Abstract
Background and aims: The mechanisms by which commensal bacteria provoke intestinal inflammation in animal models of inflammatory bowel disease (IBD) remain incompletely defined, leading to increasing interest in the innate immune response of the colonic mucosa to bacterial colonisation.
Methods: Using gene expression profiling of colonic RNA from C.B17.SCID germ free mice and those colonised with altered Schaedler’s flora, we investigated the innate immune response to bacterial colonisation in vivo. The two most consistently induced gene groups were RegIIIβ and γ as well as interferon γ (IFN-γ) response genes.
Results: Using quantitative reverse transcription-polymerase chain reaction, we showed that RegIIIβ, RegIIIγ, and IFN-γ were constitutively expressed in the colon of conventionally housed SCID mice compared with either germ free SCID or conventionally housed BALB/c mice. Induction of these genes was reproduced by chronic monoassociation of germ free SCID mice with either of two separate gut commensal bacterial species—segmented filamentous bacteria and Schaedler’s Escherichia coli. The cellular source for IFN-γ on monoassociation of SCID mice with Schaedler’s E coli was localised to a subset of intraepithelial natural killer (IENK) cells that express asialo-GM1. In vivo IFN-γ immunoneutralisation studies failed to demonstrate any alteration in RegIIIβ or γ expression.
Conclusions: Thus bacterial colonisation of the colon independently activates two distinct innate immune cell types at the mucosal interface with the colonic lumen, intestinal epithelial cells, and IENK cells, a response that may be regulated by the adaptive immune system. These innate immune responses may play a role in the pathogenesis of colitis in SCID adoptive transfer models in mice and possibly in patients with IBD.
Keywords: interferon γ, colon, SCID mice, bacteria, innate immunity
Growing evidence from clinical observations of patients with inflammatory bowel disease (IBD) as well as studies of animal models of intestinal inflammation suggests that the pathogenesis of IBD involves a dysregulated immune response to the commensal bacterial flora in the intestinal tract of genetically susceptible individuals. Interest has recently focused on the role of the innate immune system in maintaining homeostasis of the mucosal immune system in the presence of gut flora. For example, the bacterial protein flagellin has been recently identified as a dominant antigen in several murine models of intestinal inflammation as well as in patients with Crohn’s disease.1 In addition, identification of NOD2 gene mutations as a genetic factor that enhances susceptibility to the development of Crohn’s disease provides a direct link between genetic predisposition, commensal bacteria, and the innate immune system in the pathogenesis of IBD.2
The colonic epithelium represents the interface between the mucosal surface and the contents of the intestinal lumen and may be considered a component of the innate immune system. This cell type is exposed to a complex bacterial laden environment containing a vast number of toxic and proinflammatory substances that can be referred to as pathogen associated molecular patterns (PAMPs). PAMPs can bind to “Toll-like receptors” (TLRs) thereby eliciting an innate immune response. Recently, Rakoff-Nahoum et al have shown that constitutive activation of TLRs plays an important role in epithelial homeostasis and cytoprotection in the intestine.3
In common with many murine models of intestinal inflammation, the adoptive transfer of CD4+CD45RBhi T cells into syngeneic SCID or RAG−/− recipients results in the development of chronic colitis in conventionally housed animals but not in a germ free environment.4 Despite much attention on the role of specific adaptive immunity in the maintenance of immune homeostasis in the intestinal mucosa in this model system,5 little is known about the contribution of the innate immune response. In this study, we examined gnotobiotic C.B17.SCID mice to determine the effect of bacterial colonisation on the colonic innate immune response. Chronic colonisation of formerly germ free SCID mice with a combination of bacteria, altered Schaedler’s defined flora (ASF) or two individual commensal bacteria, Schaedler’s E coli or segmented filamentous bacteria (SFB), strongly and independently induced the expression of the RegIII epithelial gene family as well interferon γ (IFN-γ) by intraepithelial natural killer cells (IENKs) showing a robust host response to commensal bacteria at the mucosal interface. Interestingly, this response was absent in immunocompetent syngeneic BALB/c mice, suggesting that the adaptive immune system actively regulates the innate immune response to bacterial colonisation in the normal intestinal tract.
MATERIALS AND METHODS
Colonisation of mice and tissue harvest
Germ free BALB/c and C.B17.SCID mice were bred and housed in the germ free facility of the Department of Biology at the University of Pennsylvania (Philadelphia, Pennsylvania, USA) under sterile conditions in Trexler isolators (Standard Safety, McHenry, Illinois, USA). Colonisation of mice (mean age eight weeks) with Schaedler’s E coli (kindly provided by Russell Schaedler) was achieved by intragastric administration of 107–108 colony forming units (CFU) of bacteria. E coli colonisation levels were quantified by serial dilutions of stool plated on Schaedler’s agar. Total CFUs per gram faecal contents ×105 were then calculated. Mice were inoculated with SFB and ASF at eight weeks of age. Monoassociation of formerly germ free mice with either SFB or oligoassociated with ASF (Taconic, Germantown, New York, USA) were performed as previously described.4,6 Mice were colonised with ASF using a two step protocol described by Taconic Technical Library (www.taconic.com/library). At the time of sacrifice, the colon was removed, gently cleared of stool, and divided lengthwise. One half was immediately frozen in liquid nitrogen for RNA isolation and the other half was preserved in 10% formalin and embedded in paraffin. Additionally, faecal pellets from mice monoassociated or oligoassociated with bacteria were checked for colonisation by microscopic examination of smears.
Microarray analysis
RNA was supplied to the Penn Microarray Facility for microarray analysis on Affymetrix MG_U74A microarrays (Santa Clara, California, USA). Signal values for each gene were calculated with Microarray Suite v.5 (MAS5; Affymetrix). The resulting signal values were then analysed using Genespring 5.0 (Silcon Genetics, Redwood City, California, USA). Gene expression values were normalised to the default 50th percentile and the median for each gene. Genes that were absent by MAS5 analysis in all samples were removed from consideration. Biological samples in each experimental condition were compared, producing nine gene lists from which a list of common genes was determined. Comparisons between experimental conditions were performed, as noted in fig 1 ▶. Genes that changed by twofold or more for each pairwise comparison and were flagged as present (MAS5) in at least one sample were identified. Venn diagrams were used to identify genes in common to all nine analyses. In addition, conditional replicates were averaged and genes with a greater than twofold change between conditions and had a standard deviation of no more than half the gene’s median in one out of two conditions were identified.
Figure 1.
Diagram of the strategy used to identify genes significantly altered in expression. Sample to sample microarray analysis of germ free mice versus mice colonised with ASF (altered Schaedler’s flora). Nine individual analyses were collapsed by Venn diagrams into a single list of genes that varied either twofold up or down between conditions.
Quantitative reverse transcription-polymerase chain reaction (RT-PCR)
Reverse transcription utilising SuperScript II First Strand Synthesis Kit (Invitrogen, Carlsbad, California, USA) was followed by Syber Green quantitative PCR performed on an ABI 7000 Sequence Detector (Applied Biosystems Inc., Foster City, California, USA). Primers were designed using Primer Express software (Applied Biosystems Inc.). The absence of primer-dimer formation was verified by primer matrix analysis. Primers used in these studies were: mGAPDH: forward 5′’-GGT GGT CTC CTC TGA CTT CAA CA-3′, reverse 5′-GTT GCT GTA GCC AAA TTC GTT GT-3′; mRegIIIβ: forward 5′-TCC CAG GCT TAT GGC TCC TA-3′, reverse 5′-GCA GGC CAG TTC TGC ATC A-3′; mRegIIIγ: forward 5′-CAT CAA CTG GGA GAC GAA TCC-3′, reverse 5′-CAG AAA TCC TGA GGC TCT TGA CA-3′; IFN-γ: forward 5′-CAT TCA TGA GTA TTG CCA AGT TTG-3′, reverse 5′-GCT GGA TTC CGG CAA CAG-3′; uridine phosphorylase (Upase): forward 5′-CAT CAC CAT CAT CCG CAT TG-3′, reverse 5′-GCC TGC TGC GTG ATG ACA-3′. PCR conditions used were 50°C for two minutes, 95°C for 10 minutes, and then 40 cycles at 95°C for 15 seconds followed by 60°C for one minute. A dissociation curve was run with each PCR to ensure that primer-dimer formation did not occur. PCR results were analysed using the ΔΔCt analysis (User Bulletin #2; Applied Biosystems, Inc) with GAPDH as the housekeeping gene.
Immunohistochemistry
Paraffin embedded sections were dewaxed, rehydrated in 10 mmol/l citric acid buffer (pH 6.0), and microwaved for 10 minutes 30 seconds. Tissue sections were blocked with serial incubations in 3% hydrogen peroxide, avidin D (Vector Laboratories, Burlingame, California, USA), biotin (Vector Laboratories), and protein blocking agent (Coulter-Immunotech, Marseille, France). Sections were then incubation with affinity purified antibody overnight at 4°C, washed with 1× phosphate buffered saline, incubated with the appropriate secondary antibody at 37°C, and exposed to horseradish peroxidase conjugated ABC reagent (Vecta Elite kit; Vector Laboratories). A DAB substrate kit for peroxidase (Vector Laboratories) was used for signal detection.
The IFN-γ antibody (#R4-6A2; BD Biosciences PharMingen, San Jose, California, USA) was used at a 1:400 dilution in PBT (1×phosphate buffered saline, 0.1% bovine serum albumin, and 0.2% Triton X-100 in water) for DAB development and at 1:600 dilution in PBT for fluorescent detection. Asialo-GM1 antibody (#986-10001; Wako, Osaka, Japan) was used at a 1:8000 dilution in PBT. A Cy3 conjugated antirat secondary antibody and a Cy2 conjugated antirabbit secondary antibody (Jackson Immuno Research Laboratories, Ltd, West Grove, Pennsylvania, USA) were used at 1:200 dilutions in PBT. These slides were treated with Sudan Black B (0.2% SBB in 70% ethanol; Gallard-Schlesinger Industries, Plainview, New York, USA) and counterstained with DAPI (Molecular Probes, Eugene, Oregon, USA).
Immunoneutralisation of IFN-γ
Germ free C.B17.SCID mice were immunodepleted for IFN-γ by intraperitoneal injection of anti-IFN-γ purified from ascites fluid (XMG.6; Harlan Biosciences, Madison, Wisconsin, USA). A second set of SCID germ free mice were treated with an isotype control antibody, rat IgG (#535865; Sigma, St. Louis, Missouri, USA). The antibody was delivered by intraperitoneal injection at 1–2 mg per animal every four days for the duration of the experiment. One day after the first treatment with antibody, mice were colonised with Schaedler’s E coli. After 14 days of colonisation, mice were sacrificed. Tissue was harvested as noted above. A set of germ free mice were harvested as negative controls. Robust monoassociation of C.B17.SCID mice with Schaedler’s E coli was verified by surveillance cultures of the stool grown in Schaedler’s broth (Becton, Dickinson and Company, Sparks, Maryland, USA).
RESULTS
Microarray gene profiling of SCID mice reveals that ASF and SFB activate the expression of the pancreatitis associated protein (PAP)/Reg gene family in the colon
To identify intestinal genes that are regulated by the innate immune system in response to chronic bacterial colonisation under defined conditions, we initially evaluated the effects of a commercially available flora on intestinal gene expression. Cognisant that the composition of the commensal gut flora undoubtedly varies from facility to facility, we attempted to control for variability using a commercially available defined flora as a surrogate for normal commensal colonisation. ASF,7 a combination of eight commensal bacteria, was used to colonise germ free C.B17.SCID mice for six months after which time total RNA was isolated from the colon and caecum of three colonised mice and three germ free controls. Using Affymetrix MG_U74A murine gene microarrays followed by data mining using the GeneSpring bioinformatics platform, sample to sample comparison between experimental conditions was performed, as noted in fig 1 ▶. This nineway comparison revealed that only eight genes were upregulated by greater than twofold (table 1 ▶).
Table 1.
List of genes expressed in the colon that varied by twofold or more between C.B17.SCID germ free and altered Schaedler’s flora (ASF) monoassociated mice
| (1) Defensin related cryptdin 2 |
| (2) RegIIIγ |
| (3) Pancreatitis associated protein |
| (4) RegIIIβ |
| (5) UPase |
| (6) Phospholipase A2, group IIA |
| (7) Defensin related cryptdin 16 |
| (8) Single repeat sequence-containing transcript |
Results based on analysis described in fig 1 ▶.
Upase, uridine phosphorylase.
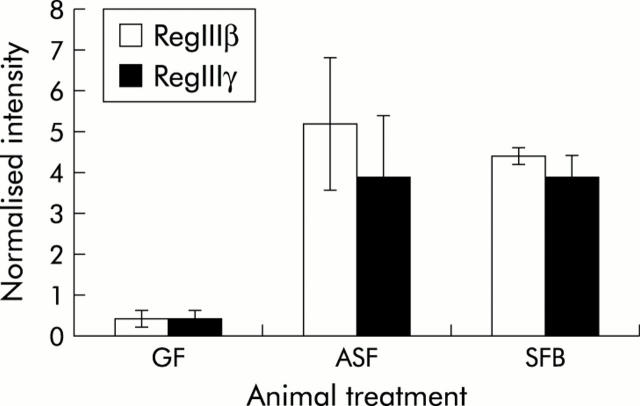
To determine if similar alterations in gene expression could be induced by single bacterial species, we used quantitative RT-PCR to examine expression of these genes on monoassociation of formerly germ free SCID mice with a single Gram positive obligate anaerobic gut commensal known as SFB8 as well as a facultative anaerobic Gram negative bacterium, Schaedler’s E coli.9 Additionally, colonisation of mice with bacteria that have very different characteristics would help to determine if alterations in intestinal gene expression are dependent on specific characteristics of the bacterial species or if it is a more general feature of commensal colonisation. Neither of these two bacterial species are components of ASF. Remarkably, this analysis showed that chronic colonisation with ASF for six months, SFB for four months, or Schaedler’s E coli, were all able to induce robust expression of PAP/Reg III genes in formerly germ free SCID mice (figs 2 ▶, 3 ▶).
Figure 2.
Colon mRNA expression of RegIIIβ and RegIIIγ in germ free C.B17.SCID mice as well as in those colonised with either altered Schaedler’s flora (ASF) or segmented filamentous bacteria (SFB). Quantification of mRNA expression with Affymetrix MG_U74A microarrays was performed using standard normalisations and average gene expression between conditions. Data (mean (SD)) are expressed as fold induction relative to germ free (GF) mRNA expression; n = 3.
Figure 3.
Relative mRNA expression of RegIIIβ, RegIIIγ, and interferon γ (IFN-γ) in BALB/c and C.B17.SCID mice in germ free, Escherichia coli monoassociated, or conventional environments, quantified by reverse transcription-polymerase chain reaction. Duration of monoassociation is indicated. Quantification of gene expression expressed as the mean (SEM) of 3–14 mice per experimental condition. GF, germ free; Conv, conventional; and E coli, Schaedler’s E coli.
The PAP/Reg family of genes are associated with the inflammatory response and are induced to high levels in patients with IBD.10,11 Importantly, PAP/Reg gene expression can be induced by IFN-γ in vitro.12 Indeed, genes that have been shown to be activated by IFN-γ are induced on chronic colonisation with either ASF or SFB (table 2 ▶).
Table 2.
Fold induction of RegIII and interferon γ (IFN-γ) inducible genes in the colon of either altered Schaedler’s flora (ASF) or segmented filamentous bacteria (SFB) colonised C.B17.SCID mice relative to those housed in a germ free environment
| Gene | ASF | SFB |
| RegIIIβ | 11.48 | 9.82 |
| RegIIIγ | 8.64 | 8.64 |
| IRG 47 | 1.84 | 2.10 |
| Rab6 | 2.43 | 2.15 |
| UPase | 5.64 | 1.79 |
| Guanylate nucleotide binding protein | 2.47 | 1.41 |
Total RNA isolated from the colon of SCID mice housed in the designated environments was analysed using the Affymetrix MG_U74A microarray. After employment of standard normalisations, the average gene expression values between conditions were used to determine fold induction (n = 3 for each condition).
Upase, uridine phosphorylase.
Monoassociation with Schaedler’s E coli results in activation of Reg III and IFN-γ in the colon of SCID mice
We sought to identify a model system which could be used to investigate the relationship between these genes and colonisation with a single bacterial species that could be easily genetically manipulated and cultured in vitro. Thus, for further evaluation, we chose to examine the effects of Schaedler’s E coli on intestinal gene expression as SFB is an obligate anaerobe that cannot be cultured in vitro8 and ASF is a combination of eight commensal organisms.7 Schaedler’s E coli effectively colonised the intestinal tract of both BALB/c and SCID mice, with the greatest amount of bacteria found in the caecum and colon (table 3 ▶).
Table 3.
Quantification of bacterial load in the gut of formerly germ free BALB/c and C.B17.SCID mice monoassociated with Schaedler’s E coli
| Mice | Days after colonisation | Average/CFU/g ×105 | ||
| Small intestine | Caecum | Large intestine | ||
| BALB/c | 14 | 264.0 (12.7) | 2000.0 (200.0) | 5860.8 (100.0) |
| 30 | 49.3 (2.5) | 1420.0 (40.0) | 2133.3 (70.5) | |
| SCID | 14 | 48.5 (12.0) | 120.6 (25.2) | 83.1 (30.3) |
| 30 | 7.2 (0.1) | 20.0 (7.0) | 46.8 (20.1) | |
Total CFU/g ×105 of luminal contents from the small intestine, caecum, and large intestine was calculated by determining the colony forming units (CFU)/plate × volume dilution × serial dilution (n = 3 mice/time point).
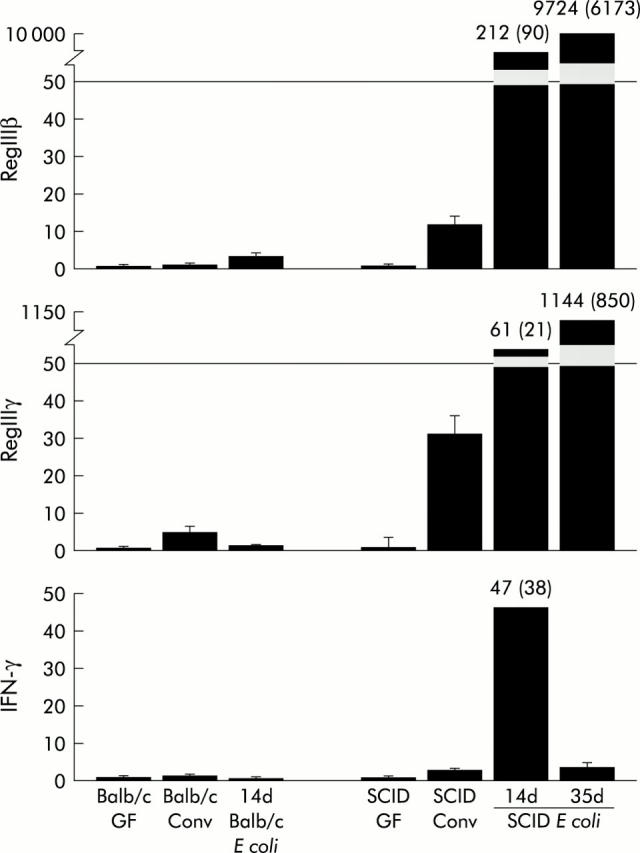
Although the number of bacteria found in these two regions of the intestine in SCID mice was less than that quantified in BALB/c mice, the number of bacteria remained stable for at least 60 days (data not shown). By contrast, there was a dramatic time dependent reduction in the number of E coli colonising the small intestine in both strains of mice. Using quantitative RT-PCR of colonic RNA, we show in fig 3 ▶ that Schaedler’s E coli induces Reg III and IFN-γ expression in SCID mice to levels equivalent to or exceeding those observed in SCID mice housed in a conventional environment. Interestingly, although greater levels of E coli can be identified in the caecum and colon of BALB/c mice compared with SCID mice (table 3 ▶), there was minimal induction of Reg III and no induction of IFN-γ in BALB/c mice, suggesting that components of the adaptive immune system inhibit this response to bacterial colonisation. Importantly, monoassociation of SCID mice for over two months led to a 3.5-fold induction of IFN-γ expression in the colon relative to that in germ free SCID mice, a response equivalent to conventionally housed SCID mice (data not shown).
Schaedler’s E coli induces RegIIIβ/γ and IFN-γ expression in the caecum and proximal colon but not in the distal colon of SCID mice
The number and species of bacteria that colonise the intestinal tract varies tremendously throughout the length of the gastrointestinal tract, with the caecum, which acts as a reservoir, containing particularly high levels of bacteria. Functional studies have shown that the caecum in rodents is important for the development of intestinal inflammation in models of IBD.13,14 Interestingly, induction of RegIIIβ, RegIIIγ, and IFN-γ expression by colonisation of SCID mice with E coli was greatest in the caecum and proximal colon, with no alteration in the distal colon (fig 4 ▶). Hence this demonstrates that the innate immune response to bacterial colonisation occurs in a region specific fashion and is greatest in an organ shown to play a critical role in the pathogenesis of IBD in certain rodent models.
Figure 4.
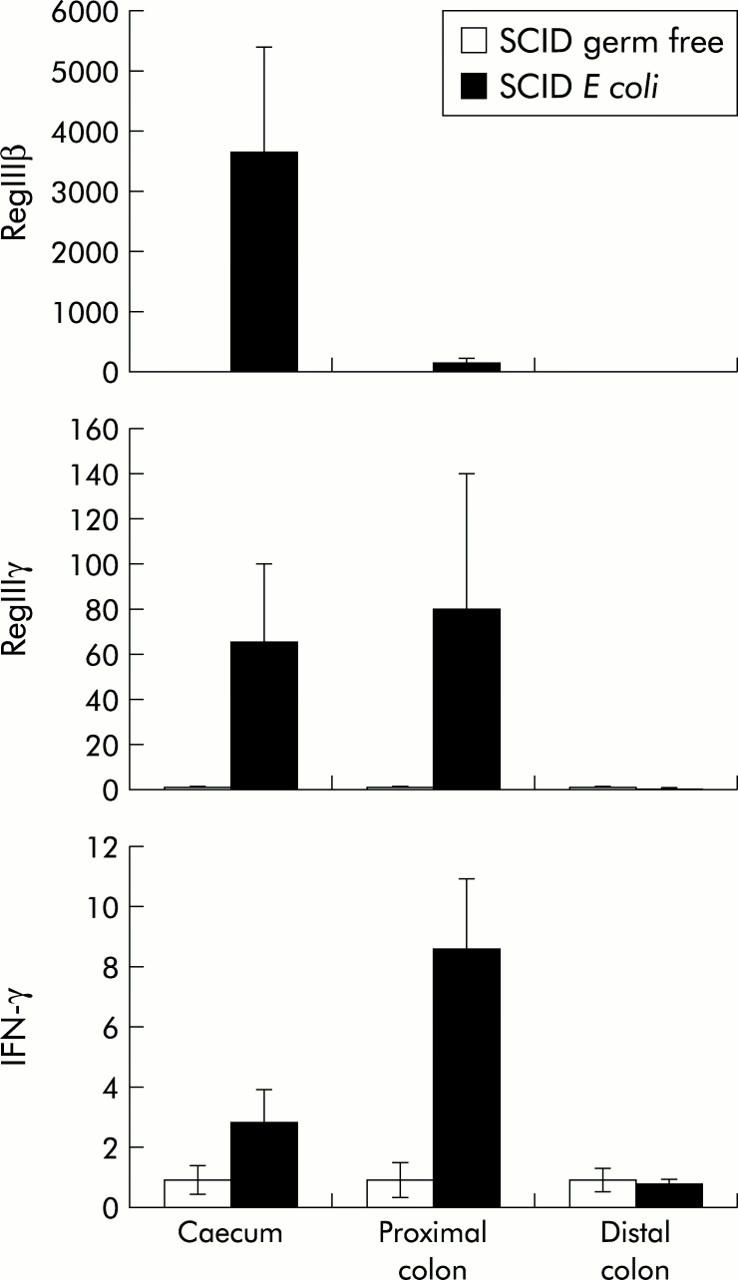
Regional expression of RegIIIβ/γ and interferon γ (IFN-γ) in C.B17.SCID germ free and E coli monoassociated mice. Quantitative reverse transcription-polymerase chain reaction for RegIIIβ, RegIIIγ, and IFN-γ in the caecum, proximal, and distal colon of SCID germ free mice and E coli monoassociated mice. Values are mean (SEM); n = 3.
Immunohistochemistry for IFN-γ identifies intraepithelial mononuclear cells as the source for IFN-γ in the colon of E coli monoassociated SCID mice
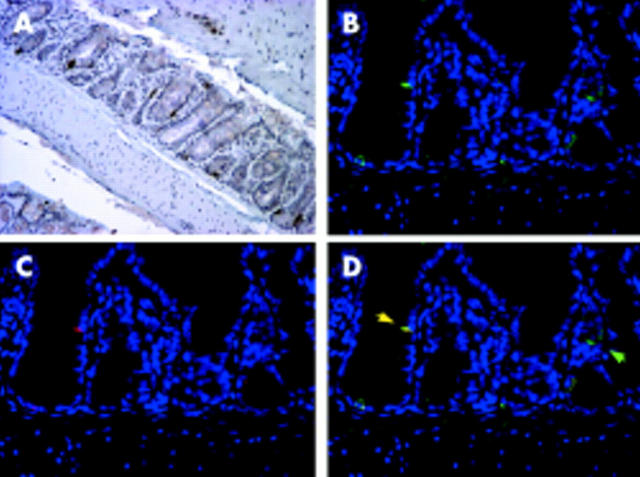
Immunohistochemistry was performed to identify the cellular source for induction of IFN-γ expression in the colon of SCID mice in response to E coli. Colonic tissues from Schaedler’s E coli monoassociated C.B17SCID mice were stained with a monoclonal antibody to IFN-γ. Staining was specific for a population of intraepithelial mononuclear cells (fig 5A ▶). As intraepithelial T lymphocytes are absent in SCID mice and natural killer (NK) cells are believed to be the sole source of IFN-γ in these mice,15 these IFN-γ producing cells are believed to be IENK cells. To confirm this, dual fluorescence immunostaining was performed using antibodies specific for asialo-GM1, a marker for NK cells and certain lymphocyte populations. In the absence of lymphocytes in SCID mice, this marker can be used to specifically identify NK cells. Asialo-GM1 cells were identified as mononuclear cells located in the intraepithelial compartment (fig 5B ▶) in the same region as cells expressing IFN-γ in E coli monoassociated SCID mice (fig 5C ▶). Dual staining confirmed that IFN-γ in the colon of these animals is produced by a subset of intraepithelial NK cells (fig 5D ▶). Interestingly, these cells were located in the lower crypt but not in the surface epithelial compartment.
Figure 5.
Immunohistochemical detection of interferon γ (IFN-γ) and asialo-GM1 in the colon of C.B17.SCID mice colonised with Escherichia coli. Colon of SCID E coli mouse stained with (A) IFN-γ antibody followed by DAB development, 20×; (B) asialo-GM1 antibody with Cy2 fluorescent secondary antibody (green), 40×; (C) IFN-γ antibody with Cy3 fluorescent secondary antibody (red), 40×; and (D) colocalisation of asialo-GM1 and IFN-γ with fluorescent secondary antibody, 40×. Yellow arrow = colocalised IFN-γ and asialo-GM1; green arrow = asialo-GM1 only. Immunofluorescent sections (B-D) costained with DAPI to indicate nuclei (blue).
Immunoneutralisation of IFN-γ in vivo does not prevent induction of REGIIIβ/γ expression in the colon of SCID mice by colonisation with Schaedler’s E coli
Previous studies have shown that expression of the RegIII gene family can be induced by IFN-γ in vitro.12 Furthermore, similar to our results shown in table 2 ▶, RegIII gene expression has been strongly associated with IFN-γ production and activation of IFN-γ response genes in the intestinal tract of mice.16 In order to directly determine if RegIII expression in the colon of SCID mice was dependent on activation of IFN-γ by E coli, RegIII expression was assessed in monoassociated SCID mice in which we attempted to deplete IFN-γ by serial administration of an immunoneutralising antibody. Immunodepletion dramatically reduced expression of IFN-γ mRNA expression in the colon of SCID mice monoassociated with Schaedler’s E coli for 14 days (fig 6 ▶). Consistent with this reduction of IFN-γ was the significant reduction in Upase expression, a well characterised IFN-γ response gene17 whose expression was enhanced by colonisation of SCID mice with either ASF or SFB (table 2 ▶). In contrast, there was no alteration in either RegIIIβ or γ expression in mice treated with anti-IFN-γ antibodies compared with those who received isotype control serum (fig 6 ▶). Thus bacterial colonisation independently activates expression of RegIIIβ/γ and IFN-γ at the mucosal interface. This notion is consistent with the observation that IFN-γ failed to induce RegIII expression in multiple intestinal cell lines in culture (data not shown).
Figure 6.
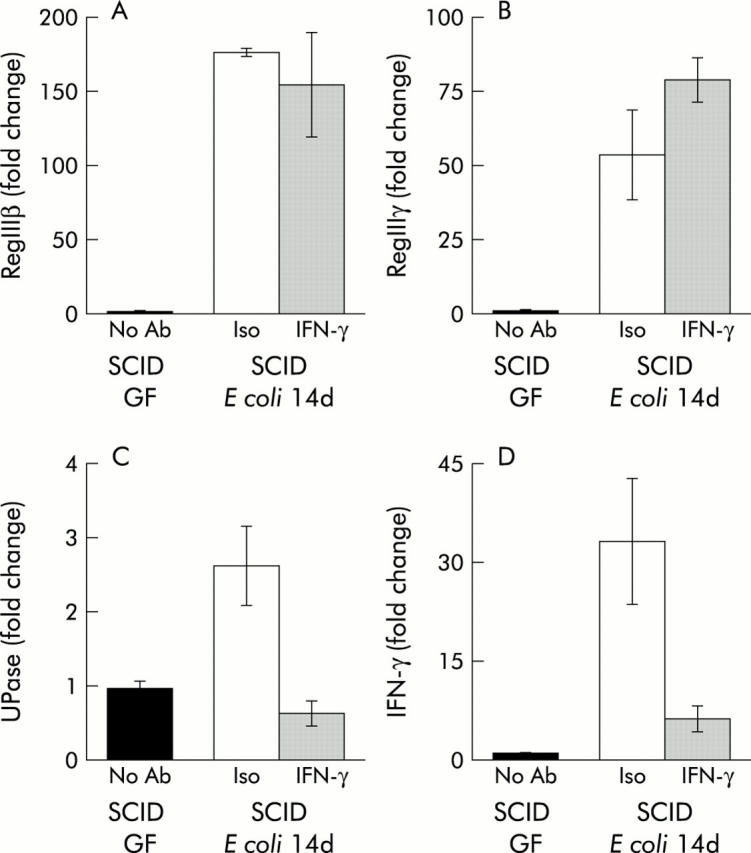
Effect of interferon γ (IFN-γ) immunoneutralisation on RegIIIβ and γ expression in the colon of C.B17.SCID mice monoassociated with E coli. SCID mice received serial intraperitoneal injections of an immunoneutralising antibody to IFN-γ prior to and during colonisation with Schaedler’s E coli. Quantification of mRNA expression for (A) RegIIIβ, (B) RegIIIγ, (C) uridine phosphorylase (Upase), and (D) IFN-γ in the colon was determined by quantitative reverse transcription-polymerase chain reaction after14 days of colonisation. GF, germ free; Ab, antibody; Iso, isotype control antibody; IFN-γ, IFN-γ antibody. Values are mean (SEM); n = 3.
DISCUSSION
The mechanisms by which commensal bacteria lead to intestinal inflammation and probiotics exhibit therapeutic benefits in various animal model systems remain to be clearly elucidated. In this report we have shown that colonisation with commensal bacteria resulted in activation of an innate immune response by two cell types at the mucosal interface with the intestinal lumen, IFN-γ production by IENKs, as well as RegIIIβ and γ expression by the epithelium.
NK cells in the intestine are located in the intraepithelial lymphoid compartment where they express markers that distinguish them from splenic NK cells.18 Thus IENKs are components of the innate immune system that are uniquely situated at the mucosal interface to allow interactions with commensal gut bacteria. Despite the importance of IFN-γ in the development of intestinal inflammation and the ability of NK cells to secrete high levels of IFN-γ, there is only limited information regarding the function of NK cells in maintaining homeostasis of the intestinal mucosal immune system. Induction of IFN-γ expression indicates that bacterial colonisation of SCID mice either activates IENKs or attracts a subset of NK cells to the intestinal epithelium that secrete high levels of IFN-γ. In this regard, a previous study has shown that NK cells help to prevent inflammation in the CD4+CD45RBhi SCID model of colitis,19 suggesting that NK cells may play an important role in preventing the development of autoimmune diseases. Indeed, human IENKs recently shown to have functional properties similar to those in mice20 are dramatically diminished in patients with active coeliac disease.21 Furthermore, depletion of NK1.1 cells in the B6 mouse leads to exacerbations of experimental autoimmune encephalomyelitis and higher mortality.22 Consistent with this notion, the two individual bacterial species used to activate IENKs to express IFN-γ in our study, SFB and E coli, do not induce colitis in animal models of IBD.4,23 In fact, E coli has been shown to exhibit beneficial probiotic effects by ameliorating intestinal inflammation in the SCID adoptive transfer model of colitis.24 In contrast, H muridarum, a commensal bacterium shown to exacerbate the development of colitis in the CD4+CD45RBhi SCID adoptive transfer model,4 does not activate the expression of IFN-γ in SCID mice (unpublished observation).
Based on the IFN-γ immunoneutralisation studies (fig 6 ▶), activation of RegIIIβ and γ gene expression appears to be an independent response of the SCID intestinal tract to bacterial colonisation. Recently, Ogawa et al showed that Reg IIIβ and γ can be induced by bacterial reconstitution of immune competent formerly germ free mice.11 This induction was only transient and was temporally related to the acute inflammatory response associated with conventionalisation. Indeed, the 1.6–3.5-fold induction of RegIIIβ and γ in immune competent BALB/c mice by acute colonisation with Schaedler’s E coli (fig 3 ▶) is consistent with these previously published results. In contrast with these results, we showed that conventionally housed SCID mice expressed levels of RegIIIβ and γ that were more than 50-fold higher than their BALB/c congenic counterparts, an enduring response reproduced by chronic colonisation with ASF, SFB, or E coli bacteria.
Originally identified as a gene expressed in regenerating rat islet after pancreatic resection,25 the Reg gene family can be classified into four subclasses (type I–IV).26 Murine Reg III genes are orthologues of human PAP whose expression is induced in patients with IBD.11,27 In situ hybridisation and immunohistochemistry have shown that these proteins are synthesised by exocrine pancreatic cells, Paneth cells in the small intestine, and in the crypt epithelium in the colon.11 The function of PAP/Reg proteins have been best characterised in multiple model systems to induce cellular proliferation and inhibit apoptosis.28–31 Therefore, it has been hypothesised that PAP/Reg proteins may act as autocrine growth factors that play a role in epithelial repair and/or protection in the setting of intestinal inflammation.27 A function for the constitutive expression of Reg proteins in the intestinal tract, however, remains to be characterised. Interestingly, Reg IIIβ and γ proteins have been shown to be secreted not only basolaterally but also apically by polarised epithelial cells32 and may therefore influence the biology of luminal bacteria. In this regard, PAP/Reg proteins have been shown to promote cellular adhesion33 or induce aggregation of E coli,34 a property perhaps related to their ability to rapidly polymerise into insoluble sedimenting structures on trypsin mediated activation.35 The influence of RegIIIβ and γ expression in SCID mice on the physical and physiological properties of Schaedler’s E coli will require further investigation.
It is clear that components of the innate immune system, particularly antigen presenting cells, play a critical role in the regulation of the specific adaptive immune system. We now present evidence that the converse is also true. Namely, that the adaptive immune system regulates activation of the innate response to bacterial colonisation of the gut. Induction of RegIIIβ and γ as well as IFN-γ expression by each of the three bacterial colonisation conditions examined in our study occurs in SCID but not BALB/c mice. Importantly, this pattern of intestinal gene expression is also observed in conventionally housed SCID compared with BALB/c mice. Activation of IENKs, as indicated by their expression of IFN-γ, and induction of RegIIIβ and γ expression to bacterial colonisation likely represents a compensatory mechanism by which the innate immune system of SCID mice increases immune surveillance and fortifies the epithelial barrier function, respectively, in the absence of an adaptive immune system. As adoptive transfer of specific T cell subsets into SCID mice results in the development of colitis only in the presence of certain commensal bacteria, the response of the adoptively transferred immune system to the innate immune alterations that we have described in this study may play an important role in determining the development of intestinal inflammation in this model system. Such aberrant interactions between the innate and adaptive immune systems induced by bacterial colonisation of the gut may have relevance to the pathogenesis of IBD.
Acknowledgments
This work was supported by grants NIH AI39368 (GDW), NIH AI37108, and the Crohn’s and Colitis Foundation of America (JJC), NIH AI061570, the Crohn’s and Colitis Foundation of America, and The Wellcome Trust (059967) (DA), and the Molecular Biology and Morphology Cores of NIH/NIDDK Center Grant DK50306.
Abbreviations
IBD, inflammatory bowel disease
IFN-γ, interferon γ
ASF, altered Schaedler’s flora
SFB, segmented filamentous bacteria
RT-PCR, reverse transcription-polymerase chain reaction
PAMPs, pathogen associated molecular patterns
TLR, Toll-like receptor
NK, natural killer
IENK, intraepithelial natural killer
CFU, colony forming units
Upase, uridine phosphorylase
PAP, pancreatitis associated protein
Conflict of interest: None declared.
REFERENCES
- 1.Lodes MJ, Cong Y, Elson CO, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest 2004;113:1296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonen DK, Cho JH. The genetics of inflammatory bowel disease. Gastroenterology 2003;124:521–36. [DOI] [PubMed] [Google Scholar]
- 3.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004;118:229–41. [DOI] [PubMed] [Google Scholar]
- 4.Jiang HQ, Kushnir N, Thurnheer MC, et al. Monoassociation of SCID mice with Helicobacter muridarum, but not four other enterics, provokes IBD upon receipt of T cells. Gastroenterology 2002;122:1346–54. [DOI] [PubMed] [Google Scholar]
- 5.Singh B, Read S, Asseman C, et al. Control of intestinal inflammation by regulatory T cells. Immunol Rev 2001;182:190–200. [DOI] [PubMed] [Google Scholar]
- 6.He W, Wang ML, Jiang HQ, et al. Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology 2003;125:1388–97. [DOI] [PubMed] [Google Scholar]
- 7.Dewhirst FE, Chien CC, Paster BJ, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol 1999;65:3287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talham GL, Jiang HQ, Bos NA, et al. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun 1999;67:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaedler RW, Orcutt RP. Gastrointestinal microflora. In: Foster HL, Small JD, Fox JG, eds. The mouse in biomedical research. New York: Academic Press, 1983:327–45.
- 10.Dieckgraefe BK, Stenson WF, Korzenik JR, et al. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics 2000;4:1–11. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa H, Fukushima K, Naito H, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis 2003;9:162–70. [DOI] [PubMed] [Google Scholar]
- 12.Dusetti NJ, Ortiz EM, Mallo GV, et al. Pancreatitis-associated protein I (PAP I), an acute phase protein induced by cytokines. Identification of two functional interleukin-6 response elements in the rat PAP I promoter region. J Biol Chem 1995;270:22417–21. [DOI] [PubMed] [Google Scholar]
- 13.Uhlig HH, Powrie F. Dendritic cells and the intestinal bacterial flora: a role for localized mucosal immune responses. J Clin Invest 2003;112:648–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rath HC, Ikeda JS, Linde HJ, et al. Varying cecal bacterial loads influences colitis and gastritis in HLA-B27 transgenic rats. Gastroenterology 1999;116:310–19. [DOI] [PubMed] [Google Scholar]
- 15.Scott P, Trinchieri G. The role of natural killer cells in host-parasite interactions. Curr Opin Immunol 1995;7:34–40. [DOI] [PubMed] [Google Scholar]
- 16.Woo AL, Gildea LA, Tack LM, et al. In vivo evidence for interferon-gamma-mediated homeostatic mechanisms in small intestine of the NHE3 Na+/H+ exchanger knockout model of congenital diarrhea. J Biol Chem 2002;277:49036–46. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe S, Hino A, Wada K, et al. Purification, cloning, and expression of murine uridine phosphorylase. J Biol Chem 1995;270:12191–6. [DOI] [PubMed] [Google Scholar]
- 18.Todd DJ, Greiner DL, Rossini AA, et al. An atypical population of NK cells that spontaneously secrete IFN-gamma and IL-4 is present in the intraepithelial lymphoid compartment of the rat. J Immunol 2001;167:3600–9. [DOI] [PubMed] [Google Scholar]
- 19.Fort MM, Leach MW, Rennick DM. A role for NK cells as regulators of CD4+ T cells in a transfer model of colitis. J Immunol 1998;161:3256–61. [PubMed] [Google Scholar]
- 20.Leon F, Roldan E, Sanchez L, et al. Human small-intestinal epithelium contains functional natural killer lymphocytes. Gastroenterology 2003;125:345–56. [DOI] [PubMed] [Google Scholar]
- 21.Camarero C, Eiras P, Asensio A, et al. Intraepithelial lymphocytes and coeliac disease: permanent changes in CD3−/CD7+ and T cell receptor gammadelta subsets studied by flow cytometry. Acta Paediatr 2000;89:285–90. [PubMed] [Google Scholar]
- 22.Zhang B, Yamamura T, Kondo T, et al. Regulation of experimental autoimmune encephalomyelitis by natural killer (NK) cells. J Exp Med 1997;186:1677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rath HC, Wilson KH, Sartor RB. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect Immun 1999;67:2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz M, Strauch UG, Linde HJ, et al. Preventive effects of Escherichia coli strain Nissle 1917 on acute and chronic intestinal inflammation in two different murine models of colitis. Clin Diagn Lab Immunol 2004;11:372–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terazono K, Yamamoto H, Takasawa S, et al. A novel gene activated in regenerating islets. J Biol Chem 1988;263:2111–14. [PubMed] [Google Scholar]
- 26.Abe M, Nata K, Akiyama T, et al. Identification of a novel Reg family gene, Reg IIIdelta, and mapping of all three types of Reg family gene in a 75 kilobase mouse genomic region. Gene 2000;246:111–22. [DOI] [PubMed] [Google Scholar]
- 27.Dieckgraefe BK, Crimmins DL, Landt V, et al. Expression of the regenerating gene family in inflammatory bowel disease mucosa: Reg Ialpha upregulation, processing, and antiapoptotic activity. J Investig Med 2002;50:421–34. [DOI] [PubMed] [Google Scholar]
- 28.Bodeker H, Vasseur S, Dusetti NJ, et al. PAP gene transcription induced by cycloheximide in AR4-2J cells involves ADP-ribosylation. Biochem Biophys Res Commun 1998;251:710–13. [DOI] [PubMed] [Google Scholar]
- 29.Nishimune H, Vasseur S, Wiese S, et al. Reg-2 is a motoneuron neurotrophic factor and a signalling intermediate in the CNTF survival pathway. Nat Cell Biol 2000;2:906–14. [DOI] [PubMed] [Google Scholar]
- 30.Moucadel V, Soubeyran P, Vasseur S, et al. Cdx1 promotes cellular growth of epithelial intestinal cells through induction of the secretory protein PAP I. Eur J Cell Biol 2001;80:156–63. [DOI] [PubMed] [Google Scholar]
- 31.Malka D, Vasseur S, Bodeker H, et al. Tumor necrosis factor alpha triggers antiapoptotic mechanisms in rat pancreatic cells through pancreatitis-associated protein I activation. Gastroenterology 2000;119:816–28. [DOI] [PubMed] [Google Scholar]
- 32.Christa L, Carnot F, Simon MT, et al. HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and pancreatic cells. Am J Physiol 1996;271:G993–1002. [DOI] [PubMed] [Google Scholar]
- 33.Valery C, Vasseur S, Sabatier F, et al. Pancreatitis associated protein I (PAP-I) alters adhesion and motility of human melanocytes and melanoma cells. J Invest Dermatol 2001;116:426–33. [DOI] [PubMed] [Google Scholar]
- 34.Iovanna JL, Keim V, Michel R, et al. Pancreatic gene expression is altered during acute experimental pancreatitis in the rat. Am J Physiol 1991;261:G485–9. [DOI] [PubMed] [Google Scholar]
- 35.Graf R, Schiesser M, Scheele GA, et al. A family of 16-kDa pancreatic secretory stress proteins form highly organized fibrillar structures upon tryptic activation. J Biol Chem 2001;276:21028–38. [DOI] [PubMed] [Google Scholar]