesis, and signal transduction. Skp1p-cullin-F-box protein (SCF) complexes are ubiquitin ligases composed of a core complex including Skp1p, Cdc53p, one of multiple F-box proteins that are thought to provide substrate specificity to the complex, and the ubiquitin-conjugating enzyme, Cdc34p. It is not understood how SCF complexes are regulated and how physiological conditions alter their levels. Here we show that three F-box proteins, Grr1p, Cdc4p, and Met30p, are unstable components of the SCF, and are themselves degraded in a ubiquitin- and proteasome-dependent manner in vivo. Ubiquitination requires all the core components of the SCF and an intact F-box, suggesting that ubiquitination occurs within the SCF complex by an autocatalytic mechanism. Cdc4p and Grr1p are intrinsically unstable, and their steady-state levels did not fluctuate through the cell cycle. Taken together, our results suggest that ubiquitin-dependent degradation of F-box proteins allows rapid switching among multiple SCF complexes, thereby enabling cells to adapt quickly to changing physiological conditions and progression through different phases of the cell cycle.
Keywords: cell cycle, Skp1p-cullin-F-box protein, Grr1p protein, proteasome
Post-translational modification by ubiquitin targets many proteins for rapid degradation by the 26S proteasome (1). Covalent attachment of ubiquitin onto lysine residues of the substrate requires the coordinated action of three classes of enzymes: the E1 ubiquitin-activating enzymes, the E2 ubiquitin-conjugating enzymes, and the E3 ubiquitin ligases (2). Whereas the E1 and E2 enzymes are primarily involved in activating and transferring ubiquitin through high-energy thioester bonds, E3 enzymes play the critical step in providing the specificity of substrate recognition. At least two multiprotein complexes function as E3-ubiquitin ligases for many cell cycle regulators (3): the anaphase-promoting complex (APC) promotes entry into anaphase and exit from mitosis, whereas the Skp1p-cullin-F-box protein (SCF) complex regulates the G1–S phase transition. In budding yeast, entry into S phase requires ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Sic1p (4). Genetic analysis has revealed four proteins, Cdc4p, Cdc34p, Cdc53p, and Skp1p, which are required for its ubiquitination (5, 6). These proteins are detected in high molecular weight complexes in vivo (7) and are sufficient in vitro to ubiquitinate phosphorylated Sic1p in the presence of ubiquitin and an E1 enzyme (5, 6). CDC34 encodes an E2 ubiquitin-conjugating enzyme (8), whereas Cdc4p contains a conserved motif called the F-box, which mediates the interaction with Skp1p (9). A large family of proteins containing F-boxes has been discovered. It is thought that these proteins are substrate-specific adaptor subunits that recruit substrates to a core ubiquitination complex composed of Cdc53p, Cdc34p, and Skp1p (9, 10). Degradation of the G1-cyclins Cln1p and Cln2p (11) and the bud emergence protein Gic2p (12) requires the SCF core subunits and the F-box protein Grr1p but not Cdc4p, whereas degradation of the Cdk-inhibitory kinase Swe1p depends on the F-box protein Met30p (13). Grr1p directly interacts with phosphorylated Cln2p through leucine-rich repeats (5, 14); Cdc4p contains WD repeats which are necessary to bind phosphorylated Sic1p (5, 6). Phosphorylation of many substrates is required for ubiquitin-dependent degradation in vivo and regulates their interaction with F-box proteins (15). Thus, multiple SCF complexes exist in vivo that target various phosphorylated substrates for ubiquitin-dependent degradation. Importantly, these SCF complexes differ in the composition of the F-box protein.
It is not known how cells regulate the level of the different SCF complexes and how these levels are altered through the cell cycle and in response to extracellular signals. The human F-box protein Skp2p is expressed in a cell cycle-dependent manner (16), suggesting that regulation of SCF complexes may mainly affect F-box proteins. Here we show that F-box proteins are themselves intrinsically unstable and are degraded by the ubiquitin-dependent pathway. Ubiquitination requires all the core components of the SCF complex and an intact F-box, suggesting that auto-ubiquitination occurs within the assembled SCF complex. Thus, our results suggest a mechanism that ensures a dynamic equilibrium between multiple SCF complexes, thereby enabling cells to rapidly adapt to changing environmental conditions and progress through the cell cycle.
METHODS
Yeast Strains and Genetic Experiments.
Yeast strains are described in Table 1. The genotypes of the yeast strains are as follows: W303, ade2-1, trp1-1, can1-100, leu2-3,112, his3-11,15, ura3, GAL+, psi+, ssd1-d2; and S288C, ade2-101, ura3-52, lys2-801, trp1-Δ1, his3Δ200, leu2-Δ. Standard yeast growth conditions and genetic manipulations were used (17). Pheromone response and mating assays were performed as described (18). Strains expressing myc- or hemagglutinin (HA) epitope-tagged versions of the proteins encoded by GRR1, MET30, and CDC4 were constructed as described (19).
Table 1.
Strains list
| Name | Relevant genotype | Background | Source |
|---|---|---|---|
| JMG101 | grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG102 | cdc4::CDC4-MYC-TRP1 | W303 | This study |
| JMG68 | met30::MET30-HA-His3-MX6 | W303 | This study |
| JMG108 | grr1::GRR1-MYC-TRP1 cln2::CLN2-HA-LEU2 | W303 | This study |
| JMG107 | cdc4::CDC4-MYC-TRP1 cln2::CLN2-HA-LEU2 | W303 | This study |
| JMG66 | erg6::LEU2 cdc4::CDC4-MYC-TRP1 | S288C | This study |
| JMG67 | erg6::LEU2 grr1::GRR1-MYC-TRP1 | S288C | This study |
| JMG53 | grr1::GRR1-MYC-TRP1 | S288C | This study |
| YMP241 | cim5-1 | S288C | C. Mann |
| JMG54 | cim5-1 grr1::GRR1-MYC-TRP1 | S288C | This study |
| JMG62 | cdc16-1 grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG28 | cdc34-2 grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG30 | cdc53-1 grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG26 | cdc4-1 grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG27 | cdc34-1 cdc4::CDC4-MYC-TRP1 | W303 | This study |
| JMG29 | skp1-12 grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG31 | skp1-11 grr1::GRR1-MYC-TRP1 | W303 | This study |
| JMG69 | skp1-12 cdc4::CDC4-MYC-TRP1 | W303 | This study |
| JMG70 | skp1-11 cdc4::CDC4-MYC-TRP1 | W303 | This study |
| JMG61 | grr1::GRR1-AAA-MYC-TRP1 | W303 | This study |
| YM2957 | grr1::LEU2 | S288C | C. Mann |
| JMG24 | doa4::LEU2 cdc4::CDC4-MYC-TRP1 | W303 | This study |
| JMG63 | GAL1-SIC1V5V33A76 grr1::GRR1-MYC-TRP1 | W303 | This study |
C. Mann is at the Commissariat à l’Energie Atomique Saclay, Gif-sur-Yvette, France.
DNA Manipulations.
Standard procedures were used for recombinant DNA manipulations (20). PCRs were performed with the Expand polymerase kit as recommended by the manufacturer (Boehringer Mannheim). Oligonucleotides were synthesized by Genset (Paris, France). Mutations in the F-box of Grr1p were introduced by PCR and changed Leu-320, Pro-321, and Glu-323 into Ala residues. The mutant construct was introduced by homologous recombination into the GRR1 locus (19). To express Grr1p-myc under the control of the inducible GAL promoter, the full-length GRR1-myc coding sequence was amplified by PCR to introduce KpnI sites on each side of the ORF. The PCR product was digested with KpnI and ligated into the yeast expression vector pRD53 (21). The full-length coding sequences of GRR1, GRR1-AAA, and SKP1 were amplified by PCR to introduce BamHI and XhoI (GRR1 and GRR1-AAA) or NcoI and XhoI sites (SKP1) on each side of the ORF and cloned as a BamHI–XhoI or NcoI–XhoI fragment into the two-hybrid vectors pEG203 and pJG4–6 (22). Plasmids encoding wild-type or dominant-negative ubiquitin (Ub3R) were constructed by inserting the 1200-bp BamHI–ClaI fragment isolated from YEp96 or YEp96 RRR (23) into the yeast vector pRS426 (21).
Antibodies, Western Blots, and Microscopy.
Standard procedures were used for yeast cell extract preparation and immunoblotting (24). Immunoblots against ubiquitin were performed using a polyclonal antibody raised against ubiquitin (Sigma) as described previously (25). Antibodies against glutathione S-transferase (GST) were obtained from Qiagen; monoclonal (HA11) and polyclonal antibodies against the HA epitope were purchased from Babco (Berkeley, CA) and used as recommended by the manufacturer. 9E10 antibodies were produced by the Swiss Institute for Experimental Cancer Research (ISREC) antibody facility, and polyclonal antibodies specific for Cdc34p (26) and Gic2p (12) were kindly provided by M. Tyers (Samuel Lunenfeld Research Institute, Toronto) and M. Jaquenoud (ISREC), respectively. For microscopy, cells were grown to early logarithmic phase in rich medium at 30°C, fixed with 3.7% formaldehyde, and photographed on a Zeiss Axiophot fluorescence microscope with a Photometrics charge-coupled device camera. Images were analyzed with PhotoShop 4.0 (Adobe).
Determination of Half-Lives and Ubiquitination Assays.
Cultures were grown to early logarithmic phase in rich medium at 30°C (25°C for temperature-sensitive mutants), at which time cycloheximide (CX; Sigma) was added to a final concentration of 50 μg/ml (stock solution, 10 mg/ml). Temperature-sensitive strains were shifted to 37°C 3 hr before addition of CX. The proteasome inhibitor MG132 (Sigma) was solubilized in DMSO and added to a final concentration of 50 μM 90 min before addition of CX. Aliquots were collected at the times indicated, and protein levels were analyzed by immunoblotting with specific antibodies. The half-life of Grr1p-myc was also determined in strains harboring a plasmid bearing GRR1-myc under the control of the inducible GAL promoter as described previously (27). W303 cells carrying multicopy plasmids encoding Grr1p-GST and Grr1p-dF-GST (14) under control of the GAL promoter were grown to early logarithmic phase in selective medium containing raffinose (2% final concentration) and induced for 1 hr by addition of galactose (2% final concentration). The half-life of the proteins was determined by using CX as described above, and the protein levels were analyzed by immunoblotting with specific antibodies against GST.
Coimmunoprecipitation and Two-Hybrid Assays.
W303 cells carrying either a plasmid encoding an epitope-tagged (HA) version of Skp1p (MT1511) or an empty plasmid (pRS315) were grown in selective medium at 30°C to mid-logarithmic phase. Cells were pelleted (100 OD600 units), resuspended in cold PBS (137 mM NaCl/2.7 mM KCl/4.3 mM Na2HPO4/1.4 mM KH2PO4, pH 7.3) containing protease inhibitors (Complete, Boehringer Mannheim), and lysed with a One Shot cell extractor (Constant Systems, Warwick, U.K.) as recommended by the manufacturer. Lysates typically contained 7–10 mg/ml total protein as determined by a Bradford assay (Bio-Rad). The lysate was incubated on a rocker for 1 hr at 4°C with 100 μl of protein G-Sepharose (33% slurry, Pharmacia) coupled with 9E10 antibodies. The beads were washed four times with lysis buffer, and bound proteins were eluted with gel sample buffer and immunoblotted with 9E10 antibodies to control for the presence of Grr1p-myc or with polyclonal antibodies against HA to visualize HA-Skp1p. Two-hybrid assays were performed as described above in the yeast strain W303 containing the lacZ reporter plasmid pSH18.34 (22). Miller units of β-galactosidase are averages from three independent experiments with standard deviations.
Cell Cycle Arrest and α-Factor Release Experiments.
Strains carrying CLN2-HA and either GRR1-myc (JMG108) or CDC4-myc (JMG107) were grown to early logarithmic phase in rich medium at 30°C, at which time cells were arrested for 3 hr by addition of hydroxyurea (200 mM; Sigma), nocodazole (15 μg/ml; Sigma) or α-factor (50 μg/ml final concentration). Arrest at the appropriate cell cycle stage was monitored microscopically. The half-lives of the proteins were determined with CX as described above. Cell synchronization experiments by release from α-factor arrest were performed as described (28); synchrony was monitored by fluorescence-activated cell sorting (FACS) analysis and microscopic determination of the budding index. FACS analysis was carried out as described by Epstein and Cross (29).
RESULTS
The F-Box Proteins Grr1p, Met30p, and Cdc4p Are Unstable.
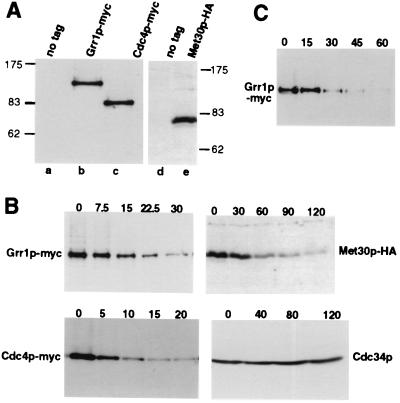
To study the regulation of F-box proteins, we replaced the genomic locus coding for Grr1p, Cdc4p, and Met30p with versions containing multiple copies of the 9E10 (myc)- or 11HA (HA)-epitope (19). The epitope-tagged proteins were functional (Fig. 5 and data not shown) and migrated at the predicted molecular masses on SDS/PAGE (Fig. 1A). We used the translation inhibitor CX to determine the half-lives of the F-box proteins (Fig. 1B): cells expressing epitope-tagged Grr1p-myc (Upper Left), Cdc4p-myc (Lower Left), or Met30p-HA (Upper Right) were grown to mid-logarithmic phase, at which time CX was added (time 0). Aliquots were removed after the times indicated (in minutes) and analyzed for the presence of the F-box proteins or Cdc34p (Lower Right) by immunoblotting. Strikingly, all three F-box proteins were unstable, with a half-life of approximately 5 min for Cdc4p, 15 min for Grr1p, and 30 min for Met30p. In contrast, Cdc34p was stable: its levels did not significantly decrease during the time course. To exclude the possibility that degradation of the F-box proteins was artificially caused by the use of CX, we confirmed the half-life of Grr1p by an independent experimental protocol: Grr1p was expressed from the GAL1 promoter, at which time synthesis was turned off by addition of glucose. As shown in Fig. 1C, Grr1p was rapidly degraded with a half-life of less then 20 min. Thus, we conclude that F-box proteins are unstable components of the SCF.
Figure 5.
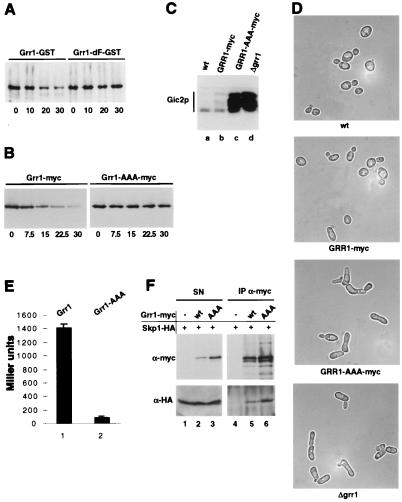
An intact F-box is required for degradation and function of Grr1p. (A) The stability of a GST-tagged Grr1p mutant (Grr1p-dF-GST) deleted for the F-box (right four lanes) was compared with wild-type Grr1p-GST (left four lanes) by using CX. Expression of Grr1p-GST was followed by immunoblotting with antibodies specific for GST. (B) The stability of a Grr1p mutant (Grr1p-AAA-myc) harboring mutations in three conserved residues in the F-box (JMG61; Right) was compared with wild-type Grr1p-myc (JMG101; Left). Note that Grr1p-AAA-myc is stable. (C) Cells expressing Grr1p-AAA-myc were unable to degrade Gic2p. Accumulation and phosphorylation of the SCFGrr1 target Gic2p was analyzed in the following strains: lane a, wild-type (W303); lane b, GRR1-myc (JMG101); lane c, GRR1-AAA-myc (JMG61); and lane d, Δgrr1 (YM2957). (D) Morphology of cells expressing Grr1p-AAA-myc (JMG61; second panel from the top), Grr1p-myc (JMG101; third panel), or wild-type Grr1p (W303; top panel) and cells deleted for GRR1 (YM2957; bottom panel). Note that cells expressing Grr1p-AAA-myc exhibit an aberrant morphology characteristic for cells lacking SCFGrr1 function (29). (×2,000.) (E) The interaction between Skp1p and wild-type Grr1p (column 1) or Grr1p-AAA (column 2) was analyzed by two-hybrid assay. Bars show Miller units with standard deviations determined. (F) Extracts prepared from cells expressing Skp1p-HA and either wild-type Grr1p-myc (JMG101; lanes 2 and 5), Grr1p-AAA-myc (JMG61; lanes 3 and 6), or untagged Grr1p (W303; lanes 1 and 4) were immunoprecipitated (IP α-myc; right panels) with 9E10 antibodies and analyzed for the presence of Grr1p-myc (upper panels) or Skp1p-HA (lower panels) by immunoblotting. An aliquot of the supernatant (SN; left panels) before to immunoprecipitation was included as a control. Note that Grr1p-AAA-myc is able to co-immunoprecipitate with Skp1p-HA.
Figure 1.
The F-box proteins Grr1p, Cdc4p, and Met30p are unstable components of the SCF. (A) Extracts prepared from cells expressing epitope-tagged Grr1p, Cdc4p, and Met30p were immunoblotted with 9E10 (lanes a–c) or HA11 antibodies (lanes d and e). The following strains were analyzed: lanes a and d, no tag (W303); lane b, Grr1p-myc (JMG101); lane c, Cdc4p-myc (JMG102); and lane e, Met30p-HA (JMG68). The positions of molecular mass markers are indicated (in kDa). (B) The half-lives of Cdc34p (Lower Right) and the F-box proteins Grr1p-myc (Upper Left), Cdc4p-myc (Lower Left), and Met30p-HA (Upper Right) were determined by using the translation inhibitor CX. Cells were grown to mid-logarithmic phase, at which time CX was added (time 0) to 50 μg/ml final concentration. Aliquots were removed at the times indicated (in minutes) and analyzed by immunoblotting with 9E10 or HA11 antibodies, or polyclonal antibodies against Cdc34p. (C) The half-life of Grr1p was determined after repression of GRR1 expressed from the GAL promoter. Cells were grown in medium containing galactose (2% final concentration) to mid-logarithmic phase, at which time glucose was added (2% final concentration) to repress the GAL promoter (time 0). Aliquots were removed at the times indicated (in minutes) and analyzed for the presence of Grr1p-myc by immunoblotting.
Degradation of Grr1p and Cdc4p Occurs Constitutively Throughout the Cell Cycle.
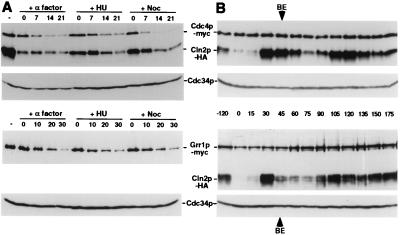
Several SCF substrates are degraded in a cell cycle-dependent manner (3, 15). To test whether degradation of F-box proteins may control the activity of SCF complexes, we determined whether the half-lives of F-box proteins change in a cell cycle-dependent manner. We found that the half-lives of Cdc4p and Grr1p were comparable irrespective of whether cells were arrested in G1 by α-factor, in S-phase by hydroxyurea, or in mitosis by nocodazole (Fig. 2A). In addition, Grr1p was rapidly degraded in cells arrested before DNA replication by expression of a nondegradable Sic1p mutant (Sic1p-Δ3P; ref. 30) (data not shown). Thus, these results demonstrate that degradation of the F-box proteins Cdc4p and Grr1p was not influenced by the cell cycle position, suggesting that F-box proteins are intrinsically unstable. To confirm these findings, we determined the protein levels of Cdc4p and Grr1p through the cell cycle. Cells were synchronized by an α-factor block/release protocol, and the levels of Grr1p, Cdc4p, and Cdc34p were analyzed by immunoblotting (Fig. 2B). Synchrony of cells was monitored by expression of HA-tagged Cln2p (Cln2p-HA) and by fluorescence-activated cell sorting analysis (data not shown). We found that the levels of Cdc4p and Grr1p remained constant throughout the cell cycle, supporting the idea that expression and degradation of these F-box proteins are not cell cycle regulated. Taken together, these results suggest that the inability of SCFCdc4 and SCFGrr1 to degrade specific substrates in the G1 phase of the cell cycle is not caused by cell cycle-specific degradation of the corresponding F-box protein. Consistent with these findings, several SCF substrates have been shown to be unstable when ectopically expressed at various cell cycle stages (9, 12, 27).
Figure 2.
Cdc4p and Grr1p are intrinsically unstable and their levels do not change during the cell cycle. (A) The half-life of Cdc4p-myc (JMG107; Upper two panels) and Grr1p-myc (JMG101; Lower two panels) was determined in cells arrested in G1 by α-factor, in S-phase by hydroxyurea (HU), and in mitosis by nocodazole (Noc). CX was added at time 0, and aliquots were removed at times indicated (minutes) and analyzed by immunoblotting for the presence of the F-box proteins Cdc4p-myc or Grr1p-myc (upper panel of each pair) or Cdc34p (lower panel of each pair). Where indicated Cln2p-HA was used as an internal control (top panel). An extract prepared from exponentially growing cells is shown in the first lane. (B) Cells expressing Cln2p-HA and either Cdc4p-myc (JMG107; Upper two panels) or Grr1p-myc (JMG108; Lower two panels) were released from an α-factor block (time 0); aliquots were removed at the times indicated (minutes) and analyzed by immunoblotting for the levels of Cdc4p-myc or Grr1p-myc, Cln2p-HA, and Cdc34p as indicated. BE marks the time of bud emergence as determined microscopically. Note that the levels of the F-box proteins Cdc4p-myc and Grr1p-myc remain constant throughout the cell cycle.
F-Box Proteins Are Degraded by Ubiquitin-Mediated Degradation.
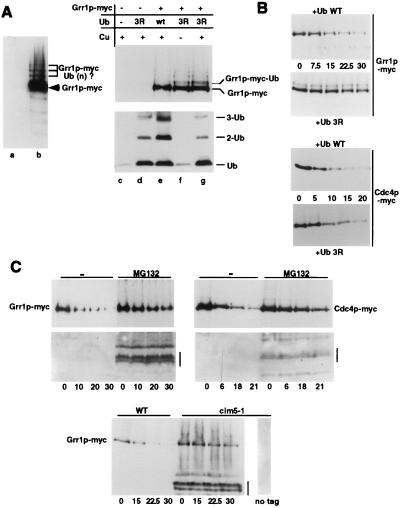
We observed that Grr1p accumulated a ladder of slower-migrating forms on SDS gels that were separated by approximately 10 kDa, characteristic of modification by ubiquitin or ubiquitin-like proteins (Fig. 3A). To examine whether F-box proteins are degraded by a ubiquitin-mediated mechanism, we first examined Grr1p immunoprecipitates for the presence of ubiquitin. However, we were unable to detect ubiquitinated species of Grr1p by this assay, presumably because these forms are rapidly cleaved by ubiquitin-deconjugation enzymes (data not shown). To circumvent this problem, we used a dominant-negative ubiquitin (Ub3R), which is efficiently ligated onto substrates but can no longer be extended into multi-ubiquitin chains (23, 31). Interestingly, cells overexpressing Ub3R (Fig. 3A, lanes d, f, and g) but not wild-type Ub (lane e) from the Cu2+-inducible promoter accumulated mono-ubiquitinated Grr1p, which migrated in SDS gels with a characteristic shift of approximately 10 kDa. A single mono-ubiquitinated form of Grr1p was detected predominantly, indicating that Grr1p is ubiquitinated in vivo preferentially on a single lysine residue. Importantly, overexpression of Ub3R stabilized Grr1p and Cdc4p (Fig. 3B), suggesting that multi-ubiquitination is required for subsequent degradation. These results demonstrate that both Grr1p and Cdc4p are ubiquitinated in vivo and suggest that they are degraded by a ubiquitin-mediated pathway. To determine whether F-box proteins are degraded by the 26S proteasome, we measured the half-life of Grr1p in cim5–1 cells, which exhibit a strong defect in proteasome function (32). As shown in Fig. 3 C Lower, Grr1p was clearly stabilized in cim5–1 mutant cells; several degradation products of Grr1p that are not present in untagged controls became apparent. Consistent with these results, Grr1p and Cdc4p were also stabilized in Δerg6 cells treated with the proteasome inhibitor MG132 (Fig. 3 C Upper) (33); deletion of ERG6 was necessary to enable uptake of the drug into cells (34). Finally, Cdc4p was partially stabilized in cells lacking the deubiquitination enzyme Doa4p (data not shown), which is associated with the proteasome and involved in recycling ubiquitin (35). Taken together, these results demonstrate that Grr1p and Cdc4p are degraded by the 26S proteasome.
Figure 3.
Grr1p and Cdc4p are ubiquitinated in vivo and degraded by the proteasome. (A) Grr1p is ubiquitinated in vivo. (Left) Immunoblot with 9E10 antibodies of extracts prepared from cells expressing Grr1p-myc (JMG101; lane b) or for control untagged Grr1p (W303; lane a) demonstrating a ladder of distinct slower-migrating species separated by approximately 10 kDa, characteristic for ubiquitinated forms. The arrowhead indicates the position of unmodified Grr1p-myc; the bracket marks ubiquitinated species of Grr1p. (Right) Extracts from Grr1p-myc cells (lanes e–g) or untagged control cells (lanes c and d) overexpressing wild-type ubiquitin (Ub; lane e) or dominant-negative Ub3R (lanes d, f, and g) from a Cu2+-inducible promoter were immunoblotted with 9E10 (Upper) or anti-ubiquitin antibodies (Lower). Reduced levels of Ub3R are detected in cells that were not treated with Cu2+ (lane f). Note that expression of Ub3R interferes with the formation of multi-ubiquitin species (Lower) and results in accumulation of mono-ubiquitinated Grr1p (Upper). (B) Mono-ubiquitinated Grr1p-myc and Cdc4p-myc are stabilized in vivo. The half-life of Grr1p-myc (Upper pair) and Cdc4p-myc (Lower pair) were determined in cells overexpressing either wild-type ubiquitin (upper panel of each pair) or Ub3R (lower panel of each pair). Note that overexpression of Ub3R stabilizes Grr1p-myc and Cdc4p-myc. (C) (Upper) Grr1p-myc and Cdc4p-myc are stabilized in the presence of the proteasome inhibitor MG132. The half-life of Grr1p-myc (JMG67; Left) or Cdc4p-myc (JMG66; Right) was determined with CX in the presence of MG132 (right lanes) or the solvent DMSO (−; left lanes). Note the accumulation of partially degraded forms of Grr1p-myc and Cdc4p-myc in the presence of MG132 (bar). (Lower) The half-life of Grr1p-myc was determined as described in wild-type (JMG53; left lanes) or cim5–1 cells (JMG54; right lanes) which are defective for proteasome function. An isogenic cim5–1 strain (YMP241) expressing untagged Grr1p is included for control (right lane). Note the accumulation of partially degraded forms of Grr1p-myc in cim5–1 cells (bar).
Ubiquitin-Dependent Degradation of Grr1p and Cdc4p Requires Components of the SCF and an Intact F-Box.
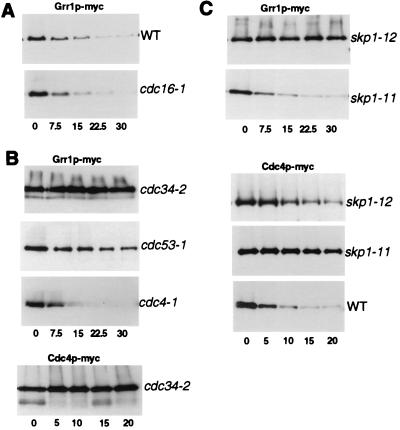
To examine which E3-ubiquitin ligase was responsible for degradation of F-box proteins, we measured the half-life of Cdc4p and Grr1p in cells defective in components of the APC or SCF. Grr1p was efficiently degraded in cdc16–1 cells (Fig. 4A), suggesting that the APC is not involved in ubiquitination of F-box proteins. In contrast, Grr1p was stable in the cdc53–1 mutant, and both Grr1p and Cdc4p were stable in the cdc34–2 mutant (Fig. 4B), demonstrating that the SCF is essential for their degradation. Grr1p was unstable in cdc4–1 mutants, indicating that Cdc4p is not the F-box protein required to target Grr1p. Interestingly, Grr1p was efficiently degraded in skp1–11 cells but was stable in skp1–12 mutant cells. Conversely, Cdc4p was stable in skp1–11 cells but partially degraded in skp1–12 cells (Fig. 4C). It is thought that Skp1p-11 is specifically defective in associating with the SCFCdc4 complex, whereas Skp1p-12 is unable to assemble functional SCFGrr1 complexes (9, 12, 27). Thus, these results indicate that Grr1p is ubiquitinated by the SCFGrr1 complex, whereas Cdc4p may be ubiquitinated by the SCFCdc4 complex, suggesting that F-box proteins are targeted for degradation by auto-ubiquitination within their own SCF complex. According to this model F-box proteins that are not assembled into functional SCF complexes would be stable. The F-box motif mediates the interaction with Skp1p and is required for assembly of F-box proteins into SCF complexes (9, 16). In support of this concept, a Grr1 mutant protein deleted for the F-box (Grr1p-dF) is unable to bind Skp1p (ref. 14 and data not shown). Importantly, the Grr1p-dF protein was stable and was no longer degraded in a ubiquitin-dependent manner (Fig. 5A). In addition, we mutated three conserved residues (L320, P321, E323) in the F-box of Grr1p to alanine residues (Grr1p-AAA); GRR1-AAA cells exhibited an aberrant morphology (Fig. 5D) characteristic for Δgrr1 cells (36) and accumulated Gic2p (Fig. 5C), demonstrating that Grr1p-AAA was defective for assembling functional SCFGrr1 complexes. As expected, Grr1p-AAA was unable to bind Skp1p by two-hybrid analysis (Fig. 5E), but surprisingly, Grr1p-AAA was still able to co-immunoprecipitate with Skp1p (Fig. 5F). In addition, GST pull-down experiments confirmed that Grr1p-AAA was able to bind Skp1p-GST (data not shown). These results suggest that, when expressed at physiological levels, some additional protein may bridge the interaction between Grr1p-AAA and Skp1p in vivo. Consistent with this idea, previous studies showed that Grr1p is able to bind Cdc53p in the absence of Skp1p (5). Importantly however, the Grr1p-AAA protein was stable and was no longer degraded in a ubiquitin-dependent manner (Fig. 5B), indicating that ubiquitination of Grr1p requires its ability to assemble into functional SCF complexes. Grr1p-AAA was also stable when expressed in cells containing wild-type Grr1p, demonstrating that Grr1p is not able to mediate degradation of Grr1p-AAA in trans (data not shown). Thus, we conclude from these results that ubiquitination of Grr1p occurs within its own SCFGrr1 complex.
Figure 4.
Degradation of Grr1p and Cdc4p requires components of the SCF. (A) Degradation of Grr1p-myc is independent of an intact APC. The half-life of Grr1p-myc was determined as above in cdc16–1 cells defective for APC function (JMG62; Lower) or wild-type cells (JMG101; Upper). (B) Degradation of Grr1p-myc and Cdc4p-myc requires an intact SCF. The half-life of Grr1p-myc (upper three panels) or Cdc4p-myc (bottom panel) was determined in cells defective for the indicated SCF components. (C) The stability of Grr1p-myc (upper two panels) and Cdc4p-myc (lower three panels) was analyzed in cells harboring different alleles of SKP1: skp1–11 cells are defective for SCFCdc4 function, whereas skp1–12 cells become arrested in G2 and are defective for SCFGrr1 function (12). Note that Grr1p-myc is efficiently degraded in skp1–11 cells (JMG31), but stable in skp1–12 cells (JMG29). Conversely, Cdc4p-myc is stable in skp1–11 cells (JMG70) but partially degraded in skp1–12 cells (JMG69).
DISCUSSION
Our results demonstrate that three F-box proteins are unstable components of SCF complexes. We thus confirm and extend a recent study by Zhou and Howley (37), which has independently found that F-box proteins are degraded by a ubiquitin-mediated mechanism. Degradation of Grr1p and Cdc4p was mediated by the 26S proteasome and required the core components of the SCF complex, Cdc53p and Skp1p, and the E2 ubiquitin-conjugating enzyme Cdc34p. Our experiments suggest that ubiquitination of F-box proteins occurs within their own complex and is not dependent on the presence of other F-box proteins. First, degradation of Grr1p and Cdc4p depended on specific skp1 alleles; Grr1p was stable in skp1-12 cells, which are defective for SCFGrr1 function, whereas Cdc4p was stable in skp1-11 cells, which are defective for SCFCdc4 function. Second, Grr1p mutant proteins that fail to assemble functional SCF complexes are stable in vivo. It will be important to determine whether Cdc4p and Grr1p can be ubiquitinated in vitro by reconstituted SCF complexes. Taken together, these results suggest that F-box proteins are targeted for degradation by auto-ubiquitination within their own SCF complex. Because F-box proteins are intrinsically short lived, these observations also imply that F-box proteins must assemble rapidly into functional SCF complexes, suggesting a very dynamic turnover of SCF complexes in vivo. The half-life of F-box proteins may thus be a measure of their ability to assemble into functional SCF complexes and may reflect regulation of assembly or activity of SCF complexes by environmental signals.
A recent study identified a small motif (the R-motif) in Cdc4p adjacent to the F-box that controls its stability (38). It is possible that this domain contains sequences required for ubiquitination or assembly into functional SCF complexes. The authors of ref. 38 propose that Skp1p is required to protect Cdc4p from degradation, whereas the results shown here suggest hat Skp1p is needed for degradation of Cdc4p. The reasons for this discrepancy are not yet understood, but it is clear that an intact F-box is required for degradation of both Cdc4p (37) and Grr1p (this study).
Phosphorylation of many targets is required for ubiquitination by the SCF complex (15), but at present it is not clear whether phosphorylation of F-box proteins is necessary for their degradation. Degradation of Cdc4p and Grr1p was independent of Cdk activity, suggesting that Cdks are not involved in triggering degradation of these F-box proteins. Phosphorylation of substrates is required for binding to the SCF complex, but at least in vitro, functional SCFCdc4 complexes can be assembled from presumably unphosphorylated SCF components expressed from baculovirus, suggesting that phosphorylation of Cdc4p may not be required for assembly (5, 6). However, until reconstitution has been achieved with purified components expressed in Escherichia coli, a phosphorylation-dependent assembly mechanism cannot be excluded.
What is the physiological role of rapid degradation of F-box proteins? F-box proteins serve as critical substrate recognition subunits of the SCF complex, which mediates degradation of many proteins required for cell cycle progression and adaptation to changes in the cellular environment. To achieve this task, the core complex composed of Cdc53p, Cdc34p, and Skp1p must constantly associate with different F-box proteins, which target specific substrates for degradation. Thus, it may be important to maintain a dynamic equilibrium among multiple SCF complexes. Consistent with this model, overexpression of Grr1p or Met30p dramatically impairs growth of cdc4-1 cells, but only mildly interferes with growth of cdc34-2 and cdc53-1 strains (26), suggesting that F-box proteins compete with each other for binding to the core complex. Overexpression of Grr1p-AAA did not interfere with degradation of Gic2p, suggesting that it cannot function in a dominant-negative manner by sequestering Gic2p into inactive complexes (data not shown). Limiting the half-life of F-box proteins allows turnover of assembled SCF complexes, thereby providing the ability to rapidly change the balance among SCF complexes by regulating the levels or assembly of specific F-box proteins during the cell cycle or in response to extracellular signals. In support of this idea, the interaction between Grr1p and Skp1p is enhanced in response to high glucose levels by a post-translational mechanism that involves the C-terminal domain of Grr1p, thereby increasing the number of SCFGrr1 complexes (39). Interestingly, we have observed that overexpression of a substrate appears to stabilize its F-box protein, suggesting that high levels of substrates may alter the equilibrium among SCF complexes (unpublished results). Such a mechanism would effectively adjust the levels of specific F-box proteins to their need in a given environment. In human cells, the level of the F-box protein Skp2p peaks in S phase and is controlled by transcriptional and post-translational mechanisms (16). Interestingly, Skp2p levels are increased in many transformed cells (40), suggesting that its degradation may be defective in tumors. It will be interesting to determine whether the stability or assembly of other F-box proteins may similarly be regulated during the cell cycle or in response to extracellular signals.
In summary, we propose that a limited half-life of already assembled SCF complexes may be required to allow rapid alterations between different SCF complexes during the cell cycle and in response to environmental changes. The model is reminiscent of cyclin-dependent kinases (Cdk): because the rate of spontaneous dissociation of assembled Cdk–cyclin complexes is low, it is thought that rapid turnover of cyclins ensures the necessary changes in subunit composition at specific cell cycle stages (41). Analogous to F-box proteins, the G1 cyclins Cln1p and Cln2p are intrinsically unstable subunits of Cdks and are constitutively targeted for ubiquitin-dependent degradation by the SCFGrr1 complex (11, 42).
Acknowledgments
We thank M. Tyers, T. Kishi, M. Jaquenoud, M. Longtime, A. Heese-Peck, C. Mann, and E. Schwob for providing antibodies, plasmids, and strains; M. Blondel and W. Krek for helpful suggestions; N. Perrinjaquet for excellent technical assistance; and B. Amati, R. Iggo, and V. Simanis for critical reading of the manuscript. J.-M.G. is supported by a Federation of European Biochemical Societies Postdoctoral Fellowship; M.P. is supported by the Swiss National Science Foundation, the Swiss Cancer League, and a Helmut Horten Incentive Award.
ABBREVIATIONS
- APC
anaphase-promoting complex
- SCF
Skp1p-cullin-F-box protein
- HA
hemagglutinin
- GST
glutathione S-transferase
- CX
cycloheximide
References
- 1.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Heller H, Elias S, Ciechanover A. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 3.Peters J M. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 4.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 5.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 6.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 7.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 9.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 10.Patton E E, Willems A R, Tyers M. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 11.Barral Y, Jentsch S, Mann C. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 12.Jaquenoud M, Gulli M-P, Peter K, Peter M. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser P, Sia R A, Bardes E G, Lew D J, Reed S I. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishi T, Yamao F. J Cell Sci. 1998;111:3655–3661. doi: 10.1242/jcs.111.24.3655. [DOI] [PubMed] [Google Scholar]
- 15.Deshaies R J. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 16.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 18.Valtz N, Peter M. Methods Enzymol. 1997;283:350–365. doi: 10.1016/s0076-6879(97)83029-7. [DOI] [PubMed] [Google Scholar]
- 19.Longtine M S, McKenzie A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;10:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene and Wiley–Interscience; 1991. [Google Scholar]
- 21.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 23.Galan J-M, Hagenauer-Tsapis R. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 25.Springael J-Y, Galan J-M, Tsapis R, André B. J Cell Sci. 1999;112:1365–1373. doi: 10.1242/jcs.112.9.1375. [DOI] [PubMed] [Google Scholar]
- 26.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breeden L, L. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- 29.Epstein C B, Cross F R. Genes Dev. 1992;6:1695–1703. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 30.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 31.Arnason T, Ellison M J. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghislain M, Udvardy A, Mann C. Nature (London) 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 33.Lee D H, Goldberg A L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 34.Lee D H, Goldberg A L. J Biol Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 35.Papa F R, Amerik A Y, Hochstrasser M. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flick J S, Johnston M. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Howley P M. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 38.Mathias N, Johnson S, Byers B, Goebl M. Mol Cell Biol. 1999;19:1758–1767. doi: 10.1128/mcb.19.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F N, Johnston M. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Kobayashi R, Galaktionov K, Beach D. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 41.Morgan D O. Curr Opin Cell Biol. 1996;6:767–772. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- 42.Lanker S, Valdivieso M H, Wittenberg C. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]