Abstract
Background: Surveillance colonoscopy is widely recommended in patients with longstanding and extensive ulcerative colitis (UC) in order to detect colorectal neoplasia at an early stage. However, it still remains questionable whether surveillance colonoscopy effectively enables early detection of UC associated neoplasia. There is a great need for sensitive markers to identify individuals at increased risk of neoplasia. The oestrogen receptor (OR) gene shows age related methylation in the colorectal epithelium and is methylated frequently in sporadic colorectal neoplasia, suggesting that OR methylation may predispose to colorectal neoplasia.
Aim: To clarify whether analysis of methylation of the OR gene in non-neoplastic epithelium can contribute to prediction of increased neoplasia risk in UC patients.
Materials and methods: A total of 165 non-neoplastic colorectal epithelia from 30 patients with longstanding and extensive UC, including 13 UC patients with neoplasia and 17 patients without, were evaluated. Methylation specific polymerase chain reaction was performed to determine the methylation status of the OR gene.
Results: Methylation of the OR gene was detected in 54 of 70 (77.1%) non-neoplastic colorectal epithelia in UC with neoplasia but in only 23 of 95 (24.2%) without neoplasia. Methylation of the OR gene was significantly more frequent in non-neoplastic epithelium from UC with neoplasia than in chronic colitic epithelium from UC without neoplasia. Furthermore, in UC with neoplasia, the OR gene was extensively methylated in non-neoplastic epithelia throughout the colorectum compared with those in UC without neoplasia.
Conclusion: These results suggest that analysis of OR gene methylation may have potential as a useful marker for identifying individuals at increased risk of neoplasia among those with longstanding and extensive UC.
Keywords: ulcerative colitis, neoplasia, oestrogen receptor, methylation
Patients with ulcerative colitis (UC) show an increased incidence of colorectal neoplasia, and UC associated colorectal neoplasia represents a major cause of increased mortality in such patients.1,2 In order to improve the prognosis of patients with UC associated neoplasia, diagnosis at an early or precancerous stage is crucial. Predisposition to colorectal neoplasia in UC is generally considered to depend on two risk factors—namely, the presence of longstanding disease and extensive colitis.3–5 Therefore, surveillance colonoscopy with multiple step biopsy has been widely recommended for patients with longstanding and extensive UC.6–8 However, studies examining the efficacy of surveillance in UC have produced conflicting results, and have suggested that surveillance achieves detection of early stage neoplasia in only a minority of patients and cannot guarantee cancer detection at a curable stage.9–14 In order to improve the efficacy of surveillance, there is a great need for sensitive and specific markers to identify individuals at increased risk of neoplasia among those with longstanding and extensive UC.
In several neoplasias, aberrant methylation of promoter region CpG islands, as an epigenetic modification of DNA, is associated with transcriptional inactivation of tumour suppressor genes and plays a crucial role in the development and progression of neoplasia. In normal colorectal epithelium, some genes are methylated with aging, and this alteration is known as age related methylation.15 Issa and colleagues16 reported that methylation of the oestrogen receptor (OR) CpG island increased with age in non-neoplastic colorectal epithelium, and that the same methylation occurred in most sporadic colorectal neoplasias. They concluded that methylation of the OR gene in aging colorectal epithelium could represent one of the earliest events predisposing to sporadic colorectal tumorigenesis.
In this study, to clarify whether analysis of OR gene methylation in non-neoplastic epithelium can contribute to the prediction of increased risk for UC associated neoplasia, we investigated the incidence of OR methylation in non-neoplastic epithelium from longstanding and extensive UC patients with and without colorectal neoplasia.
MATERIALS AND METHODS
Patients and samples
We studied non-neoplastic colorectal epithelia from 30 patients with longstanding (>7 years) and extensive (proximal to the splenic flexture) UC. Thirteen UC patients had neoplasia and 17 patients had no neoplasia. In all patients, whenever possible, multiple samples were taken from six regions (rectum, sigmoid colon, descending colon, transverse colon, ascending colon, and caecum) of the colorectum. Non-neoplastic samples of UC with neoplasia were retrieved from colectomy specimens, which were resected at Dokkyo University School of Medicine Hospital and our collaborative institutions between 1999 and 2003. Samples of UC without neoplasia were obtained from biopsy specimens in surveillance colonoscopy between 2002 and 2003. The ethics committee of Dokkyo University School of Medicine approved the study, and all patients gave informed consent for inclusion.
Histological evaluation
Histologically, we classified the inflammatory activity of each specimen into the following categories: no inflammation, mild to moderate inflammation, or severe inflammation. We confirmed that all of the samples were negative for neoplasia in accordance with the Vienna classification of gastrointestinal epithelial neoplasia.17 This new classification is practical, and has been recommended for resolving the large discrepancies between Western and Japanese pathologists in the diagnosis of gastrointestinal epithelial neoplasias and to better aid understanding of research data in the field of gastroenterology. We defined both dysplasia and carcinoma as neoplasia in accordance with this classification.
DNA extraction
Formalin fixed paraffin embedded samples were cut serially at a thickness of 5 or 10 µm. Based on histological findings, the tissue of each region was microdissected and DNA was then extracted using a DNA isolator PS kit (Wako Pure Chemical, Osaka, Japan) according to the supplied protocol.
Methylation analysis
DNA methylation status of the OR gene was determined by methylation specific polymerase chain reaction (MSP).18 Briefly, 500 ng of genomic DNA were modified by sodium bisulfite (Nacarai Tesque, Inc., Kyoto, Japan) using low melting agarose (SeaPlaque agarose; BioWhittaker Molecular Applications, Rockland, Maine, USA), as described previously.19 DNA from the unmethylated breast cancer cell line MCF-7 and the methylated colon cancer cell line HT29 were used as negative and positive controls, respectively.16 Agarose beads were incubated for 18 hours at 50°C to ensure complete sodium bisulfite modification. A quarter of the beads containing modified DNA were amplified in a total volume of 25 µl of polymerase chain reaction (PCR) mixture. The reaction mixture contained 2.5 µl of 10×PCR Gold Buffer, 2 µl of 25 mM MgCl2, 2.5 µl of dNTP mixture, 0.75 units of AmpliTaq Gold (Applied Biosystems, Tokyo, Japan), 10 pmol of forward primer, and 10 pmol of reverse primer. The primer sequences of OR for the methylated reaction were 5′- GTG TAT TTG GAT AGT AGT AAG TTC GTC -3′ (forward primer) and 5′- CGT AAA AAA AAC CGA TCT AAC CG -3′ (reverse primer), which amplify a 118 bp product, and the primer sequences for the unmethylated reaction were 5′- GGT GTA TTT GGA TAG TAG TAA GTT TGT -3′ (forward primer) and 5′- CCA TAA AAA AAA CCA ATC TAA CCA -3′ (reverse primer), which amplify a 120 bp product. MSP was carried out using the following amplification profile: five minutes at 94°C once; 30 seconds at 94°C, 30 seconds at 55°C, 30 seconds at 72°C for 40 cycles; then four minutes at 72°C. PCR products were electrophoresed on 2% agarose gels and visualised under UV illumination using ethidium bromide staining.
Statistical analysis
Age and disease duration of UC patients with and without neoplasia are expressed as mean (SD), and differences between groups were analysed using Welch’s t test; differences at p<0.05 were considered significant. The χ2 test or Fisher’s exact test was used to compare the inflammatory activity of colitis and the incidence of methylation in UC patients with and without neoplasia, and to determine the correlation between the inflammatory activity of colitis and methylation status; differences at p<0.05 were considered significant.
RESULTS
Clinical and pathological features
The clinical and pathological features of the patients with and without UC neoplasia are summarised in table 1 ▶. In a total of 13 patients with UC neoplasia, six patients were men and seven were women, and mean age was 54.5 (SD 11.8) years (range 41–79). All patients except one (case 8) had longstanding UC; known duration of the disease was mean 16.5 (SD 4.7) years (range 6–23). Twelve patients had pancolitis and the remaining patient had left sided colitis. Eleven of the patients underwent resections because of complications related to cancer, and two of the patients underwent resection because of dysplasia progression. There were 17 patients without neoplasia: 12 were men and five were women, and mean age was 47.6 (SD 8.0) years (range 41–66). The known duration of the disease was mean 18.5 (SD 8.4) years (range 9–36) and 16 patients had pancolitis and the remaining patient left sided colitis. There were no significant differences in mean age or known disease duration between patients with neoplasia and those without.
Table 1.
Clinical and pathological features of patients with and without ulcerative colitis (UC) neoplasia
| Patient No | Age (y) | Sex (M/F) | Duration of disease (y) | Extent of disease† | Location of neoplasia | Histology of neoplasia‡ | Dukes’ stage |
| UC with neoplasia | |||||||
| 1 | 58 | F | 22 | Total | Rectum | Muc | C |
| 2 | 41 | F | 15 | Left side | Sigmoid | Poor | C |
| 3 | 60 | F | 19 | Total | Sigmoid | Dysplasia | |
| 4 | 57 | F | 12 | Total | Rectum | Well | A |
| 5 | 51 | M | 17 | Total | Rectum | Well | C |
| 6 | 79 | F | 23 | Total | Rectum | Moderate | C |
| 7 | 44 | M | 18 | Total | Rectum | Dysplasia | |
| 8 | 48 | F | 6 | Total | Rectum | Well | A |
| 9 | 74 | F | 14 | Total | Transverse | Well | A |
| 10 | 44 | M | 19 | Total | Sigmoid | Well | C |
| 11 | 45 | M | 14 | Total | Sigmoid | Moderate | A |
| 12 | 50 | M | 14 | Total | Rectum | Moderate | B |
| 13 | 45 | M | 22 | Total | Rectum | Well | A |
| UC without neoplasia | |||||||
| 14 | 43 | M | 12 | Total | |||
| 15 | 44 | M | 25 | Total | |||
| 16 | 44 | M | 10 | Total | |||
| 17 | 41 | M | 12 | Total | |||
| 18 | 50 | F | 24 | Left side | |||
| 19 | 49 | M | 13 | Total | |||
| 20 | 57 | M | 9 | Total | |||
| 21 | 51 | M | 15 | Total | |||
| 22 | 66 | F | 14 | Total | |||
| 23 | 42 | M | 16 | Total | |||
| 24 | 53 | M | 36 | Total | |||
| 25 | 59 | F | 31 | Total | |||
| 26 | 51 | M | 33 | Total | |||
| 27 | 42 | F | 17 | Total | |||
| 28 | 42 | M | 19 | Total | |||
| 29 | 42 | M | 19 | Total | |||
| 30 | 33 | F | 10 | Total | |||
†Total, total colitis; Left side, left sided colitis; Well, well differentiated adenocarcinoma; moderate, moderately differentiated adenocarcinoma; poor, poorly differentiated adenocarcinoma; muc, mucinous carcinoma.
Incidences of OR gene methylation
A total of 165 non-neoplastic epithelia obtained from 30 patients were available for the MSP study. At some regions, because of lack of an available tissue specimen, no methylation analysis was performed. OR methylation was present in 54 of 70 (77.1%) non-neoplastic epithelia from patients with neoplasia (fig 1A ▶) compared with 23 of 95 (24.2%) non-neoplastic epithelia from patients without neoplasia (fig 1B ▶). The incidence of OR methylation in non-neoplastic epithelium was higher in patients with neoplasia than in those without (p<0.001).
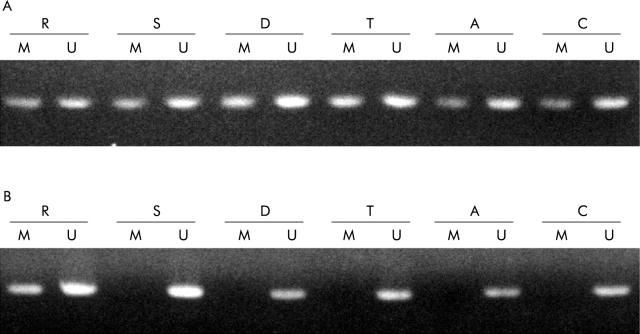
Figure 1.
Methylation analysis of the oestrogen receptor (OR) gene in each region of the colorectum. Methylation specific polymerase chain reaction products in lanes marked U show the presence of unmethylated templates whereas products in lanes marked M show the presence of methylated templates. R, rectum; S, sigmoid colon; D, descending colon; T, transverse colon; A, ascending colon; C, caecum. (A) Representative samples of non-neoplastic epithelia from a patient with neoplasia. The OR gene was methylated in all regions of the non-neoplastic colorectal epithelium. (B) Representative samples of non-neoplastic epithelia from a patient without neoplasia. The OR gene was methylated in only rectal epithelium whereas no methylation of the OR gene was detected in other regions.
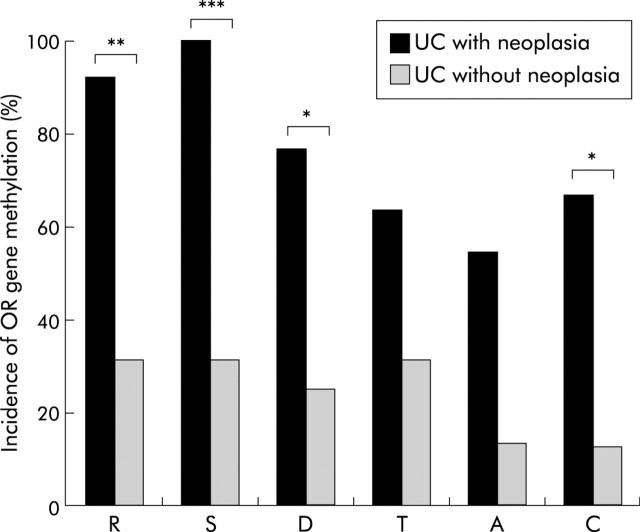
OR methylation status in each region of the colorectum is shown schematically in fig 2 ▶. OR methylation in UC patients with neoplasia occurred extensively in the non-neoplastic epithelium throughout the colorectum, and OR gene methylated in both non-neoplastic epithelia from UC patients with carcinoma and dysplasia. Eleven of 13 patients with neoplasia had at least three regions with methylation and three of 13 had all analysed regions with methylation, whereas 15 of 17 patients without neoplasia had less than two regions with methylation and five of 17 had no region with methylation. Figure 3 ▶ demonstrates the incidence of OR gene methylation in non-neoplastic epithelium from UC patients with and without neoplasia in each region of the colorectum. In the rectum, sigmoid colon, descending colon, and caecum, the incidences of OR methylation in UC with neoplasia were significant higher than in those without neoplasia (rectum 92.3% v 31.2%, p<0.01; sigmoid colon 100% v 31.2%, p<0.001; descending colon 76.9% v 25.0%, p<0.05; transverse colon 63.6% v 31.3%, p = 0.20; ascending colon 54.5% v 13.3%, p = 0.069; caecum 66.7% v 12.5%, p<0.05).
Figure 2.
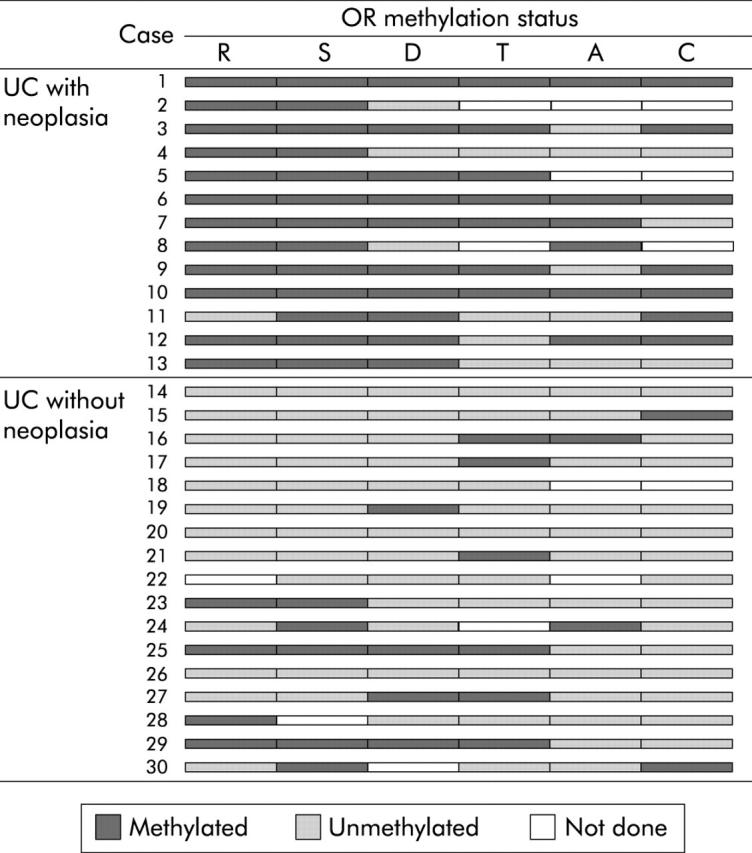
Oestrogen receptor (OR) methylation status in each region of the colorectum is shown schematically. Methylated and unmethylated regions are indicated (blank regions, not done). R, rectum; S, sigmoid colon; D, descending colon; T, transverse colon; A, ascending colon; C, caecum.
Figure 3.
Incidence of oestrogen receptor (OR) gene methylation in non-neoplastic epithelium from ulcerative colitis (UC) patients with and without neoplasia in each region of the colorectum. In the rectum, sigmoid colon, descending colon, and caecum, the incidence of OR methylation in UC with neoplasia was significant higher than in those without neoplasia. *p<0.05, ** p<0.01, ***p<0.001. R, rectum; S, sigmoid colon; D, descending colon; T, transverse colon; A, ascending colon; C, caecum.
Relationship between OR gene methylation and inflammatory activity of colitis
With regard to inflammatory activity in the 70 samples from UC patients with neoplasia, 22 (31.4%) had no inflammation, 26 (37.1%) had mild inflammation, 15 (21.4%) had moderate inflammation, and seven (10.0%) had severe inflammation. Among 95 samples from UC patients without neoplasia, 20 (21.1%) had no inflammation, 51 (53.7%) had mild inflammation, 16 (16.8%) had moderate inflammation, and eight (8.4%) had severe inflammation. There were no significant differences in inflammatory activity between patients with and without neoplasia.
The relationship between OR gene methylation status and the inflammatory activity of colitis is shown in table 2 ▶. The incidence of OR gene methylation was 52.4% in non-neoplastic epithelium without inflammation, 40.3% in that with mild inflammation, 48.4% in that with moderate inflammation, and 60.0% in that with severe inflammation. There was no relationship between OR gene methylation status and the inflammatory activity of colitis.
Table 2.
Relationship between oestrogen receptor (OR) gene methylation status and the inflammatory activity of colitis
| Inflammatory activity of colitis | |||||
| None (n = 42) | Mild (n = 77) | Moderate (n = 31) | Severe (n = 15) | ||
| OR methylation status | |||||
| Methylated | 22 (52.4%) | 31 (40.3%) | 15 (48.4%) | 9 (60.0%) | |
| Unmethylated | 20 (47.6%) | 46 (59.7%) | 16 (51.6%) | 6 (40.0%) | |
DISCUSSION
In this study, we have shown that the incidence of OR gene methylation in non-neoplastic epithelium was higher in patients with neoplasia than in those without. In UC with neoplasia, OR gene methylation was detected not only in regions in which neoplasia was located but also in other regions widely throughout the colorectum. Furthermore, these epigenetic alterations occurred in both non-neoplastic epithelia from UC patients with carcinoma and dysplasia. These results suggest that an increase in the incidence of age related methylation—in other words, progression of aging in the colorectal epithelium—is preceding or is a relatively early event in UC associated tumorigenesis. Analysis of OR gene methylation in non-neoplastic colorectal epithelium may be a useful adjunct for identifying individuals at increased risk of neoplasia among those with longstanding and extensive UC, and contribute to more effective cancer surveillance.
In order to detect neoplasia at a surgically curative and preferably preinvasive stage, periodic colonoscopy combined with extensive biopsy sampling throughout the colorectum is recommended. According to several studies that have discussed the efficacy of surveillance, however, an appreciable number of cancers were detected at an advanced stage despite surveillance colonoscopy, and many of these cases had less than ideal outcomes. Thus it still remains questionable whether surveillance colonoscopy with multistep biopsy effectively enables early detection of UC associated neoplasia.
The unsatisfactory efficacy of current surveillance colonoscopy for early detection of UC associated neoplasia can be attributed to difficulties in endoscopic and histological diagnoses of UC associated neoplasia at an early stage. Endoscopically, early stage UC associated neoplasias show various macroscopic changes. These changes are not clear, and are sometimes missed in areas of chronically inflamed epithelium.20–22 Therefore, detection of UC associated neoplasias at the precancerous and early stages is difficult by endoscopy, and physicians have no choice but to depend on histological evaluation of multiple step biopsies obtained by surveillance colonoscopy. Histologically, UC associated neoplasia often involves one part of the crypt, a few crypts in the epithelium, or is found throughout the inflamed epithelium.6 Therefore, it is not unusual for pathologists to experience difficulty in discriminating between neoplastic lesions and inflammatory regenerative epithelium using biopsy specimens stained with haematoxylin and eosin. Furthermore, different pathologists may use different diagnostic criteria for determining neoplasia.6,23
In order to overcome such difficulties, adjunctive diagnostic modalities such as chromoendoscopy for identifying neoplasia in non-neoplastic inflamed epithelium and analysis of p53 alteration for distinguishing neoplastic lesion from regenerative epithelium have been reported.24–28 However, it would be impractical to perform these adjunctive modalities for surveillance of all UC patients with longstanding and extensive colitis because of their labour intensive nature and expense. If refinement of high and low risk subgroups of UC patients with longstanding and extensive colitis were possible, it would enable physicians to conduct more intensive surveillance with these modalities (chromoendoscopy and analysis of p53 alteration) for patients at higher risk of developing colorectal neoplasia.
Recently, numerous studies have revealed higher frequencies of molecular alterations to non-neoplastic epithelium in UC patients with neoplasia than in non-neoplastic epithelium in UC patients without neoplasia, suggesting that these molecular alterations may be used as new markers for identifying individuals with UC at increased risk of neoplasia.27–33 In the present study, we observed age related methylation of the OR gene in non-neoplastic epithelium throughout the entire colorectum of patients with longstanding and extensive UC. Age related methylation has been demonstrated for several genes, including OR, insulin-like growth factor II, MYOD1, N33, and E-cadherin in the colorectal epithelium, and patients with a high level of methylation in their colorectal epithelium may be at higher risk of developing sporadic colorectal neoplasia.16,34,35 In this study, we selected and analysed the OR gene because OR gene methylation is predominantly a phenomenon of the epithelium. Ahuja and colleagues34 examined differences in the methylation status of the OR, MYOD, and N33 genes between epithelial and stromal components of colorectal mucosa, and clarified that the OR gene showed four times higher methylation in the epithelial components than in the stromal components. In contrast, the N33 and MYOD genes showed equivalent levels of methylation in both components. Therefore, we considered that OR gene methylation status would not be susceptible to the influence of contamination by stromal cells.
In UC associated neoplasia, Issa and colleagues36 first reported that the OR, MYOD, and CSPG2 genes, and p16 gene exon 1, which behave as age related methylation in colorectal epithelium, were intensively methylated in neoplastic epithelium from high grade dysplasia/cancer patients with UC. Furthermore, they showed that these genes were highly methylated in non-neoplastic epithelium from UC patients with high grade dysplasia/cancer compared with non-neoplastic epithelium from UC patients without dysplasia. They suggested that age related methylation could be used as a molecular marker for identifying individuals with ulcerative colitis at increased risk of developing neoplasia but they did not mention methylation status by region in the colorectum. In our present study, we analysed OR gene methylation in multiple samples taken from six regions (rectum, sigmoid colon, descending colon, transverse colon, ascending colon, and caecum) throughout the colorectum, and showed that OR methylation in UC patients with neoplasia occurred more frequently and extensively in the non-neoplastic epithelium throughout the colorectum than in those without neoplasia. From these findings it is expected that analysis of a single biopsy sample (for example, a rectal biopsy) may have the possibility of helping identify UC patients at particularly high risk of developing neoplasia in contrast with the large number of biopsy samples needed for surveillance colonoscopy at present.
Although the mechanism of age related methylation is not well known, there may be several contributory factors, such as exogenous carcinogens, reactive oxygen species, and host genetic differences.15,37–39 Chronic inflammation in UC would be associated with an elevated level of exogenous carcinogens and reactive oxygen species, and UC colonocytes would be subjected to a high level of epigenetic damage. In the present study, the inflammatory activity of colitis did not have any relationship with the incidence of OR gene methylation, suggesting that OR gene methylation does not simply reflect transient inflammation and that cumulative and repetitive inflammation may lead to the occurrence of methylation.
In our study, although there were no significant differences in age and disease duration between UC patients with and without neoplasia, suggesting that all UC patients would theoretically have been exposed to an equal degree of damage by exogenous carcinogens and reactive oxygen species, why was there a wide difference in the incidence of OR gene methylation between non-neoplastic epithelium from UC patients with and without neoplasia? Possible reasons include differences in cumulative inflammation among patients in the two groups despite the similarity of disease duration, differences between actual disease duration and those listed in the hospital records, and individual differences in susceptibility to gene methylation after exposure to exogenous carcinogens and reactive oxygen stress. Clarification of the reasons for this difference awaits further study.
Our results show that OR methylation is worthy of study to explore its potential as a biomarker in the surveillance of UC patients. We demonstrated the occurrence of gene methylation; quantitative information on the status of methylation was lacking. Therefore, we are currently conducting a quantitative analysis of age related methylation using a laser capture microdissection technique and the combined bisulfite restriction analysis (COBRA) method. Further longitudinal studies are needed to address the predictive power and clinical utility of the methylation analysis. It is anticipated that analysis of OR gene methylation using rectal biopsy specimens may make it possible to identify UC patients at high risk of future neoplastic progression, and that in future it could be realistic, on the basis of OR methylation status, to adjust a patient’s surveillance interval and to select UC patients who should undergo intensive surveillance that includes adjunctive modalities such as chromoendoscopy and analysis of p53 alteration.
Acknowledgments
This study was supported by a grant in aid (No 16790390) for Scientific Research from the Japan Society for the Promotion of Science (JSPS). The authors greatly thank Dr S Kameoka and Dr M Itabashi (Department of Surgery II, Tokyo Women’s Medical college), Dr B Iizuka (The Institute of Gastroenterology, Tokyo Women’s Medical college), Dr H Mitooka (Division of Gastroenterology, Kobe Kaisei Hospital), Dr T Tanaka (Division of Gastroenterology, Shizuoka City Shizuoka Hospital) and Dr N Kitajima (Division of Gastroenterology, Kasai City Hospital) for kindly supplying the tissue materials. The authors also thank Dr H Kawamata (Department of Oral and Maxillofacial Surgery, Dokkyo University School of Medicine) for critical reading and valuable advice on this manuscript, and Ms C Sato-Matsuyama, A Shimizu, T Ohtsuki, M Katayama, and S Miyahara (Department of Surgical and Molecular Pathology, Dokkyo University School of Medicine) for technical assistance.
Abbreviations
UC, ulcerative colitis
OR, oestrogen receptor
MSP, methylation specific polymerase chain reaction
PCR, polymerase chain reaction
Published online first 3 May 2005
Conflict of interest: None declared.
REFERENCES
- 1.Ekbom A, Helmick C, Zack M, et al. Ulcerative colitis and colorectal cancer. A population-based study. N Engl J Med 1990;323:1228–33. [DOI] [PubMed] [Google Scholar]
- 2.Langholz E, Munkholm P, Davidsen M, et al. Colorectal cancer risk and mortality in patients with ulcerative colitis. Gastroenterology 1992;103:1444–51. [DOI] [PubMed] [Google Scholar]
- 3.Greenstein AJ, Sachar DB, Smith H, et al. Cancer in universal and left-sided ulcerative colitis: factors determining risk. Gastroenterology 1979;77:290–4. [PubMed] [Google Scholar]
- 4.Gyde SN, Prior P, Allan RN, et al. Colorectal cancer in ulcerative colitis: a cohort study of primary referrals from three centres. Gut 1988;29:206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilat T, Fireman Z, Grossman A, et al. Colorectal cancer in patients with ulcerative colitis. A population study in central Israel. Gastroenterology 1988;94:870–7. [DOI] [PubMed] [Google Scholar]
- 6.Riddell RH, Goldman H, Ransohoff DF, et al. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol 1983;14:931–68. [DOI] [PubMed] [Google Scholar]
- 7.Kornbluth A, Sachar DB. Ulcerative colitis practice guidelines in adults. American College of Gastroenterology, Practice Parameters Committee. Am J Gastroenterol 1997;92:204–11. [PubMed] [Google Scholar]
- 8.Farrell RJ, Peppercorn MA. Ulcerative colitis. Lancet 2002;359:331–40. [DOI] [PubMed] [Google Scholar]
- 9.Lynch DA, Lobo AJ, Sobala GM, et al. Failure of colonoscopic surveillance in ulcerative colitis. Gut 1993;34:1075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axon AT. Cancer surveillance in ulcerative colitis—a time for reappraisal. Gut 1994;35:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi PM, Nugent FW, Schoetz DJ Jr, et al. Colonoscopic surveillance reduces mortality from colorectal cancer in ulcerative colitis. Gastroenterology 1993;105:418–24. [DOI] [PubMed] [Google Scholar]
- 12.Connell WR, Lennard-Jones JE, et al. Factors affecting the outcome of endoscopic surveillance for cancer in ulcerative colitis. Gastroenterology 1994;107:934–44. [DOI] [PubMed] [Google Scholar]
- 13.Connell WR, Talbot IC, Harpaz N, et al. Clinicopathological characteristics of colorectal carcinoma complicating ulcerative colitis. Gut 1994;35:1419–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii S, Fujimori T, Chiba T, et al. Efficacy of surveillance and molecular markers for detection of ulcerative colitis-associated colorectal neoplasia. J Gastroenterol 2003;38:1117–25. [DOI] [PubMed] [Google Scholar]
- 15.Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol 2000;249:101–18. [DOI] [PubMed] [Google Scholar]
- 16.Issa JP, Ottaviano YL, Celano P, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 1994;7:536–40. [DOI] [PubMed] [Google Scholar]
- 17.Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000;47:251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res 1996;24:5064–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook MG, Goligher JC. Carcinoma and epithelial dysplasia complicating ulcerative colitis. Gastroenterology 1975;68:1127–36. [PubMed] [Google Scholar]
- 21.Butt JH, Konishi F, Morson BC, et al. Macroscopic lesions in dysplasia and carcinoma complicating ulcerative colitis. Dig Dis Sci 1983;28:18–26. [DOI] [PubMed] [Google Scholar]
- 22.Schneider A, Stolte M. Clinical and pathomorphological findings in patients with colorectal carcinoma complicating ulcerative colitis. Z Gastroenterol 1993;31:192–7. [PubMed] [Google Scholar]
- 23.Dixon MF, Brown LJ, Gilmour HM, et al. Observer variation in the assessment of dysplasia in ulcerative colitis. Histopathology 1988;13:385–97. [DOI] [PubMed] [Google Scholar]
- 24.Fujii S, Fujimori T, Chiba T. Usefulness of analysis of p53 alteration and observation of surface microstructure for diagnosis of ulcerative colitis-associated colorectal neoplasia. J Exp Clin Cancer Res 2003;22:107–15. [PubMed] [Google Scholar]
- 25.Kiesslich R, Fritsch J, Holtmann M, et al. Methylene blue-aided chromoendoscopy for the detection of intraepithelial neoplasia and colon cancer in ulcerative colitis. Gastroenterology 2003;124:880–8. [DOI] [PubMed] [Google Scholar]
- 26.Rutter MD, Saunders BP, Schofield G, et al. Pancolonic indigo carmine dye spraying for the detection of dysplasia in ulcerative colitis. Gut 2004;53:256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lashner BA, Shapiro BD, Husain A, et al. Evaluation of the usefulness of testing for p53 mutations in colorectal cancer surveillance for ulcerative colitis. Am J Gastroenterol 1999;94:456–62. [DOI] [PubMed] [Google Scholar]
- 28.Holzmann K, Weis-Klemm M, Klump B, et al. Comparison of flow cytometry and histology with mutational screening for p53 and Ki-ras mutations in surveillance of patients with long-standing ulcerative colitis. Scand J Gastroenterol 2001;36:1320–6. [DOI] [PubMed] [Google Scholar]
- 29.Rabinovitch PS, Dziadon S, Brentnall TA, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res 1999;59:5148–53. [PubMed] [Google Scholar]
- 30.Andersen SN, Lovig T, Clausen OP, et al. Villous, hypermucinous mucosa in long standing ulcerative colitis shows high frequency of K-ras mutations. Gut 1999;45:686–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh CJ, Klump B, Holzmann K, et al. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res 1998;58:3942–5. [PubMed] [Google Scholar]
- 32.Sato F, Harpaz N, Shibata D, et al. Hypermethylation of the p14(ARF) gene in ulcerative colitis-associated colorectal carcinogenesis. Cancer Res 2002;62:1148–51. [PubMed] [Google Scholar]
- 33.Brentnall TA, Crispin DA, Bronner MP, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res 1996;56:1237–40. [PubMed] [Google Scholar]
- 34.Ahuja N, Li Q, Mohan AL, et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 1998;58:5489–94. [PubMed] [Google Scholar]
- 35.Waki T, Tamura G, Tsuchiya T, Sato K, et al. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol 2002;161:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Issa JP, Ahuja N, Toyota M, et al. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res 2001;61:3573–7. [PubMed] [Google Scholar]
- 37.Lee YW, Klein CB, Kargacin B, et al. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol Cell Biol 1995;15:2547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obst B, Wagner S, Sewing KF, et al. Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis 2000;21:1111–15. [PubMed] [Google Scholar]
- 39.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst 2002;94:755–61. [DOI] [PubMed] [Google Scholar]