Abstract
Background
Gastric adenocarcinomas comprise one of the common types of cancers in Asian countries including Japan. Comprehensive protein profiling of paired surgical specimens of primary gastric adenocarcinomas and nontumor mucosae derived from Japanese patients was carried out by means of two-dimensional gel electrophoresis (2D-EP) and liquid chromatography-electrospray ionic tandem mass spectrometry (LC-ESI-MS) to establish gastric cancer-specific proteins as putative clinical biomarkers and molecular targets for chemotherapy.
Results
Relatively common alterations in protein expression were revealed in the tumor tissues. Increases in manganese dismutase and nonhistone chromosomal protein HMG-1 (HMG-1) were observed, while decreases in carbonic anhydrases I and II, glutatione-S-transferase and foveolin precursor (gastrokine-1) (FOV), an 18-kDa stomach-specific protein with putative tumor suppressor activity, were detected. RT-PCR analysis also revealed significant down-regulation of FOV mRNA expression in tumor tissues.
Conclusion
A possible pathological role for down-regulation of FOV in gastric carcinogenesis was demonstrated. Evaluation of the specific decreases in gene and protein expression of FOV in patients may be utilized as clinical biomarkers for effective diagnosis and assessment of gastric cancer.
Background
Gastric adenocarcinomas comprise one of the common types of cancers in Asian countries including Japan, being second only to lung cancer as to the number of deaths it causes. In spite of the recent development of diagnostic techniques, most gastric cancer patients are diagnosed at an advanced stage and have a very low five-year survival rate (less than 10%) [1]. This is partially due to a lack of specific and sensitive biomarkers for the diagnosis and monitoring of disease progress at an early stage, although some gastric tumor markers, including the carcinoembryonic antigen, have been used and are partly effective. As gastric carcinogenesis is a multistep process, comprehensive analysis is also required for individual cases, in which different molecular events occur in each carcinogenic process.
Recently, proteomic analysis was utilized to comprehensively examine protein expression in bodily fluids, tissues and cells [2-4]. This approach, as clinical proteomics, is very useful for identifying disease-associated proteins that show changes in expression and modification corresponding to a disease condition [5,6]. These disease-related proteins are expected to be biomarkers for diagnosis and putative targeted proteins for treatment [7-9]. On the other hand, comprehensive analyses of transcriptomes in tumor tissues from various cancer patients using DNA microarrays and gene chips have been performed in recent years [10]. However, a lack of correlation between changes in mRNAs and carcinogenesis has been demonstrated, and quantitative and qualitative changes of post-translationally modified proteins as final gene products are considered to be more informative than those of mRNAs in tumor tissues for studying the molecular events in carcinogenesis. Proteomic studies for the identification of tumor-associated proteins in gastric cancer are increasing, and proteome databases for gastric tissues [11] and cell lines [12] have been constructed. Most of them concern specific proteins or antigens that reflect the chemo- and thermo-resistant properties of stomach cancer [13-15], and that are associated with Helicobactor pylori [16,17]. In the present study, we performed comprehensive proteome analysis of tumor and nontumor tissues in Japanese patients with gastric carcinomas, and identified several proteins of which the expression levels are commonly altered in clinical cases. In particular, the expression of gastrokine-1 (GKN-1) was suggested to be under both transcriptional and translational control.
Results
Protein separation and identification
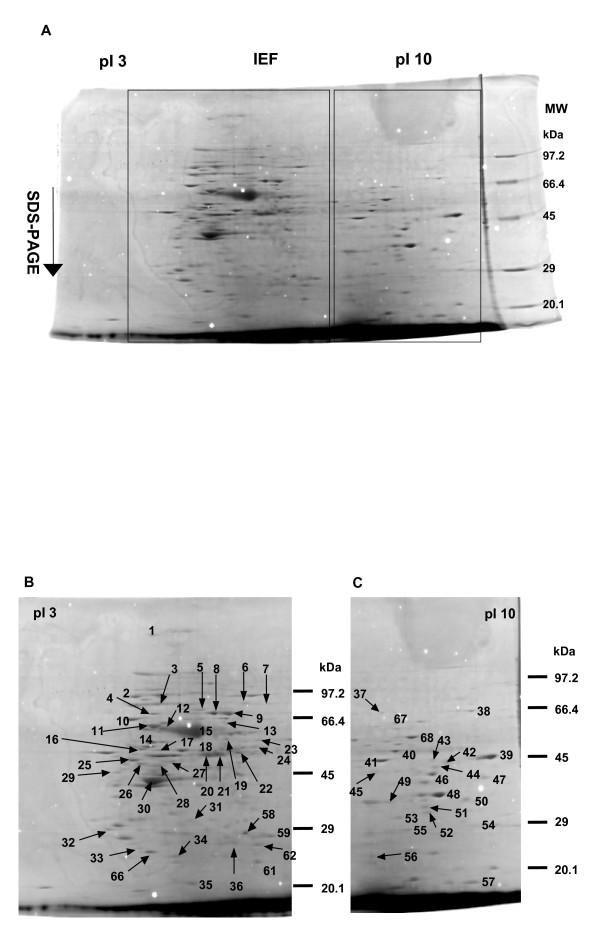
Figure 1A shows an image overview of a typical master gel for a gastric tumor tissue. Around 200 protein spots stained with Coomassie brilliant blue (CBB) R-250 were well separated in the gels. The numbered spots in Figure 1B and 1C were excised from a gel, treated with trypsin and then subjected to liquid chromatography-electronic spray ionization tandem mass spectrometer (LC-ESI-MS/MS) analysis. Seventy-two of them representing 69 different protein species were identified. Table 1 lists all of the proteins identified through peptide matching with the Mascot search algorithm. The accuracy in protein profiling was evaluated as the score value (above 37).
Figure 1.
(A) An overview of a master 2D gel image for tumor tissue derived from a patient with a gastric adenocarcinoma. (B) and (C) The numbered protein spots were identified by LC-ESI-MS/MS and protein matching, as shown as enlarged figures.
Table 1.
Protein profile detected in tumor tissue derived from a Japanese patient with a gastric adenocarcinoma.
| Spot No. | Accession number | Protein identification | Mass | S.C. (%) | pI | Score |
| 1 | 24308169 | axonemal heavy chain dynein type 3 | 473776 | 0 | 6.04 | 37 |
| 2 | 15010550 | heat shock protein gp96 precursor | 90309 | 7 | 4.73 | 62 |
| 3 | 72222 | heat shock protein 90-β | 83584 | 14 | 4.97 | 176 |
| 4 | 5729877 | heat shock 70 kDa protein 8 isoform 1 | 71082 | 19 | 5.37 | 283 |
| 5 | 38044288 | gelsolin isoform b | 80876 | 5 | 5.58 | 63 |
| 6 | 4826960 | glutaminyl-tRNA synthetase | 88655 | 1 | 6.71 | 44 |
| 7 | 4501867 | aconitase 2 | 86113 | 10 | 7.36 | 231 |
| 8 | 119717 | ezrin | 69470 | 7 | 5.94 | 75 |
| 9 | 4557871 | transferrin | 79280 | 9 | 6.81 | 97 |
| 10 | 16507237 | heat shock 70 kDa protein 5 | 72402 | 15 | 5.07 | 165 |
| 11 | 24234686 | heat shock protein 70 kDa protein 8 | 53598 | 16 | 5.62 | 160 |
| 12 | 4885431 | heat shock 70 kDa protein 1B | 70267 | 11 | 5.48 | 143 |
| 13 | 1082886 | tumor necrosis factor type 1 receptor-associated protein TRAP-1 | 75694 | 13 | 8.43 | 288 |
| 14 | 31542947 | chaperonin, mitochondrial matrix protein P1, P60 lymphocyte protein | 61187 | 13 | 5.7 | 299 |
| 15 | 28592 | serum albumin | 71316 | 23 | 6.05 | 292 |
| 16 | 340219 | vimentin | 53738 | 18 | 5.03 | 79 |
| 17 | 4503729 | similar to FK506-binding protein 4 | 49031 | 5 | 5.6 | 36 |
| 18 | 32709 | IFP53 | 53559 | 21 | 5.93 | 277 |
| 19 | 4502643 | chaperonin-containing TCP1, subunit6A (zeta 1) | 58444 | 6 | 6.23 | 79 |
| 20 | 7106439 | tubulin, β5 | 50095 | 14 | 4.78 | 125 |
| 21 | 37492 | α-tubulin | 50810 | 13 | 5.02 | 94 |
| 22 | 125604 | pyruvate kinase, M2 isozyme | 58447 | 5 | 7.95 | 234 |
| 23 | 4557014 | catalase | 59947 | 6 | 6.9 | 120 |
| 24 | 4503483 | eukaryotic translation elongation factor 2 | 96246 | 4 | 6.41 | 56 |
| 25 | 179279 | ATP synthase β subunit | 56861 | 5 | 5.39 | 59 |
| 26 | 7657041 | down-regulated in metastasis | 320846 | 2 | 7.07 | 40 |
| 27 | 4504169 | glutathione synthetase, GSH synthetase | 52523 | 5 | 5.67 | 99 |
| 28 | 12017952 | GE36 | 73327 | 2 | 5.2 | 37 |
| 29 | 34234 | laminin-binding protein | 31888 | 12 | 4.84 | 51 |
| 30 | 16359158 | actin, beta | 42078 | 14 | 5.29 | 171 |
| 31 | 6635125 | KIAA0284 protein | 161473 | 2 | 6.36 | 47 |
| 32 | 5803225 | 14-3-3 epsilon | 29326 | 8 | 4.63 | 49 |
| 33 | 4504707 | inositol polyphosphate-4-phosphatase, type II, 105 kD | 105749 | 1 | 5.87 | 37 |
| 34 | 35038601 | hypothetical protein DKFZp761A078 | 74864 | 2 | 7.27 | 47 |
| 35 | 12017952 | GE36 | 73327 | 1 | 5.2 | 38 |
| 36 | 4505877 | plectin 1 isoform 1 | 520111 | 1 | 5.57 | 44 |
| 37 | 38158018 | centrosomal protein 1, centriole associated protein, centriolin | 269874 | 1 | 5.44 | 39 |
| 38 | 30157438 | CTD-binding SR-like protein rA9 | 180240 | 1 | 9.15 | 41 |
| 39 | 4503471 | eukaryotic translation elongation factor 1 α1 | 50451 | 12 | 9.1 | 191 |
| 40 | 4757810 | ATP synthase | 59828 | 17 | 9.16 | 178 |
| 41 | 693933 | 2-phosphopyruvate-hydratase α-enolase | 47421 | 22 | 7.01 | 240 |
| 42 | 123576 | 47 kDa heat shock protein precursor | 46352 | 14 | 8.27 | 177 |
| 43 | 5032069 | splicing factor 3b, subunit 4 | 44414 | 3 | 8.54 | 58 |
| 44 | 4503471 | eukaryotic translation elongation factor 1 α1 | 50451 | 4 | 9.1 | 47 |
| 45 | 5921789 | citrate synthase, mitochondrial precursor | 51959 | 12 | 8.13 | 114 |
| 46 | 4505763 | phosphoglycerate kinase 1 | 44985 | 14 | 8.3 | 87 |
| 47 | 4504069 | aspartate aminotransferase 2 precursor | 47844 | 6 | 9.14 | 52 |
| 48 | 7669492 | glyceraldehyde-3-phosphate dehydrogenase | 36201 | 13 | 8.57 | 49 |
| 49 | 4757756 | annexin A2 | 38808 | 9 | 7.57 | 87 |
| 50 | 6648067 | malate dehydrogenase, mitochondrial precursor | 35965 | 7 | 8.92 | 60 |
| 51 | 5031857 | lactate dehydrogenase A | 36950 | 6 | 8.44 | 43 |
| 52 | 238427 | porin 31 HM | 30737 | 29 | 8.63 | 124 |
| 53 | 5174447 | guanine nucleotide-binding protein | 35511 | 8 | 7.6 | 65 |
| 54 | 34740329 | heterogeneous nuclear ribonucleoprotein A3 | 39799 | 7 | 9.1 | 43 |
| 55 | 4504983 | galectin-3 | 26229 | 12 | 8.58 | 42 |
| 56 | 4504447 | heterogeneous nuclear ribonucleoprotein A2/B1 isoform A2 | 36041 | 7 | 8.67 | 40 |
| 57 | 86901 | ATP-dependent DNA helicase RAP30/74 chain RAP30 | 26350 | 3 | 9.46 | 41 |
| 58 | 230445 | carbonic anhydrase I | 28903 | 12 | 6.44 | 67 |
| 59 | 455739 | carbonic anhydrase II | 29285 | 8 | 6.87 | 82 |
| 60 | 26892090 | beta-globin chain variant | 16101 | 40 | 7.86 | 121 |
| 61 | 34709 | manganese superoxide dismutase | 24891 | 10 | 8.35 | 111 |
| 62 | 4507645 | triosephosphate isomerase 1 | 26938 | 17 | 6.45 | 47 |
| 63 | 38488935 | foveolin precursor | 20546 | 16 | 5.65 | 95 |
| 64 | 478813 | nonhistone chromosomal protein HMG-1 | 25139 | 13 | 5.41 | 60 |
| 65 | 2204207 | glutathione S-transferase | 23595 | 34 | 5.43 | 73 |
| 66 | 178755 | proapolipoprotein | 28944 | 8 | 5.45 | 103 |
| 67 | 37267 | transketolase | 68435 | 4 | 7.9 | 59 |
| 68 | 189998 | M2-type pyruvate kinase | 58447 | 10 | 7.95 | 157 |
| 69 | 825605 | glutathione S-transferase | 25650 | 10 | 8.51 | 65 |
These proteins can be classified into several categories based on their functions, including cytoskeleton proteins, stress-related and chaperoning proteins, acute-phase proteins, glycolytic enzymes, enzymes involved in metabolism and cell proliferation, tumor suppressor proteins and stomach-specific proteins.
Common alterations of protein expression between tumor and nontumor tissues in gastric cancer patients
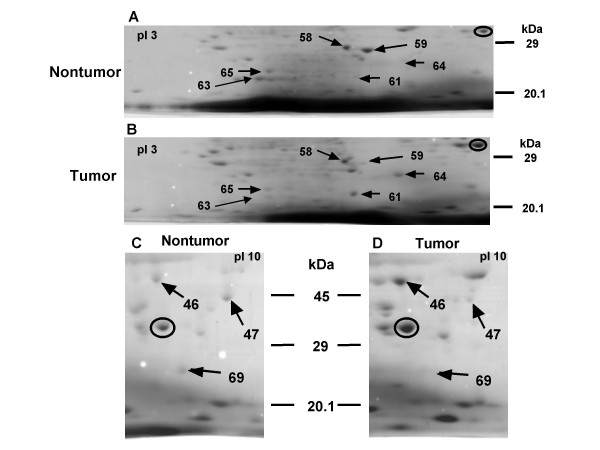
Diverse alterations in proteomes were detected between tumor and nontumor tissues from the same patients. As shown in Figure 2, the several common alterations were observed among in five Japanese gastric cancer patients (Cases A to E). Manganese superoxide dismutase (MnSOD), nonhistone chromosomal protein HMG-1 (HMG-1), phosphoglycerate kinase 1 (PGK-1), carbonic anhydrase I and II (CA I and II), foveolin precursor FOV (gastrokine-1), aspartate aminotransferase 2 precursor (AST), and glutathione S-transferase (GST) exhibited common changes in expression between tumor and nontumor tissues, including among the identified proteins. The protein expression of MnSOD and HMG-1 was demonstrated to be up-regulated in tumor tissues compared to in nontumor tissues. On the other hand, the CA I and II, FOV, AST and GST proteins were revealed to be down-regulated in tumor tissues. The fold changes in the expression of these proteins relative to that of GAPDH are summarized in Table 2. The most remarkable decrease was shown in the level of FOV in all cases. The degree of the decrease in GST protein expression was almost the same as that in FOV (Cases B, D, and E), although one was not observed in the other two cases. On the other hand, the increase in HMG-1 was marked in three cases (A, C, and D), although such a difference in expression was not observed in the other two cases. MnSOD exhibited a tendency to increase in the tumor tissues of three patients (Cases A, C, and D), but a relative decrease was also observed in Case B. As for PGK-1, a significant relationship between fold changes in protein expression and the pathological grading of tumors was hardly observed.
Figure 2.
Detailed alteration patterns of proteins. (A) Nontumor and (B) tumor proteins including CAI (No.58), CAII (No.59), MnSOD (No.61), FOV (No.63), HMG-1 (No.64), and GST (No.65). (C) Nontumor and (D) tumor proteins including PGK-1 (No.46), AST (No.47), and GST (No.69). GAPDH, circled, was used as a reference protein.
Table 2.
Fold changes in protein expression between nontumor and tumor tissues derived from five patients with gastric adenocarcinomas.
| Protein | Case A | Case B | Case C | Case D | Case E |
| CA I | -6.5 | -1.6 | -3.3 | -4.3 | -2.6 |
| CA II | -2.6 | -1.3 | -26 | -4.3 | -13 |
| FOV | -13 | -13 | -26 | * | -26 |
| MnSOD | +1.7 | -2.6 | +6.5 | +1.4 | +1.1 |
| HMG-1 | +13 | * | +3.7 | +13 | * |
| PGK1 | +1.6 | +1.1 | -1.2 | +2.6 | -1.4 |
| AST | -13 | -1.6 | -6.5 | -3.7 | -2.6 |
| GST | * | -26 | * | -13 | -13 |
CA I, carbonic anhydrase I.
CA II, carbonic anhydrase II.
FOV, foveolin precursor.
MnSOD, manganese superoxide dismutase.
HMG-1, nonhistone chromosomal protein.
PGK1, phosphoglycerate kinase 1.
AST, aspartate aminotransferase 2 precursor.
GST, glutathione S-transferase.
*, not determined.
RT-PCR analysis
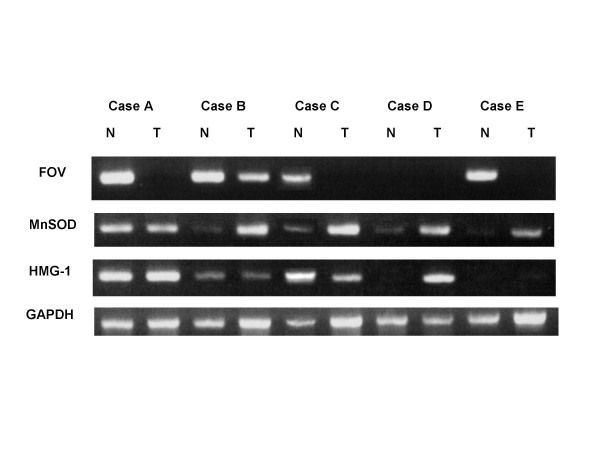
By RT-PCR, changes in mRNA expression in tumor and nontumor tissues derived from gastric cancer patients were analyzed for proteins that exhibited alterations in protein expression, including FOV, MnSOD and HMG-1. As shown in Figure 3, FOV mRNA was significantly decreased in tumor tissues in four at all patients (Cases A, B, C, and E), while it was not detected in either nontumor or tumor tissues from the other patient (Case D). On the other hand, MnSOD mRNA was markedly increased in four patients (Cases B, C, D, and E), although little difference in the mRNA level between nontumor and tumor tissues in Case A was detected. However, a relationship between HMG-1 mRNA expression and the pathological phenotypes of tumors was hardly observed.
Figure 3.
Comparison of mRNA expression of proteins possibly associated with carcinogenesis between nontumor and tumor tissues derived from five patients with gastric adenocarcinomas (cases A to E) by RT-PCR. N and T indicate nontumor and tumor tissues, respectively.
Discussion
In this study we performed proteomic analysis of tumor and nontumor tissues derived from Japanese gastric adenocarcinoma patients. Sixty-nine different proteins in the tumor tissue from a gastric cancer case were identified by 2D-EP and LC-ESI-MS/MS, which included stress proteins, Hsp70, Hsp90 and chaperonine-containing TCP1 (CCT), for self-protection; glycolytic enzymes, triose phosphate isomerase 1, α-enolase and PGK-1, for the growing energy requirement; cytoskeletal proteins, ezrin, gelsolin isoform b and vimentin; proteins involved in cell differentiation and proliferation, galectin-3 and transferrin; and proteins exhibiting putative tumor suppressor activity, FOV. Many of these proteins have been reported to be associated with the tumorigenesis of gastric adenocarcinomas involving multiple steps and factors [18,19].
We also demonstrated that the expression levels of several proteins in tumor tissues were commonly altered in five Japanese patients with gastric adenocarcinomas, compared to those in nontumor tissues. A common increase in protein expression in tumor tissues occurred for MnSOD, HMG-1 and PGK-1, whereas a common decrease in tumor tissues occurred for FOV, GST and AST.
Superoxide (O2-), a free radical, is essential for the anti-microbial action of granulocytes and monocytes. Superoxide dismutase (SOD) rapidly removes the excess amount of superoxide produced in the stress and biological reactions in vivo through catalysis of the conversion of superoxide with H+ to H2O2 and O2. MnSOD, one of the SODs, is located in mitochondria and contributes to the protection of mitochondrial DNA from damage. It has been reported that enhanced expression of MnSOD in progressive gastric cancer should be related to the 5-year survival rate after surgery [20] and sensitivity to chemotherapy [21]. In this study, protein expression of MnSOD was up-regulated in 4 of 5 gastric cancer patients, suggesting a self-protecting response. On the other hand, the up-regulation of MnSOD in tumor tissues has been considered to interfere with effective chemotherapy based on radical production, which may cause a decrease in sensitivity to anti-cancer drugs and determine the severity of the cancer.
HMG-1 regulates the transcription of various genes and the structural stabilization of chromosomes as a DNA-binding protein. HMG-1 has been reported to be associated with carcinogenesis and metastasis in colorectal and breast cancer [22].
Transcriptional up-regulation of HMG-1 in gastric cancer has also been demonstrated [23]. Expression of HMG-1 in tumor tissues was suggested to be related to resistance to cisplatin [24]. We demonstrated here an increase in this protein in three cases.
GST is a drug-metabolizing enzyme that catalyzes the conjugation of reduced glutathione to drugs and metabolites, and also contributes to the detoxification of carcinogens. Therefore, a decrease in the activity may be a risk factor for carcinogenesis. Infection by Helicobacter pylori has been reported to cause a decrease in GST expression [25], suggesting that a decrease in GST activity might be a cause of gastric carcinogenesis. In this study, a decrease in GST protein expression was observed in three cases. A decrease in GST protein expression may be utilized as a biomarker for the diagnosis of gastric tumors.
AST is an aminotransferase that acts on amino acids and α-keto acid, that is distributed widely in human organs. AST is released into the blood stream due to enhanced permeability and destruction of tissues. A elevated concentration of AST protein has been reported in the blood of patients with hepatitis, malignant tumors including hepatomas, and leuchemia. Gene expression of GST has also been demonstrated to be up-regulated in colorectal tumor tissues [26]. However, a decrease in AST protein expression was commonly observed in all tumor tissues derived from the present five patients with gastric adenocarcinomas. Further study should be performed to elucidate whether a decrease in the AST protein is specifically observed in gastric tumor tissues or not.
Elevated expression of PGK-1, which is one of the enzymes in the glycolytic pathway and which catalyzes the dephosphorylation of 1,3-bisphosphoglycerate to produce ATP, has been observed in many malignant tumor tissues that are dependent on ATP as a major energy source. Solid tumors are thought to need the overproduction of ATP to maintain the enhanced proliferation. The up-regulation of glycolytic enzymes, including PGK-1 in lung cancer [27] and M2-type pyruvate kinase in colorectal cancer [28], has been reported and suggested to be useful for cancer screening. However, in the present study, a significant relation between PGK-1 protein expression and the phenotypes of gastric adenocarcinomas was hardly detected. Therefore, PGK-1 was considered to be not suitable for the diagnosis of gastric cancer.
Carbonic anhydrases (CAs) are zinc-containing enzymes that are distributed widely in various organs and that comprise a large family including CA-I to CA-IX. They catalyze the hydration of CO2 for intermediate metabolism, and maintain the pH and ion equilibrium in the body. So far a direct relationship has been demonstrated between malignant transformation and protein expression for CAs I through VII [29]. Two earlier studies revealed that the expression of both CA-I and CA-II was significantly reduced in colorectal tumors compared to in normal colorectal epithelia or mucosae [30]. Another report presented results showing that reduced expression of CA-I and CA-II was correlated with the biological aggressiveness of colorectal cancer and synchronous distant metastasis [31]. As suggested previously [32], gastric and colorectal carcinomas may share a similar mechanism of cell proliferation and mucosal malignancy, and may become a biomarker for these carcinomas, because common decreases in the protein expression of CA-I and CA-II were also observed in this study.
FOV, which is identical to a stomach-specific 18-kDa antrum mucosa protein! (AMP-18) [33,34], GKN1 [35], and trefoil factor interactions(z) (TFIZ) [36], was demonstrated to be dramatically down-regulated or absent in the gastric adenocarcinoma patients in this study. The human AMP-18 and human carcinoantigenic CA11 gene product, which encodes an amino acid sequence that differs from that of AMP-18 in only a single residue [37,38], function as secreted growth factors partly responsible for maintaining a normal functional gastric epithelium [33]. Both AMP-18 and the CA11 gene product have been reported to be intensively expressed in normal stomach tissue but not in most gastric cancers [33,37,38]. Immunohistochemical studies demonstrated that AMP-18 appears to be present in mucosal epithelial cells of the normal human gastric antrum and duodenum [33]. Recently, TFIZ was demonstrated to act as a growth regulator of gastric epithelial cells through the formation of a heterodimer with trefoil factor 1 (TFF1) [36]. Our present data further confirmed the high expression of FOV in nontumor gastric tissues and the significant suppression of the protein in gastric adnocarcinomas, indicating that FOV play roles in the maintenance of normal differentiation of epithelial cells and tumor suppression but not in tumor tissues. Furthermore, we demonstrated that transcription of FOV mRNA was also commonly down-regulated in gastric cancer patients, indicating that a marked decrease in FOV protein was caused by suppression of FOV gene expression. Furthermore, in one patient the protein was not detected in nontumor tissues, suggesting that the expression level of FOV protein in individuals may determine the gastric adenocarcinoma phenotype. Accordingly, the expression level of FOV as a biomarker may be informative for the assessment of gastric cancer.
Conclusion
Protein profiling of tumor and nontumor tissues derived from Japanese patients with gastric adenocarcinomas was performed using 2D-EP and LC-ESI-MS/MS. The identified proteins included molecular chaperones, energy-producing enzymes, cytoskeletal proteins, and so on. Common protein alterations were detected in the gastric cancer patients. Protein expression of MnSOD and HMG-1 was up-regulated while that of GST, AST and FOV was down-regulated in gastric tumor tissues. A correlation between the alteration of these proteins and their transcriptional expression in gastric cancer was hardly observed in this study, except for in the case of FOV. Both the protein and gene expression of FOV, a stomach-specific secretory growth factor for normal gastric epithelial cells, was markedly down-regulated in tumor tissues derived from Japanese patients with gastric adenocarcinomas. Monitoring of the expression levels of this stomach-specific protein in clinical samples may provide useful information for the diagnosis of gastric cancer as a specific biomarker and for better understanding of gastric carcinogenesis.
Methods
Materials
DNase I, RNase A, 2-mercaptoethanol (2-ME), glass beads (212–300 μm), Nonidet P-40, acrylamide, N,N,N',N'-tetramethylenediamine (TEMED), sodium dodecylsulfate (SDS), iodoacetamide and dithiothreitol (DTT) were purchased from Sigma (St. Louis, MO). Agarose for isoelectronic focusing (IEF), and Pharmalyte pI 3–10, 4–6.5 and 8–10.5 were from Amersham Bioscience (Piscataway, NJ). Trypsin (sequencing grade) was from Roche (Manheim, Germany). Phenylmethylsulfonyl fluoride (PMSF), thiourea, sorbitol, sodium pyrophosphate, ammonium persulfate, D,L-aspartic acid, trichloroacetic acid, sulfosalicylic acid dihydrate, acetic acid, acetonitrile, formic acid and trifluoroacetic acid (TFA) were from Wako Pure Chemicals (Osaka, Japan). Urea was from Katayama Chemicals (Osaka, Japan). Pepstatin A and leupeptin were from the Peptide Institute (Osaka, Japan). NH4HCO3 and N,N'-methylenebisacrylamide were from Nacalai Tesque (Kyoto, Japan). CBB R-250 was from ICN Biomedicals Inc. (Aurora, OH). Molecular mass standards were from APRO Science, Inc. (Tokushima, Japan). TRIZOL reagent was from Life Technologies (Frederick, MD). Oligo(dT)12–18 primer, deoxynucleotides (dNTPs), and RNaseOUT were from Invitrogen (Carlsbad, CA). M-MLV reverse transcriptase and Taq DNA polymerase were from Promega (Madison, WI).
Tissues and sample preparation
Primary gastric adenocarcinomas and adjacent nontumor mucosae were collected on gastrectomy and provided by the Dept. of Surgery, Graduate School of Medicine, The University of Tokushima, Tokushima, Japan. The research was carried out in accordance with the Declaration of Helsinki of the World Medical Association, and was approved by the ethical committee of the University of Tokushima. Informed consent was also given by all of the patients who provided the clinical samples. Tissues were frozen in a dry ice-methanol bath as soon as possible after dissection and stored in a deep freezer (-80°C) before use. For mRNA analysis, tissues were first immersed in RNAlater (Takara, Tokyo, Japan) before freezing. Detailed clinicopathological data including the tumor stage (according to the AJCC system), site and differentiation, and histological data on the tissue samples are listed in Table 3. None of these cases were classified in the scirrhous type category, and tumor tissue was clearly distinguishted from non-tumor one in each case. For two-dimensional gel electrophoresis (2D-EP), protein extraction from tissues was carried out by the following procedure. Frozen blocks (20–30 mg wet weight) were homogenized with a plastic pestle (Toyobo, Tokyo, Japan) in the presence of glass beads in 10 vol/wet weight of dialysis buffer comprising 5 M urea, 1 M thiourea, 10 mM NaPPi, 1.67 μL/mL 2-ME, 0.005% DNase I, 0.05 mg/ml RNase A, 20 μM leupeptin, 1 mM EDTA, 2 mM PMSF and 20 μM pepstatinA, and then centrifuged at 50,000 rpm for 30 min at 4°C (Beckman -Coulter, Fullerton, CA). The resultant supernatant was used as the tissue extract. Protein concentrations were determined with a Bradford protein assay kit (Bio-Rad, Hercules, CA) using bovine gamma-globulin as a standard.
Table 3.
Clinical features of the patients with gastric adenocarcinomas.
| Case | Age | Sex | Locationa | Gradeb | Stage | |
| A | 63 | Male | UM | G3 | IIIA | T2N2M0H0 |
| B | 68 | Female | L | G2 | II | T2N2M0 |
| C | 76 | Male | ML | G2 | IV | T4N3M0 |
| D | 78 | Female | L | G2 | III | T2N0M0H0 |
| E | 77 | Male | ML | G2 | II | T2N1M0 |
aU: upper, M: middle, L: lower
bG2: moderately differentiated, G3: poorly differentiated.
Two-dimesional gel electrophoresis
2D-EP was carried out as described previously [39]. The first-dimensional isoelectric focusing was performed in an 1% (w/v) agarose gel (ϕ 2.6 × 180 mm) with a pH 3–10 gradient at 700 V for 18 hr at 4°C, and the second-dimensional SDS gel electrophoresis was performed with a 5–15% (w/v) acrylamide gradient (Mr range, 6–200 kDa) in a standard slab gel (20 × 13 cm) at 15 mA for 3 h, and then at 70 mA for 2 h at room temperature. Gels were stained with CBB R-250.
Protein samples (500 μg) extracted from the tumor center and surrounding histologically normal mucosa were subjected to 2D-EP and run in pairs side by side.
Some of the stained spots were excised from the 2D-gel, in-gel digested with trypsin and then subjected to LC-ESI-MS/MS analysis as described previously [40]. The peptide mixture was separated with a reversed phase nanoLC system (Famous, Swichos II, Ultimate, LC Packings, Sunnyvale, CA). The eluted peptides were sprayed directly into an ESI mass spectrometer (Esquire3000 Plus, Bruker-Daltonics, Fremont, CA).
A large volume of MS/MS data was acquired with DataAnalysis 3.1 software (Bruker-Daltonics), converted to text files listing the mass values of the parent ions, and intensities and masses of fragment ions, and then processed with the MASCOT algorithm (Matrix Science Ltd, London, U.K.) to assign peptides in the NCBI non-redundant sequence database using a taxonomic restriction, 'human'. The database search was performed with the parameters described by Yoshimura et al. [40].
Image analysis and MS peptide sequencing
Image acquisition and analysis were performed with Molecular Imager FXProPlus (Bio-Rad) and ImageMaster software (Bio-Rad). Comparisons were made between gel images of tumor and matched nontumor samples pair by pair. Normalized volume differences were statistically calculated for all five cases. The content of glyceraldehyde 3-phosphodehydrogenase (GAPDH) protein was used as a reference to evaluate the fold alteration of protein expression between tumor and matched nontumor tissues as the levels of GAPDH mRNA and protein were not changed between tissues derived from patients. Consistently and significantly different spots were selected for analysis by LC-ESI-MS/MS. Protein spots!were cut out from gels in small pieces, and subjected to in-gel tryptic digestion overnight [40]. Peptide mass spectra were recorded, and parameters for spectra acquisition were used as stated previously [40]. Accuracy in database protein matching using Mascot Search [41] was judged as a score over 37, which was obtained in most of the analyses.
RNA isolation and RT-PCR analysis
Tumor and matched nontumor samples (approx. 50 mg wet weight) were minced, and then homogenized manually in 1 ml of TRIZOL reagent (Invitrogen) on ice. RNA was isolated without DNase I treatment according to the manufacturer's protocol. Briefly, 0.2 ml of CHCl3 was added to the homogenate, followed by centrifugation at 20,600 × g for 15 min. An equal volume of 2-propanol was added to the resultant supernatant to precipitate RNA. After centrifugation, the pellet was rinsed with 75% ethanol/diethylpyridylchloride (DEPC)-treated water, followed by drying. The pellet was dissolved in an appropriate volume of DEPC-treated water as the total RNA fraction. For reverse transcription (RT), 2 μg of RNA from each sample was transcribed at 37°C for 1 h in the presence of 200U of Molony leukemia virus reverse transcriptase (Promega), oligo(dT)12–18 primer, 0.5 mM dNTPs and 50U of RNaseOUT. The PCRs for carbonic anhydrase-I and II, glutatione-S-transferase, FOV, and GAPDH were performed within a linear range of amplification using the selected primer set and conditions, and expected size of products, as summarized in Table 4. The PCR products were analyzed by 1.5% agarose gel electrophoresis and stained with ethidium bromide.
Table 4.
Primer sets used for RT-PCR analysis.
| Primers | Primer sequences | |
| MnSOD | sense | 5' - acgcggcctacgtgaacaacctgaa - 3' |
| antisense | 5' - aaccccaacctgagccttggacacc - 3' | |
| HMG-1 | sense | 5' - cgggaggagcataagaagaagcacc - 3' |
| antisense | 5' - caatggacaggccaggatgttctcc - 3' | |
| GAPDH | sense | 5' - gtcatccatgacaactttgg - 3' |
| antisense | 5' - tgctgtagccaaattcgttg - 3' |
MnSOD, manganese superoxide dismutase.
HMG-1, nonhistone chromosomal protein.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Abbreviations
2D-EP, Two-dimensional gel electrophoresis; 2-ME, 2-Mercaptoethanol; AMP-18, Antrum mucosa protein-18; AST, Aspartate aminotransferase 2 precursor; CA I and II, Carbonic anhydrase I and II; CAs, Carbonic anhydrases; CBB, Coomassie brilliant blue; CCT, Chaperonine-containing TCP1; DEPC, Diethylpyridylchloride; dNTPs, Deoxynucleotides; DTT, Dithiothreitol; FOV, Foveolin precursor (gastrokine-1); GAPDH, Glyceraldehyde 3-phosphodehydrogenase; GKN1, Gastrokine-1; GST, Glutathione S-transferase; HMG-1, Nonhistone chromosomal protein HMG-1; IEF, Isoelectronic focusing; LC-ESI-MS/MS, Liquid chromatography-electronic spray ionization-tandem mass spectrometer; MnSOD, Manganese superoxide dismutase; PGK-1, Phosphoglycerate kinase 1; PMSF, Phenylmethylsulfonyl fluoride; RT, Reverse transcription; SDS, Sodium dodecylsulfate, SOD, Superoxide dismutase; TEMED, N,N,N',N'-Tetramethylenediamine; TFA, Trifluoroacetic acid; TFF1, Trefoil factor 1; TFIZ, Trefoil factor interactions(z).
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
TY mainly carried out the molecular studies, participated in the proteomic determination and RT-PCR analysis. YK carried out the proteomic studies, partly participated in the proteomic determination. YY supported the proteomic studies by TY. YT partly participated in the RT-PCR for gastric cancer-related protein. NT parly participated in the proteomic determination. HO carried out the operation of gastric cancer patiens, participated in providing the cancer tissues. AU carried out the operation of gastric cancer patients, participated in providing clinical information. TY supported the proteomic studies and advised the methods. KI conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by grants from Taiho Pharmaceutical Co., Ltd. and the Suzuken Memorial Foundation.
Contributor Information
Takahiro Yoshihara, Email: yoshihara-y@sakai-chem.co.jp.
Yoshito Kadota, Email: c400441003@stud.tokushima-u.ac.jp.
Yoshiyuki Yoshimura, Email: yosimura@ph.tokushima-u.ac.jp.
Yutaka Tatano, Email: tatanoy@med.shimane-u.ac.jp.
Naohiro Takeuchi, Email: kitoh@ph.tokushima-u.ac.jp.
Hiroshi Okitsu, Email: umemoto@clin.med.tokushima-u.ac.jp.
Atsushi Umemoto, Email: umemoto@clin.med.tokushima-u.ac.jp.
Takashi Yamauchi, Email: yamauchi@ph.tokushima-u.ac.jp.
Kohji Itoh, Email: kitoh@ph.tokushima-u.ac.jp.
References
- Peddanna N, Holt S, Verma RS. Genetics of gastric cancer. Anticancer Res. 1995;15:2055–2064. [PubMed] [Google Scholar]
- Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, 2nd, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics. 2005;4:409–418. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Snyder M. Proteomics. Annu Rev Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- McGregor E, Dunn MJ. Proteomics of heart disease. Hum Mol Genet. 2003;12:R135–144. doi: 10.1093/hmg/ddg278. [DOI] [PubMed] [Google Scholar]
- Alaiya A, Al-Mohanna M, Linder S. Clinical cancer proteomics: promises and pitfalls. J Proteome Res. 2005;4:1213–1222. doi: 10.1021/pr050149f. [DOI] [PubMed] [Google Scholar]
- He QY, Chiu JF. Proteomics in biomarker discovery and drug development. J Cell Biochem. 2003;89:868–886. doi: 10.1002/jcb.10576. [DOI] [PubMed] [Google Scholar]
- Calvo KR, Liotta LA, Petricoin EF. Clinical proteomics: from biomarker discovery and cell signaling profiles to individualized personal therapy. Biosci Rep. 2005;25:107–125. doi: 10.1007/s10540-005-2851-3. [DOI] [PubMed] [Google Scholar]
- Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, Arteaga CL, Caprioli RM. Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res. 2004;64:9093–9100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- Baak JP, Path FR, Hermsen MA, Meijer G, Schmidt J, Janssen EA. Genomics and proteomics in cancer. Eur J Cancer. 2003;39:1199–1215. doi: 10.1016/S0959-8049(03)00265-X. [DOI] [PubMed] [Google Scholar]
- Ha GH, Lee SU, Kang DG, Ha NY, Kim SH, Kim J, Bae JM, Kim JW, Lee CW. Proteome analysis of human stomach tissue: separation of soluble proteins by two-dimensional polyacrylamide gel electrophoresis and identification by mass spectrometry. Electrophoresis. 2002;23:2513–2524. doi: 10.1002/1522-2683(200208)23:15<2513::AID-ELPS2513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Seike M, Kondo T, Fujii K, Yamada T, Gemma A, Kudoh S, Hirohashi S. Proteomic signature of human cancer cells. Proteomics. 2004;4:2776–2788. doi: 10.1002/pmic.200300795. [DOI] [PubMed] [Google Scholar]
- Sinha P, Hutter G, Kottgen E, Dietel M, Schadendorf D, Lage H. Increased expression of annexin I and thioredoxin detected by two-dimensional gel electrophoresis of drug resistant human stomach cancer cells. J Biochem Biophys Methods. 1998;37:105–116. doi: 10.1016/S0165-022X(98)00020-7. [DOI] [PubMed] [Google Scholar]
- Sinha P, Poland J, Schnolzer M, Celis JE, Lage H. Characterization of the differential protein expression associated with thermoresistance in human gastric carcinoma cell lines. Electrophoresis. 2001;22:2990–3000. doi: 10.1002/1522-2683(200108)22:14<2990::AID-ELPS2990>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18:505–509. doi: 10.3346/jkms.2003.18.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson CL, Larsson T, Gustafsson E, Karlsson KA, Davidsson P. Identification of protein vaccine candidates from Helicobacter pylori using a preparative two-dimensional electrophoretic procedure and mass spectrometry. Anal Chem. 2000;72:2148–2153. doi: 10.1021/ac9912754. [DOI] [PubMed] [Google Scholar]
- Haas G, Karaali G, Ebermayer K, Metzer WG, Lamer S, Zimny-Arndt U, Diescher S, Goebel UB, Vogt K, Roznowski AB, Wiedenmann BJ, Meyer TF, Aebischer T, Jungblut PR. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics. 2002;2:313–324. doi: 10.1002/1615-9861(200203)2:3<313::AID-PROT313>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Durany N, Joseph J, Campo E, Molina R, Carreras J. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase and enolase activity and isoenzymes in lung, colon and liver carcinomas. Br J Cancer. 1997;75:969–977. doi: 10.1038/bjc.1997.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QY, Cheung YH, Leung SY, Yuen ST, Chu KM, Chiu JF. Diverse proteomic alterations in gastric adenocarcinoma. Proteomics. 2004;10:3276–3287. doi: 10.1002/pmic.200300916. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Chae SW, Hur GC, Cho SJ, Kim MK, Choi J, Nam SY, Kim WH, Yang HK, Lee BL. Manganese superoxide dismutase expression correlates with a poor prognosis in gastric cancer. Pathobiology. 2003;70:353–360. doi: 10.1159/000071276. [DOI] [PubMed] [Google Scholar]
- Hur GC, Cho SJ, Kim CH, Kim MK, Bae SI, Nam SY, Park JW, Kim WH, Lee BL. Manganese superoxide dismutase expression correlates with chemosensitivity in human gastric cancer cell lines. Clin Cancer Res. 2003;9:5768–5775. [PubMed] [Google Scholar]
- Evans A, Lennard TW, Davies BR. High-mobility group protein 1(Y): metastasis-associated or metastasis-inducing? J Surg Oncol. 2004;88:86–99. doi: 10.1002/jso.20136. [DOI] [PubMed] [Google Scholar]
- Xiang YY, Wang DY, Tanaka M, Suzuki M, Kiyokawa E, Igarashi H, Naito Y, Shen Q, Sugimura H. Expression of high-mobility group-1 mRNA in human gastrointestinal adenocarcinoma and corresponding non-cancerous mucosa. Int J Cancer. 1997;74:1–6. doi: 10.1002/(SICI)1097-0215(19970220)74:1<1::AID-IJC1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Nagatani G, Nomoto M, Takano H, Ise T, Kato K, Imamura T, Izumi H, Makishima K, Kohno K. Transcriptional activation of the human HMG1 gene in cisplatin-resistant human cancer cells. Cancer Res. 2001;61:1592–1597. [PubMed] [Google Scholar]
- Baek HY, Lim JW, Kim H, Kim JM, Kim JS, Jung HC, Kim KH. Oxidative-stress-related proteome changes in Helicobacter pylori-infected human gastric mucosa. Biochem J. 2004;379:291–299. doi: 10.1042/BJ20031208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Kato M, Yoshikawa T, Chen H, Brown EJ, Masuho Y, Omata M, Seki N. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem Biophys Res Commun. 2001;289:876–881. doi: 10.1006/bbrc.2001.6047. [DOI] [PubMed] [Google Scholar]
- Chen G, Charib TG, Wang H, Huang CC, Kuick R, Thomas DG, Shedden KA, Misek DE, Taylor JM, Giordano TJ, Kardia SL, Iannettoni MD, Yee J, Hogg PJ, Orringer MB, Hanash SM, Beer DG. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci USA. 2003;100:13537–13542. doi: 10.1073/pnas.2233850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt PD, Mazurek S, Toepler M, Schlierbach P, Bretzel RG, Eigenbrodt E, Kloer HU. Faecal tumor M2 pyruvate kinase: a new, sensitive screening tool for colorectal cancer. Br J Cancer. 2004;91:980–984. doi: 10.1038/sj.bjc.6602033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogradi A. The role of carbonic anhydrases in tumors. Am J Pathol. 1998;153:1–4. doi: 10.1016/S0002-9440(10)65537-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Staniunas RJ, Barnard GF, Jessup JM, Steele GD, Jr, Chen LB. The significance of carbonic anhydrase expression in human colorectal cancer. Gastroenterlogy. 1993;105:820–826. doi: 10.1016/0016-5085(93)90900-w. [DOI] [PubMed] [Google Scholar]
- Bekku S, Mochizuki H, Yamamoto T, Ueno H, Takayama E, Tadakuma T. Expression of carbonic anhydrase I or II and correlation to clinical aspects of colorectal cancer. Hepatogastroenterology. 2000;47:998–1001. [PubMed] [Google Scholar]
- Leppilampi M, Saarnio J, Karttunen TJ, Kivela J, Pastorekova S, Pastorek J, Waheed A, Sly WS, Parkkila S. Carbonic anhydrase isozymes IX and XII in gastric tumor. World J Gastroenterol. 2003;9:1398–1403. doi: 10.3748/wjg.v9.i7.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TE, Powell CT, Wang Z, Bhattacharyya S, Walsh-Reitz MM, Agarwal K, Toback FG. A novel mitogenic protein that is highly expressed in cells of the gastric antrum mucosa. Am J Physiol Gastrointest Liver Physiol. 2003;285:G332–343. doi: 10.1152/ajpgi.00453.2002. [DOI] [PubMed] [Google Scholar]
- Toback FG, Walsh-Reitz MM, Musch MW, Chang EB, Del Valle J, Ren H, Huang E, Martin TE. Peptide fragments of AMP-18, a novel secreted gastric antrum mucosal protein, are mitogenic and motogenic. Am J Physiol Gastrointest Liver Physiol. 2003;285:G344–353. doi: 10.1152/ajpgi.00455.2002. [DOI] [PubMed] [Google Scholar]
- Oien KA, McGregor F, Butler S, Ferrier RK, Downie I, Bryce S, Burns S, Keith WN. Gastrokine 1 is abundantly and specifically expressed in superficial gastric epithelium, down-regulated in gastric carcinoma, and shows high evolutionary conservation. J Pathol. 2004;203:789–797. doi: 10.1002/path.1583. [DOI] [PubMed] [Google Scholar]
- Westley BR, Griffin SM, May FE. Interaction between TFF1, a gastric tumor suppressor trefoil protein, and TFIZ1, a brichos domain-containing protein with homology to SP-C. Biochemistry. 2005;44:7967–7975. doi: 10.1021/bi047287n. [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y, Mukai H, Hino F, Asada K, Kato I. Isolation of two novel genes, down-regulated in gastric cancer. Jpn J Cancer Res. 2000;91:459–463. doi: 10.1111/j.1349-7006.2000.tb00967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Nakamori S, Tsujie M, Okami J, Yamamoto H, Nagano H, Dono K, Umeshita K, Sakon M, Furukawa H, Hiratsuka M, Kasugai T, Ishiguro S. Human stomach-specific gene, CA11, is down-regulated in gastric cancer. Int J Oncol. 2001;19:701–707. doi: 10.3892/ijo.19.4.701. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Shinkawa T, Taoka M, Kobayashi K, Isobe T, Yamauchi T. Identification of protein substrates of Ca2+/calmodulin-dependent protein kinase II in the postsynaptic density by protein sequencing and mass spectrometry1. Biochem Biophys Res Commun. 2002;290:948–954. doi: 10.1006/bbrc.2001.6320. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88:759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- ProteinProspector http://prospector.ucsf.edu/