Figure 2.
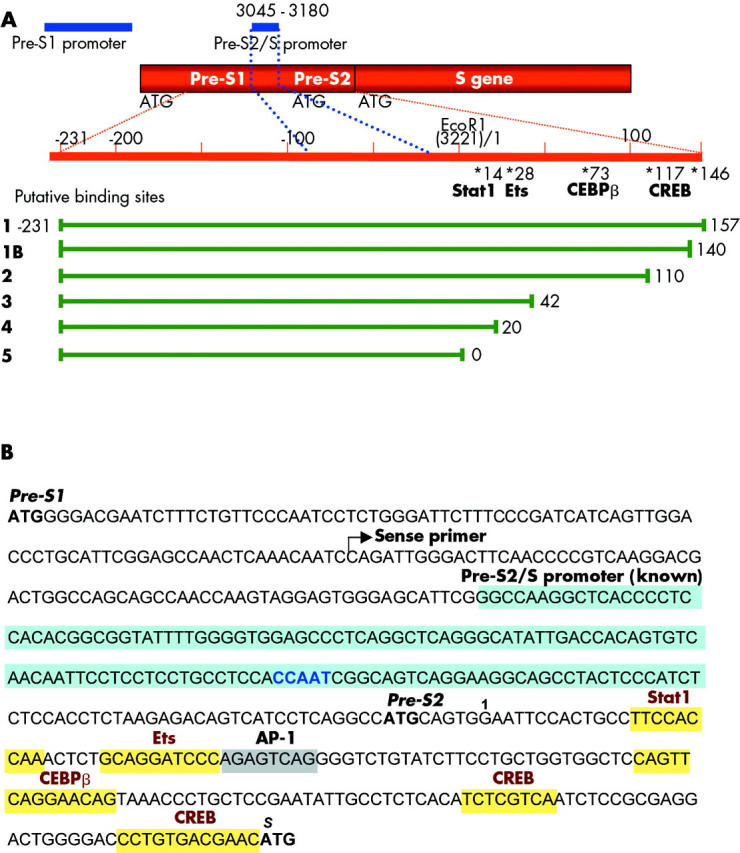
Organisation of the hepatitis B virus (HBV)-S gene, its promoters, and putative transcription factor binding sites. (A) Schematic diagram of the organisation of the HBV-S gene and its promoters (EcoR1 = 1/3221). The known region of the pre-S2/S promoter (blue) is located within the pre-S1 region from nt 3045–3180. Upstream of the S-ATG, the location of putative transcription factor binding sites for Stat1, Ets, CEBPβ, and cAMP response element binding protein (CREB) are indicated, as revealed by computer based analysis. A putative activator protein 1 (AP-1) site partially overlaps with an Ets motif. Promoter fragments of different lengths (constructs 1–5) were cloned in front of the luciferase reporter gene into the pGL2-plasmid. (B) Sequence of the pre-S1 and pre-S2 region of HBV (genotype A, subtype adw2). ATGs, the start of the applied sense primer (for luciferase constructs) and the known pre-S2/S promoter (blue) with the CCAAT binding motif are highlighted. In addition, upstream binding sites for HNF3 and NF1 plus a downstream binding site for Sp1 have been previously identified to contribute to the known pre-S2/S promoter (not shown). In the pre-S2 region, several transcription factor binding sites were predicted, and are emphasised by yellow boxes.
