Abstract
Background and aims: Expression of inducible nitric oxide synthase (iNOS) is greatly upregulated in the colonic mucosa of patients with collagenous and ulcerative colitis. As the transcription factor nuclear factor κB (NFκB) is a major inducer of iNOS gene expression, we compared activation and transcriptional activity of NFκB in colonic mucosal biopsies from these patients.
Patients: Eight patients with collagenous colitis, six with relapsing ulcerative colitis, and eight with uninflamed bowel were studied.
Methods: NFκB DNA binding activity was assessed by electrophoretic mobility shift assay and inhibitor of NFκB (IκB) kinase (IKK) activity by immunocomplex kinase assay. In vivo recruitment of NFκB to the iNOS promoter was determined by chromatin immunoprecipitation analysis and transcriptional activity by NFκB gene expression profiling arrays. Cells showing NFκB activation were identified by immunohistochemistry.
Results: In collagenous and ulcerative colitis, as opposed to uninflamed bowel, IKKβ activity and strong NFκB DNA binding gave rise to activation of identical NFκB subunits and recruitment of transcriptionally active p65 to the iNOS promoter. In collagenous colitis, activated NFκB was observed only in epithelial cells while up to 10% of lamina propria macrophages showed activation in ulcerative colitis.
Conclusions: In collagenous and ulcerative colitis, colonic mucosal NFκB is activated and recruited to the iNOS promoter in vivo via an IKKβ mediated pathway. As collagenous colitis is not associated with tissue injury, these data challenge the prevailing view that activation of NFκB per se mediates tissue injury. Our results suggest that downstream inflammatory reactions leading to tissue damage originate in lamina propria immune cells, as increased NFκB activity in collagenous colitis was localised solely in epithelial cells, but present also in macrophages in ulcerative colitis.
Keywords: collagenous colitis, inflammatory bowel disease, nitric oxide synthase, nuclear factor κB, ulcerative colitis
Collagenous colitis is an inflammatory bowel disease of unknown aetiology characterised by chronic watery diarrhoea in the absence of mucosal injury. Endoscopic appearance is usually normal but colonic mucosal biopsies reveal infiltration of lymphocytes and plasma cells in the lamina propria, thickened subepithelial layer of collagen, and excess of intraepithelial lymphocytes. Recently, we have observed greatly increased production rates of nitric oxide (NO) into the colonic lumen of patients with collagenous colitis1 and provided evidence for the hypothesis that the enzyme, inducible NO synthase (iNOS), is the source of excess NO production.2 As surgical exclusion of the colon by split ileostomy induces clinical and histological remission3 and re-establishment of gut continuity causes a rapid relapse, one or more unknown luminal factor(s) may be responsible for induction of iNOS at the luminal border of the colonic epithelium.
One of the major transcriptional inducers of iNOS gene expression is the transcription factor nuclear factor κB (NFκB).4,5 Signal transduction through NFκB is initiated on binding of ligands to cell membrane receptors (for example, Toll-like receptor 2 and 4, tumour necrosis factor (TNF) receptor, and interleukin (IL)-1 receptor) and intracellular recognition receptors (for example, nucleotide oligomerisation domain (NOD)-1 and NOD-2) leading to activation of the IκB kinase (IKK) complex and subsequently phosphorylation of the inhibitor of NFκB (IκBα or IκBβ). Phosphorylation targets the inhibitor to polyubiquitination and proteasomal degradation, thus activating NFκB. Active NFκB translocates into the nucleus where it binds to the promoter region of multiple genes with predominantly proinflammatory actions. In addition to stimulating expression of IL-1, TNF-α, IL-6, IL-8, major histocompatibility complex class II, and intercellular adhesion molecule 1, activated NFκB also stimulates expression of iNOS and its own inhibitor, IκBα. Thus a single central pathway mediates activation signals from multiple bacterial and cytokine stimuli to increase production of a characteristic profile of proinflammatory molecules.
In ulcerative colitis, which is characterised by relapsing injurious inflammation of the colorectal mucosa, NFκB activation has been reported to occur both in macrophages and epithelial cells6,7 resulting in high expression levels of iNOS8 and excess production of NO.1 As similarly high levels of iNOS are observed in the apparently normal colonic mucosa from patients with collagenous colitis,2 it seems a priori unlikely that upregulation of iNOS, and thus NFκB per se, should be responsible for the tissue injury observed in active ulcerative colitis. To test this hypothesis, we compared DNA binding and transcriptional activity of NFκB in colonic mucosal biopsies from patients with active collagenous colitis, active ulcerative colitis, and uninflamed bowel, in addition to identifying the cell types responsible for NFκB activation in the named conditions.
MATERIALS AND METHODS
Patients
Permission for the study was obtained from the regional ethics committee and all participants gave informed written consent. Patients with an established diagnosis of collagenous colitis, based on typical histopathological features,9 were included if they had experienced diarrhoea (stool volume >300 ml) for at least three consecutive days during the week prior to endoscopy. All medication was discontinued at least two weeks prior to the study. Patients with relapsing ulcerative colitis were included if they had endoscopic disease activity at routine examination and had received no topical treatment or systemic corticosteroids within the past month or immunosuppressive drugs within the past three months. Oral 5-aminosalicylic acid at a daily dose of 2–4 g was allowed, if no change in medication had been made in the last two weeks prior to the investigation. Stool cultures and microscopy were performed in all patients with colitis to detect pathogens, including Clostridium difficile, and all were negative. Patients referred for endoscopy for symptoms of irritable bowel syndrome or haematochezia to exclude colorectal cancer served as controls if they had a normal colonoscopy and uninflamed mucosa at histopathological examination.
Preparation of fusion proteins
Four glutathione S-transferase (GST) fusion proteins were made: (a) GST-IκBα (1–54) WT containing the wild-type (WT) N terminal regulatory domain (residues 1–54) of the IκBα protein; (b) GST-IκBα (1–54) MUT containing two phosphorylation sites, serine 32 and 36, replaced by alanine; (c) GST-p100 (754–900) WT containing the C terminal regulatory domain (residues 754–900) of the NFκB2 protein; and (d) GST-p100 (754–900) MUT containing two phosphorylation sites, serine 866 and 870, replaced by alanine. Constructs were made by cloning polymerase chain reaction (PCR) generated fragments corresponding to IκBα or NFκB2 p100, respectively, into the pGEX-2T vector (Amersham Biosciences, Buckinghamshire, UK). Mutants were made by site directed mutagenesis with the Quickchange kit (Stratagene, La Jolla, California, USA) using the manufacturer’s recommendations. Mutations were verified by sequencing (MWG-Biotech, Ebersberg, Germany). Fusion protein was made by transforming One Shot BL21(DE3) cells (Invitrogen, Paisley, UK) with each construct and an overnight culture in 5 ml of LB broth medium with 100 μg/ml ampicillin was performed. Next day, 1 ml of each overnight culture was inoculated into 100 ml LB broth medium with 100 μg/ml ampicillin and allowed to grow to an OD600 of 0.6–0.8, before protein expression was induced by adding isopropylthio-b-d-galactoside to a final concentration of 0.4 mM. The culture was grown for an additional two hours before cells were harvested by centrifugation. Cell pellets were resuspended in 2 ml of ice cold lysis buffer (phosphate buffered saline (PBS) with 1% nonidet p-40 (NP-40) and protease inhibitor cocktail 1:500) and sonicated three times on ice in short 10 second bursts alternating with a 10 second resting on ice. After centrifugation of samples, the GST-fusion proteins were purified on a GST purification column (Amersham Biosciences) according to the manufacturer’s recommendations.
Kinase assay
The assay was performed as described by Joseph A DiDonato.10 Briefly, 300 μg of cytosol extract was precleared with protein G-Sepharose FF (Amersham Biosciences) and incubated for one hour with either 2 μg anti-IKKα (#556532; BD Pharmingen, San Diego, California, USA) or 2 μg anti-IKKβ (#sc-7329; Santa Cruz Biotechnology, Santa Cruz, California, USA) on a rotating platform at 4°C followed by addition of protein G-Sepharose and incubation for an additional hour. Protein G beads were then pelleted by centrifugation and washed twice in wash buffer (20 mM HEPES-KOH (pH 7.6), 40 mM β-glycerophosphate, 20 mM NaF, 20 mM p-nitrophenyl phosphate (PNPP), 1 mM dithiothreitol (DTT) 1 mM Na3VO4, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (#P8340; Sigma, St Louis, Missouri, USA) diluted 1:500)), twice in wash buffer with 2 M urea, twice in kinase buffer (20 mM HEPES-KOH (pH 7.6), 20 mM β-glycerophosphate, 10 mM PNPP, 50 mM NaCl, 2 mM DTT, 0.1 mM Na3VO4, and protease inhibitor cocktail), and once in kinase buffer with 10 mM MgCl2. The assay was performed in 10 μl kinase buffer with MgCl2 and supplemented with 5 μCi [γ-32P]ATP (Amersham Biosciences) and 1 μg GST fusion protein at 30°C for one hour. The reactions were separated by electrophoresis and blotted onto a PVDF membrane. Activity was visualised on a phosphor-imager (FujiFilm, Stockholm, Sweden).
Western blot analysis
Western blotting was carried out as previously described.2 Antibodies used in this analysis were: anti IKKβ (C-20) from Santa Cruz Biotechnology and anti-IKKα from BD Pharmingen.
Preparation of cytosolic and nuclear extracts
Six fresh biopsies were gently homogenised in ice cold 500 μl buffer H (10 mM HEPES-KOH (pH 7.9), 10 mM KCl, 0.1 mM ethylenediaminetetra-acetic acid (EDTA), 0.1 mM ethylene glycol-bis-(2-aminoethylether) tetra-acetic acid (EGTA), 0.75 mM spermidine, 0.15 mM spermine, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail diluted 1:500) in a Dounce homogeniser with a B-type pestle. Then, 5 μl 10% NP-40 was added and the homogenate was allowed to stand for 10 minutes on ice followed by two gentle strokes with the B-type pestle. The homogenate was transferred to a microfuge tube, underlaid with 400 μl buffer H with 30% (w/v) sucrose, and centrifuged at 1200 g for 10 minutes in a swing out rotor. After centrifugation, the upper (cytosolic) phase was transferred to a new tube, frozen in liquid nitrogen in small aliquots, and stored at −80°C until required for analysis. The nuclear pellet was resuspended in 400 μl buffer N (20 mM HEPES-KOH (pH 7.6), 20% (v/v) glycerol, 10% (w/v) sucrose, 420 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail 1:500) and incubated for 45 minutes on an end over end rotor in a cold room followed by centrifugation for 30 minutes at 20 000 g. The nuclear extract was then dialysed overnight against buffer D (20 mM HEPES-KOH (pH 7.9), 20% v/v glycerol, 0.2 mM EDTA, 0.1 M KCl, 0.5 mM PMSF, and 1 mM DTT). The formed precipitate was removed by centrifugation and the extract was concentrated by centrifugation through an Ultrafree-0.5 centrifugation tube (Millipore, Bedford, Massachusetts, USA) with a 5K cut off membrane until the sample volume was reduced to approximately 50 μl. Extracts were frozen in small aliquots in liquid nitrogen and stored at −80°C until required. Protein concentration was determined by the Bradford method (Bio-Rad, Hercules, California, USA) using bovine serum albumin (BSA) as a reference.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed by incubating 5 μg nuclear extract with 1 μg sonicated poly (dI:dC) (Amersham Bioscience), 1.5 μl mobility shift buffer (100 mM HEPES-KOH, pH 7.9, 600 mM KCl, 40 mM MgCl2, 1 mM EDTA, 1 mg/ml BSA, and 2.5 mM DTT), 1.5 μl glycerol, and water to a final volume of 14 μl, followed by addition of 5 fmol 32P labelled double stranded oligonucleotide (specific activity 15 000 cpm/fmol) corresponding to the Ig-κB promoter (5′-AGC TTC AGA GGG GAC TTT CCG AGA GGT CGA-3′). Samples were incubated at room temperature for 30 minutes and loaded on a pre-run 5% acrylamide gel containing 45 mM Tris borate and 1 mM EDTA (0.5×TBE), and run for 25 minutes at 200 V in 0.5×TBE. Then the gel was fixed in 10% acetic acid/20% methanol/70% water for 15 minutes, dried under vacuum on a piece of Whatman 3 MM paper (Whatman International, Kent, UK), exposed to a phosphor-imaging plate for at least 12 hours, and analysed on a phosphor-imager (FujiFilm). For supershift experiments, extracts were incubated with 2 μg antibody overnight at 4°C prior to addition of labelled probe.
Chromatin immunoprecipitation
This method was based on a protocol from Peggy Farnham’s Laboratory11 (http://genomcenter.ucdavis.edu/farnham/farnham). Two colonic mucosal biopsies were collected in PBS with 1% formaldehyde and allowed to stand at room temperature for 15 minutes before cross linking was stopped by addition of glycine to a final concentration of 0.125 M. Then the biopsies were washed in ice cold PBS with 1 mM PMSF and inhibitor cocktail (diluted 1:500) and transferred to a small Dounce homogeniser. Homogenisation was done with a B-type pestle and cells were swelled in lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP40, 1 mM PMSF, and inhibitor cocktail) for 10–15 minutes followed by two strokes with the pestle. The nucleus was isolated by centrifugation, resuspended in nuclear lysis buffer (50 mM Tris-Cl pH 8.1, 10 mM EDTA, 1% sodium dodecyl sulphate (SDS), 1 mM PMSF, and inhibitor cocktail) and left in ice for 30 minutes. Chromatin was sheared by sonication to an average length of 1000–500 bp and centrifuged at 15 000 rpm for 10 minutes. The chromatin solution was precleared with addition of Staphylococcus aureus Cowan 1 (SAC) for 15 minutes at 4°C. Before use, SAC was blocked with 1 μg/μl sheared herring sperm DNA and 1 μg/μl BSA and incubated overnight at 4°C. The precleared chromatin sample was split into two, diluted 10 times with IP dilution buffer (16.7 mM Tris-Cl pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton×100, and 0.01% SDS) and incubated overnight at 4°C with 2 μl anti-p65 (#SA-238; Biomol, Plymouth Meeting, Pennsylvania, USA), or with no antibody, respectively. A third sample containing only IP dilution buffer and antibody was also included. The next day, antibody was precipitated by addition of SAC for one hour followed by centrifugation. The supernatant from the sample with no antibody was set aside for total input assessment. Samples were then washed twice in cold dialysis buffer (50 mM Tris-Cl pH 8.0, 2 mM EDTA, and 0.2% Sarkosyl) and five times in cold IP wash buffer (100 mM Tris-Cl pH 9.0, 500 mM LiCl, 1% NP-40, and 1% deoxycholic acid). Thereafter, the samples were eluted from the SAC by incubation twice in 150 μl elution buffer (50 mM NaHCO3, 1% SDS) for 15 minutes. The eluates were collected into a single tube and cross linking was reversed by incubating samples with 10 μg RNase A in a 0.3 M NaCl solution at 67°C for five hours. DNA was precipitated by addition of 2.5 volumes of ethanol and left at −20°C overnight. Then, the DNA was redisolved in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, pH 8.0) and treated with 30 μg Proteinase K (Roche, Mannheim, Germany) for 90 minutes at 45°C, followed by extraction with phenol/chloroform/isoamyl alcohol and chloroform/isoamyl alcohol, respectively. DNA was then precipitated with 0.3 M NaCl, 5 μg glycogen, 5 μg tRNA, and two volumes of ethanol overnight at −20°C and redisolved the following day in 30 μl 10 mM Tris, pH 8.0. We used 1 μl per PCR reaction.
Gene expression profiling
Colonic mucosal biopsies were immediately submerged into ice cold RNAlater (Ambion, Austin, Texas, USA) and incubated at 4°C overnight. Total RNA was extracted using Trizol reagent (Invitrogen) and genomic DNA contamination was removed with DNA-free (Ambion). The integrity of purified RNA was assessed by running 1 μg RNA on a 1% agarose gel containing 2.2 M formaldehyde in 3-morpholino-propane-sulfonic acid (MOPS) electrophoresis buffer (20 mM MOPS (pH 7.0), 2 mM Na-acetate, and 1 mM EDTA (pH 8.0)). Gene expression profiling was done as described by the manufacturer using the GEArray Q Series Human NFκB Signaling Pathway Gene Array (SuperArray Bioscience Corp, www.superarray.com) and 1.5 μg total RNA. Images were recorded with FujiFilm LAS-1000 equipment and quantification of spots was done using FujiFilm ImageGauge v 4.0 software. Data were normalised to the mean of all housekeeping genes provided on the array.
Double labelled immunohistochemistry
A phospho specific antibody, phospho-NFκB p65(Ser276) (#3037; Cell Signalling Technology, Beverly, Massachusetts, USA) was employed to distinguish active from inactive NFκB. Antibodies against CD68 (a macrophage marker) and vimentin (a fibroblast marker) were purchased from DakoCytomation (Glostrup, Denmark). Parallel sections of formalin fixed and paraffin embedded tissue samples (5 μm) were dewaxed in Tissue-Clear (Sakura Finetek Europe, Zoeterwoude, the Netherlands) for 15 minutes and thoroughly hydrated through series of diluted ethanol. Sections were then subjected to an antigen retrieval step by microwave oven treatment for 15 minutes in 10 mM Tris, pH 9.0, 0.5 mM EGTA solution. Staining was performed by use of the EnVision Doublestain System (#K1395; DakoCytomation). Substrates provided by the kit were however substituted with aminoethylcarbazole (#A6926; Sigma; staining red) and Fast Blue BB (#F3378; Sigma; staining blue). Anti-phospho-NFκB (Ser276) antibody was diluted 1:40, anti-CD68 1:5000, and anti-vimentin 1:400. Stained tissue was interpreted as previously described.12 Thus red-brown nuclear cell staining was used as an index for activation induced nuclear translocation of NFκB. Concomitant intense blue-granular cytoplasmic staining was used to identify macrophages (CD68 positive cells) or fibroblasts (vimentin positive cells). The stained tissue was scored in a blinded manner by two investigators (AH and LA) using a double headed microscope, as described by Geddert and colleagues.13 The following criteria were agreed upon before analysis of NFκB staining: 0, no nuclear staining; +, nuclear staining in 1–10% of the cell type under examination; ++, nuclear staining in 11–25% of the cell type under examination; +++, nuclear staining in over 25% of the cell type under examination.
Statistics
Categorical data were analysed by χ2 test and continuous data by one way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Prior to ANOVA, Bartlett’s test was performed to test for equal variances. If positive, data were log transformed and if unequal variances persisted the final analysis was done by Kruskal-Wallis test. A p value of <0.05 (two tailed) was considered significant.
RESULTS
DNA binding activity of NFκB is enhanced in collagenous and ulcerative colitis
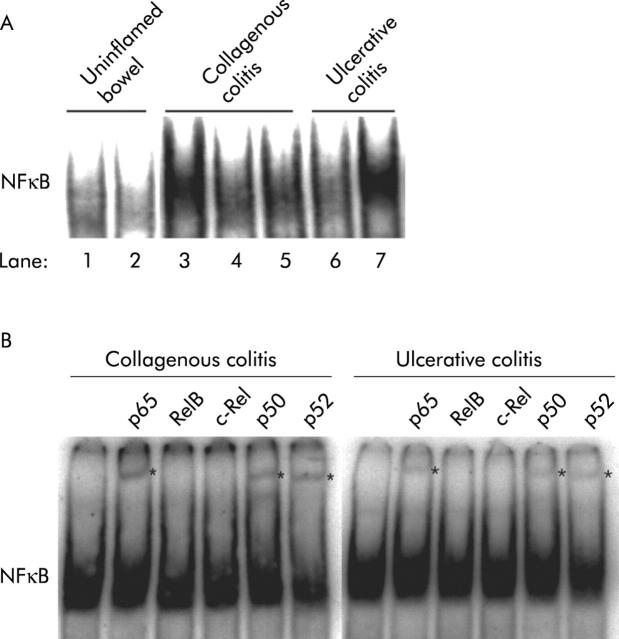
DNA binding activity in nuclear extracts of colonic mucosal biopsies from patients with uninflamed bowel showed weak binding activity (fig 1A ▶, lanes 1 and 2) while strong binding activity was observed in all patients with collagenous colitis (fig 1A ▶, lanes 3–5) and relapsing ulcerative colitis (fig 1A ▶, lanes 6 and 7).
Figure 1.
(A) Nuclear factor κB (NFκB) DNA binding activity in nuclear extracts of colonic mucosal biopsies from patients with uninflamed bowel, collagenous colitis, or ulcerative colitis. Nuclear extracts were incubated with a 32P labelled κB oligonucleotide probe and subjected to electrophoretic mobility shift assay (EMSA). Lanes 1, 2: patients with uninflamed bowel; lanes 3–5: patients with collagenous colitis; lanes 6, 7: patients with mild and severe ulcerative colitis, respectively. A representative result of two experiments is shown. (B) Nuclear extracts of colonic mucosal biopsies from patients with active ulcerative colitis or collagenous colitis were incubated with antibodies prior to EMSA analysis. An asterisk (*) denotes supershifted bands. A representative result of two experiments is shown.
Patterns of activated NFκB subunits are similar in collagenous and ulcerative colitis
Supershift analysis using antibodies specific for NFκB revealed activation of a similar subset of NFκB subunits in collagenous and ulcerative colitis, including p65, p50, and p52 (fig 1B ▶).
IKKβ is activated both in collagenous and ulcerative colitis in the absence of IKKα activation
As illustrated in fig 2A ▶, the intensity of bands in wild-type substrates and mutated substrates, respectively, showed no specific IKKα activity. In contrast, IKKβ was equally activated in patients with collagenous colitis and relapsing ulcerative colitis (p<0.05 compared with uninflamed bowel; fig 2B ▶).
Figure 2.
In vitro IκB kinase (IKK) activities in cytosolic extracts of colonic mucosal biopsies from patients with uninflamed bowel (C), active collagenous colitis (COC), or ulcerative colitis (UC). (A, B) IKKα and IKKβ activities determined by immune complex kinase assay (KA) using cytosolic extracts of colonic mucosal biopsies incubated with glutathione S-transferase (GST)-p100 (754–900) (A) or GST-IκBα (1–54) (B) substrate and radioactive ATP. The specificity of phosphorylation was determined by comparing reactions with wild-type (WT) or mutant (MUT) substrates, in which serine 866 and 870 (A) or serine 32 and 36 (B) were replaced by alanine. The reaction mixture was resolved on an acrylamide gel, transferred to a polyvinylidene difluoride membrane and subjected to phosphorimager analysis. The membrane was subsequently probed with antibody against IKKα or IKKβ as a loading control using western blot analysis (WB). A representative result of six assays is shown.
NFκB binds to the iNOS promoter in vivo in collagenous and ulcerative colitis
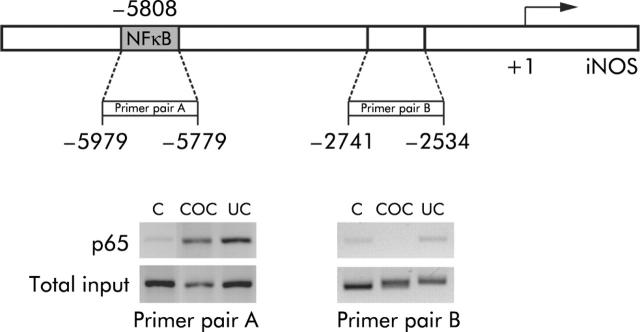
In biopsies from all patients with collagenous and ulcerative colitis, chromatin immunoprecipitation analysis showed that NFκB p65 was recruited to an NFκB regulatory element 5808 bp upstream of the transcriptional start site of the iNOS gene4 but not to an irrelevant region (fig 3 ▶). No recruitment was observed in uninflamed bowel.
Figure 3.
Chromatin immunoprecipitation assay of p65 binding to a nuclear factor κB (NFκB) regulatory region within the inducible nitric oxide synthase (iNOS) promoter in nuclear extracts of colonic mucosal biopsies from patients with uninflamed bowel (C; n = 2), collagenous colitis (COC, n = 2), or active ulcerative colitis (UC, n = 2). Soluble chromatin was prepared from formaldehyde cross linked sonicated biopsies. Specific antibody against p65 was used to precipitate protein bound DNA fragments which were subsequently amplified by polymerase chain reaction using primers flanking an NFκB regulatory region (−5979 to −5779; primer pair A) or a non-regulatory region (−2741 to −2534; primer pair B). Total input refers to amplification of 1% of the total amount of DNA prior to immunoprecipitation.
Transcriptional activity of NFκB occurs both in collagenous and ulcerative colitis
The transcriptional status of 96 selected NFκB dependent genes was assessed using an mRNA expression profiling array which showed statistically significant differences in mRNA expression between biopsies from patients with collagenous and ulcerative colitis compared with patients with uninflamed bowel. As illustrated in table 1 ▶, six genes were significantly upregulated or downregulated in collagenous colitis while 10 genes were upregulated or downregulated in ulcerative colitis compared with uninflamed bowel. Three of these genes (iNOS, complement factor B, and IL-1α) were upregulated in both groups of colitis patients, as illustrated in fig 4 ▶ using iNOS as an example.
Table 1.
Multiple mRNA expression of nuclear factor κB dependent genes in biopsies of colonic mucosa from patients with collagenous colitis, active ulcerative colitis, or uninflamed bowel
| Gene name | Uninflamed bowel (n = 7) | Collagenous colitis (n = 8) | Ulcerative colitis (n = 6) | |||
| Median | Range | Median | Range | Median | Range | |
| iNOS | 5 | (1–11) | 24.5* | (11–115) | 54.5* | (31–258) |
| Complement factor B | 26 | (23–112) | 91.5* | (62–145) | 197.5*† | (102–589) |
| IL-1α | 8 | (2–18) | 19* | (11–35) | 19* | (9–48) |
| IFN regulating factor 1 | 42 | (29–178) | 111.5* | (74–170) | 71 | (57–188) |
| IFN-β | 5 | (1–11) | 12.5* | (4–26) | 9 | (7–51) |
| IL-1 receptor associated kinase 2 | 6 | (4–51) | 4* | (1–9) | 11.5† | (4–16) |
| Serum amyloid A1 | 14 | (6–23) | 23.5 | (5–104) | 134.5*† | (68–1038) |
| Complement C3 precursor | 48 | (24–196) | 74 | (20–251) | 225.5* | (58–751) |
| IL-8 | 6 | (2–12) | 14.5 | (5–288) | 26* | (14–150) |
| Toll-like receptor 5 | 5 | (3–20) | 8 | (6–12) | 15*† | (14–44) |
| Angiotensinogen | 10 | (6–18) | 14 | (8–28) | 24.5* | (9–65) |
| IL-1 receptor type 1 | 49 | (26–51) | 43.5 | (25–67) | 69*† | (38–115) |
| IκB kinase alpha | 48 | (36–55) | 36 | (23–59) | 29.5* | (14–42) |
iNOS, inducible nitric oxide synthase; IL, interleukin; IFN, interferon; IκB, inhibitor of nuclear factor κB.
Expression was analysed by a microarray system and quantified by densitometry.
Data were normalised by dividing individual densitometric values with the mean value of all housekeeping genes provided with the array. The table shows mRNA expression from normalised data for each gene.
p<0.05 by one way ANOVA or Kruskal-Wallis test (see statistics) compared with *uninflamed bowel or †collagenous colitis.
Figure 4.
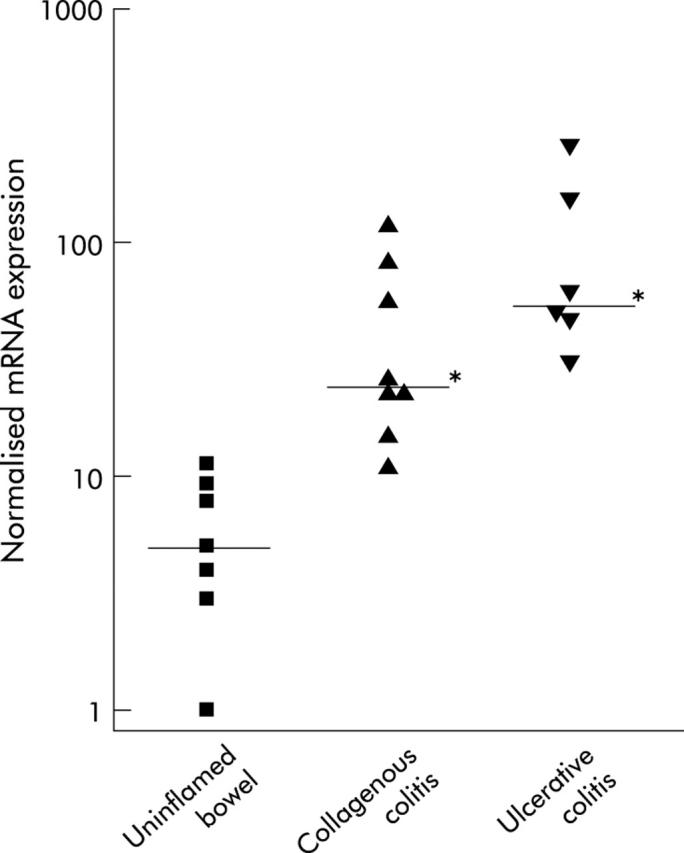
Expression of inducible nitric oxide synthase (iNOS) mRNA in biopsies from colonic mucosa of patients with uninflamed bowel, collagenous colitis, or active ulcerative colitis determined by a microarray system and quantified by densitometry. Data were normalised by dividing individual densitometric values with the mean value of all housekeeping genes provided with the array. In the scatterplot, the logarithmic y axis denotes normalised iNOS mRNA expression, with group medians displayed as horizontal lines. *p<0.05 by one way ANOVA and Tukey’s test compared with uninflamed bowel.
NFκB translocation occurs both in collagenous and ulcerative colitis but is limited to the epithelium in collagenous colitis
In biopsies from uninflamed bowel, NFκB staining (red) was negative (0) or showed weak but distinct staining in less than 10% of epithelial cells (+) (fig 5A ▶, B; table 2 ▶). No stromal cells showed positive NFκB staining. In biopsies from patients with collagenous colitis, diffuse NFκB staining was observed in superficial epithelial cells (+++) while no, or sporadic (<1%), macrophages (blue) showed staining (0) (fig 5C ▶, D; table 2 ▶). In biopsies from patients with ulcerative colitis, NFκB staining was observed both in epithelial cells(++), macrophages (+), and CD68 negative stromal cells (+) (fig 5E ▶, F; table 2 ▶). Fibroblasts (vimentin positive stromal cells) were negatively stained for NFκB activation (0) in uninflamed bowel, collagenous colitis, and ulcerative colitis (table 2 ▶).
Figure 5.
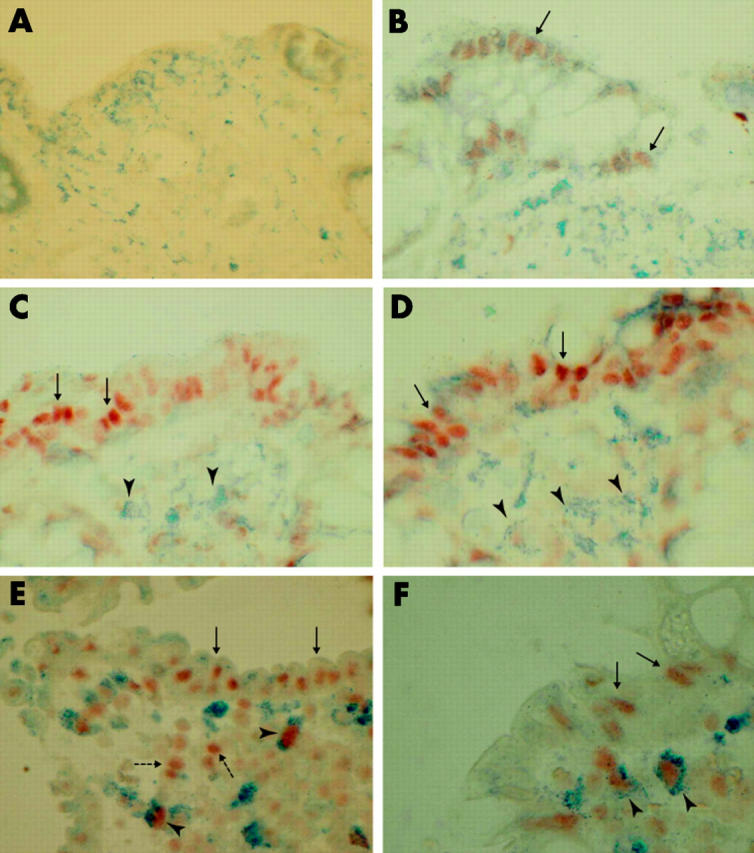
Double immunohistochemical detection of activated nuclear factor κB (NFκB) and cellular markers for macrophages (CD68) in paraffin embedded formalin fixed biopsies from patients with uninflamed bowel (A, B), collagenous colitis (C, D), or ulcerative colitis (E, F). Negative NFκB staining (A) or weak but distinct focal staining of epithelial cells (B; arrows; red staining) was seen in uninflamed bowel. In collagenous colitis, NFκB nuclear staining was predominantly seen in the epithelium (C, D; arrows) while CD68 positive macrophages (C, D; arrowheads; blue staining) and CD68 negative stromal cells were left unstained. In ulcerative colitis, epithelial cells (E, F; arrows), CD68 positive macrophages (E, F; arrowheads), and CD68 negative stromal cells (E; broken arrows) showed intense nuclear expression of NFκB. Magnification ×100 (A–E) and ×150 (F).
Table 2.
Immunohistochemical staining: scoring of nuclear factor κB staining according to cell type
| Material | Epithelial cells | Macrophages (CD68 positive cells) | Fibroblasts (vimentin positive cells) |
| Uninflamed bowel | 0/+ | 0 | 0 |
| Collagenous colitis | +++ | 0 | 0 |
| Ulcerative colitis | ++ | + | 0 |
Percentage of stained cells was scored as follows: 0, no nuclear staining; +, nuclear staining in 1–10% of the cell type under examination; ++, nuclear staining in 11–25% of the cell type under examination; +++, nuclear staining in more than 25% of the cell type examined.
In uninflamed bowel, weak but distinct and focal staining was observed in epithelial cells (0/+). In collagenous colitis, only sporadic (<1%) CD68 positive cells were seen with nuclear factor κB staining, with an overall score of zero (0).
DISCUSSION
NFκB activation in the colonic mucosa from patients with ulcerative colitis has previously been reported.6,7 The results of the present study demonstrate that NFκB is activated both in collagenous and ulcerative colitis compared with uninflamed bowel, as judged by EMSA, chromatin immunoprecipitation, and immunohistochemistry. The results of IκB kinase assays revealed that IKKβ, but not IKKα, was activated, while gene expression profiling showed markedly increased expression of iNOS and several other NFκB dependent genes. We could not demonstrate any differences between collagenous and ulcerative colitis as regards IKK activity, activation of NFκB subunits, recruitment of NFκB to the iNOS promoter, or activation of NFκB in epithelial cells. In contrast, macrophages with activated NFκB were observed only in colonic mucosa from patients with ulcerative colitis.
Supershift analysis showed DNA binding activity of three different NFκB subunits both in collagenous and ulcerative colitis—namely, p65, NFκB1 p50, and NFκB2 p52. The presence of p65 and p50 accords with our findings of IKKβ activity in both inflammatory conditions whereas the presence of NFκB2 p52 would imply activity of IKKα also.14–16 Although IKKα was present in the mucosa, as judged by immunoblotting, there was no detectable activity in our extracts (fig 2A ▶). Recently, it was reported that IKKα translocates into the nucleus on activation and phosphorylates histone H3 at serine 10, which is a requirement for induction of immediate-early genes.17,18 If only a small fraction of active IKKα is present in the cytosolic compartment, the observed discrepancy might be explained by the absence of nuclear proteins in our IKK assays, which were performed on cytosolic extracts. Experiments on whole cell extracts should therefore be performed to clarify this issue.
Colonic mucosal biopsies consist of a variety of cell types, including epithelial cells and immune cells. Although iNOS is localised primarily to the epithelium in collagenous colitis,2 the observed NFκB DNA binding activity is not necessarily associated with iNOS expression. Thus NFκB might be active in one cell type while another cell type is the source of iNOS upregulation. At any rate, our demonstration of NFκB p65 binding to the iNOS promoter in vivo indicates that NFκB is the transcriptional activator of iNOS in the epithelium of patients with collagenous colitis.
The observation of NFκB binding to DNA is insufficient for demonstration of transcriptional activity. Numerous layers of control mechanisms, such as phosphorylation and acetylation of p65 and histones,17,19–22 determine the transcriptional activity of NFκB. To exclude the possibility that NFκB is transcriptionally silent in collagenous colitis, we performed gene expression profiling on a subset of NFκB dependent genes, including iNOS, using a microarray system. Of 96 genes tested, only a few were significantly upregulated in patients with colitis but minor differences in expression pattern between collagenous and ulcerative colitis were observed, most likely reflecting the fact that NFκB was activated in different cell types. As the genes upregulated are NFκB dependent genes, it seems likely that NFκB is indeed transcriptionally active both in collagenous and ulcerative colitis.
As demonstration of in vivo p65 binding to the iNOS promoter in biopsy specimens from collagenous colitis provides no direct evidence of the cell types involved, we used double labelled immunohistochemical detection of activated NFκB and cellular markers for macrophages and fibroblasts (fig 5 ▶, table 2 ▶). These data showed activation of NFκB both in collagenous and ulcerative colitis compared with uninflamed bowel. In collagenous colitis, NFκB activation was located in epithelial cells. In contrast, activation of NFκB in ulcerative colitis was demonstrated both in macrophages and CD68 negative stromal cells, which were not fibroblasts, in addition to epithelial cells. This discrepancy suggests that activation of NFκB in epithelial cells does not lead to mucosal damage in itself. Instead, activation of NFκB in macrophages may be a prerequisite of injury in colonic inflammation. Clearly, further studies are needed to draw safe conclusions.
In summary, our results demonstrate activation of NFκB via an IKKβ mediated pathway, recruitment of NFκB to the iNOS promoter in vivo, and expression of NFκB dependent genes in colonic mucosal biopsies from patients with collagenous and ulcerative colitis. In collagenous colitis, activated NFκB was observed only in epithelial cells while lamina propria macrophages showed activated NFκB in ulcerative colitis. As collagenous colitis is never associated with tissue injury, our findings challenge the prevailing view that activation of NFκB per se mediates mucosal damage in colonic inflammation. Instead, the downstream inflammatory reactions are more likely to be determined by the cellular origin of NFκB activation.
Acknowledgments
We wish to thank Anne Hallander for excellent technical assistance and Toyota Fonden Danmark for sponsoring the FujiFilm phosphorimager and the FujiFilm LAS-1000 equipment.
Abbreviations
ANOVA, analysis of variance
BSA, bovine serum albumin
DTT, dithiothreitol
EDTA, ethylenediaminetetra-acetic acid
EMSA, electrophoretic mobility shift assay
EGTA, ethylene glycol-bis-(2aminoethylether) tetra-acetic acid
GST, glutathione S-transferase
IκB, inhibitor of NFκB
IKK, IκB kinase
IL, interleukin
iNOS, inducible nitric oxide synthase
IP, immunoprecipitation
MOPS, 3-morpholino-propane-sulfonic acid
NFκB, nuclear factor κB
NO, nitric oxide
NOD, nucleotide oligomerisation domain
NP-40, nonidet p-40
PBS, phosphate buffered saline
PCR, polymerase chain reaction
PMSF, phenylmethylsulfonyl fluoride
PNPP, p-nitrophenyl phosphate
PVDF, polyvinylidene difluoride
SDS, sodium dodecyl sulphate
SAC, Staphylococcus aureus Cowan 1
TBE, tris borate EDTA
TNF, tumour necrosis factor
WT, wild-type
Conflict of interest: None declared.
REFERENCES
- 1.Perner A, Nordgaard I, Matzen P, et al. Colonic production of nitric oxide gas in ulcerative colitis, collagenous colitis and uninflamed bowel. Scand J Gastroenterol 2002;37:183–8. [DOI] [PubMed] [Google Scholar]
- 2.Perner A, Andresen L, Normark M, et al. Expression of nitric oxide synthases and effects of L-arginine and L-NMMA on nitric oxide production and fluid transport in collagenous colitis. Gut 2001;49:387–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Järnerot G, Tysk C, Bohr J, et al. Collagenous colitis and fecal stream diversion. Gastroenterology 1995;109:449–55. [DOI] [PubMed] [Google Scholar]
- 4.Taylor BS, de Vera ME, Ganster RW, et al. Multiple NF-kappaB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J Biol Chem 1998;273:15148–56. [DOI] [PubMed] [Google Scholar]
- 5.Taylor BS, Shao L, Gambotto A, et al. Inhibition of cytokine-induced nitric oxide synthase expression by gene transfer of adenoviral I kappa B alpha. Surgery 1999;126:142–7. [PubMed] [Google Scholar]
- 6.Rogler G, Brand K, Vogl D, et al. Nuclear factor kappaB is activated in macrophages and epithelial cells of inflamed intestinal mucosa. Gastroenterology 1998;115:357–69. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber S, Nikolaus S, Hampe J. Activation of nuclear factor kappa B in inflammatory bowel disease. Gut 1998;42:477–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer II, Kawka DW, Scott S, et al. Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology 1996;111:871–85. [DOI] [PubMed] [Google Scholar]
- 9.Lindström CG. ‘Collagenous colitis’ with watery diarrhoea—a new entity? Pathol Eur 1976;11:87–9. [PubMed] [Google Scholar]
- 10.DiDonato JA. Assaying for I kappa B kinase activity. Methods Enzymol 2000;322:393–400. [DOI] [PubMed] [Google Scholar]
- 11.Boyd KE, Wells J, Gutman J, et al. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci U S A 1998;95:13887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkegaard T, Hansen A, Bruun E, et al. Expression and localisation of matrix metalloproteinases and their natural inhibitors in fistulae of patients with Crohn’s disease. Gut 2004;53:701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geddert H, Heep HJ, Gabbert HE, et al. Expression of cyclin B1 in the metaplasia-dysplasia-carcinoma sequence of Barrett esophagus. Cancer 2002;94:212–18. [DOI] [PubMed] [Google Scholar]
- 14.Senftleben U, Cao Y, Xiao G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 2001;293:1495–9. [DOI] [PubMed] [Google Scholar]
- 15.Dejardin E, Droin NM, Delhase M, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 2002;17:525–35. [DOI] [PubMed] [Google Scholar]
- 16.Müller JR, Siebenlist U. Lymphotoxin beta receptor induces sequential activation of distinct NF-kappa B factors via separate signaling pathways. J Biol Chem 2003;278:12006. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y, Verma UN, Prajapati S, et al. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 2003;423:655–9. [DOI] [PubMed] [Google Scholar]
- 18.Anest V, Hanson JL, Cogswell PC, et al. A nucleosomal function for IkappaB kinase-alpha in NF-kappa B-dependent gene expression. Nature 2003;423:659–63. [DOI] [PubMed] [Google Scholar]
- 19.Zhong H, May MJ, Jimi E, et al. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell 2002;9:625–36. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Fischle W, Verdin E, et al. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 2001;293:1653–7. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J 2002;21:6539–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiernan R, Bres V, Ng RWM, et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem 2003;278:2758–66. [DOI] [PubMed] [Google Scholar]