Abstract
Theories of evolution that state natural selection acts on individuals have been modified to include multiple levels of selection. Here we demonstrate in chimeric protochordates that primitive germ cell (pgc) and somatic cell (psc) lineages have traits that also make them likely units of natural selection. Specifically, by using microsatellites to determine the genetic identity of various somatic and gametic tissues within vascularly fused Botryllus schlosseri chimeras, we show that genetically distinct pgc and psc can compete for access to developing gonads and somatic organs, and that this competition is hierarchical, reproducible, and heritable. Given that a single, highly polymorphic locus (Fu/HC) controls whether two contacting colonies fuse or reject, our findings also support a leading hypothesis for why the highly polymorphic histocompatibility loci common to many metazoa may have arisen or been maintained: to limit supercompetitor lineages to histocompatible kin.
Among the most basic questions in evolutionary biology is at what levels of organization are genomes competing in the processes of natural selection (1, 2). In the modern synthesis of Fisher, Wright, and Haldane, it was assumed that selection only acted at the level of the individual (3–5). More recently, it has been proposed that selection can also act at the level of groups of genes, cells, groups of individuals, and even species, and that there can be interplay and conflicts between these different levels of selection (6–10). Buss (7) has argued that the early focus on the individual as the unit of selection was to a considerable extent a legacy of the Weismann–Nussbaum doctrine that asserts that heritable variation can only be passed on through a select set of cells known as the germ line, which are set aside during early ontogeny (11, 12). The doctrine has been confirmed for a number of solitary organisms, including roundworms, flies, and vertebrates (13, 14). In vertebrates, primordial germ cells (pgc) arise in early embryogenesis, migrate to extraembryonic sites, and then return during early fetal development to colonize the genital ridges and eventually form the gonads (14).
While somatic variants are not incorporated into the germ line of solitary organisms, the genome producing the soma and the genome that is transmitted by the germ line are not necessarily identical in a large class of colonial organisms (6, 7, 15, 16). Many invertebrate species exist as colonies of asexually derived individuals, some with interconnecting transport and/or vascular systems. In these colonial species, it could be said that although the whole colony is functioning as a somatic unit, both the colony and the component individuals could be considered animals (17). Because growth in colonial invertebrates occurs through several rounds of asexual reproduction, there must be a pool of pgc and/or their precursors that can delay differentiation until the start of sexual reproduction (6, 7). As a consequence, germ line variants could theoretically arise anytime after the early development of the organism (when presumably the pgc or their precursors first appear) and throughout the rounds of asexual reproduction; these pgc variants could then compete for germ-line niches in the developing gonads. In colonial invertebrates, germ-line variants can arise through mutation, but another avenue for the introduction of genomically distinct germ lines is through vascular fusion with a non-genetically identical colony, wherein pgc can migrate from one colony to the next (18–20). It is this latter method that is considered in this paper. In colonial invertebrates that have both sexual and asexual phases of reproduction, there is also a separation in time between the formation of the first primitive somatic cells (psc) that participate in embryonic organogenesis and those that participate in organogenesis during asexual development. Such a separation could allow genetically distinct psc lineages to compete for somatic niches and distinct pgc to compete for germ-line niches (18, 19). As Buss (6, 7) and others (21–25) have pointed out, these competitions among genetically distinct cell lineages within a single somatic entity can lead to substantial fitness costs or benefits as well as the emergence of histocompatibility barriers to the indiscriminate intercolony passage of pgc and psc.
The colonial urochordate Botryllus schlosseri produces zygotes that mature into tadpole larvae with a chordate body plan. On release from the maternal colony, these tadpoles swim to a nearby subtidal surface and undergo a metamorphosis to a sessile juvenile individual (oozooid). Metamorphosis precipitates the loss of the chordate body plan of notochord, neural tube, and segmented musculature and results in the production of a temporarily solitary organism with its own neural, gastrointestinal, cardiovascular, hematopoietic, respiratory, glandular, surface epithelial, and gonadal components. After settlement, the oozooid commences a series of asexual budding cycles, beginning with a small focus of undifferentiated cells that eventually gives rise to a multiindividual colony composed of morphologically and genetically identical blastozooids. Within such a colony, the blastozooids are connected to one another by both a common tunic and a network of anastomosed extracorporeal blood vessels (26). Eventually, ovaries and testes form within blastozooids, and sexual reproduction commences (26).
When two genetically distinct oozooids or colonies come into contact, they either anastomose their peripheral blood vessels to form vascular and blood chimeras (26) or develop an inflammatory rejection that permanently separates the colonies (27). In B. schlosseri, the processes of fusion and rejection are controlled by a single, highly polymorphic locus, now called Fu/HC (27–29). Such a genetically based self–nonself-recognition system in colonial species has been suggested by Burnet, Buss, and others to be an adaptation for limiting fusion and, therefore, the risk of cell lineage competition to kin (6, 7, 22–25, 30, 31).
Using microsatellite markers, we demonstrated previously that in FuHC, compatible fused but genetically distinct colonies (i) the blood, the buds, and the germ cells can be chimeric and (ii) in some cases, the cells of tissues of one individual can be derived from cells from the other individual (18, 19). These previous studies did not test whether somatic and germ-line competitions were reproducible and heritable. In organisms that have both sexual and asexual reproductive and/or developmental pathways, sexual inheritance is determined by the passage of traits through a pedigree and asexual inheritance by passage of traits to asexually derived progeny. Here we show that the pgc and psc competitive relationships among genotypes is reproducible, hierarchical, and asexually heritable. The proposal that pgc competitive ability is a heritable trait was also supported by a pedigree analysis of germ-line “winners” and “losers.” These results, therefore, provide substantive evidence that cell lineage selection may play a role in shaping the life history of B. schlosseri and, in so doing, support the idea that natural selection can operate at multiple levels of organization (6–11).
MATERIALS AND METHODS
Microsatellite Analyses of Genotypes.
As described below, 2–5 months after fusion, somatic and gametic tissues from the fused colonies were harvested and genetically assayed by using a PCR-based technique that used primers amplifying unique microsatellite loci (18, 19, 32, 33). At 5–6 representative sites, tissue samples were taken of a single bud (to determine somatic cell origin) and sperm was liberated from the testes of an adjacent adult bud (to determine germ cell origin) and blood. In most cases, two independent microsatellite loci were examined, and in all cases where distinct alleles were present, results were concordant (Fig. 1). To determine whether the genetic patterns we observed at 2 months were stable or would change after many additional asexual cycles, we also genetically analyzed the tissues from one replicate of each bi- and trichimera from experimental set I and one replicate of the trichimeras from sets III–V after 5 months.
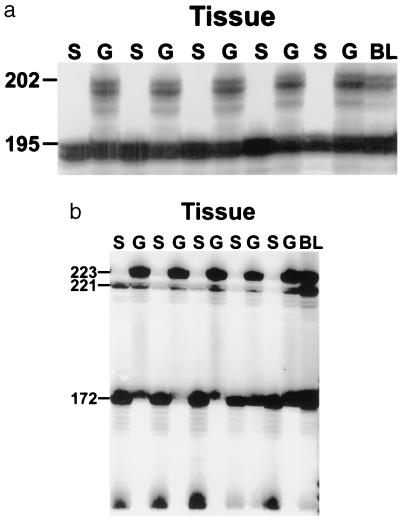
Figure 1.
Genetic analysis of buds (S), sperm (G), and blood (BL) from one of the replicate bichimeras of experimental set 3 using two microsatellite loci. The bichimera was formed through the fusion of colonies G and F. (A) The results presented are these derived from the use of microsatellite locus PB41. The Upper band (202 bp) is unique to the gametic winner, (colony F) and the Lower band (195 bp) is shared by both genotypes. The presence of only the Lower band in the somatic tissues suggest that all of the somatic tissues are derived from colony G, while the presence of both the Upper and Lower band in the gametic tissues suggests that these tissues are derived either just from colony F or from both colonies. (B) Results using microsatellite locus PBC1. The Top band (223 bp) is unique to the gametic winner (colony F), while the Bottom two bands (172 and 221 bp) are unique to colony G. The presence of only the Bottom two bands in the somatic tissues suggest that all of the somatic tissues are derived from colony G, while the presence of just the Top band in the gametic tissues suggests that these tissues are derived from colony F.
Crosses.
Botryllus colonies were maintained in the laboratory as described (34). Laboratory crosses between two trichimeras (sets I and III) and a wild colony were undertaken to test the extent to which microsatellite analysis of sperm was a good predictor of progeny production by gametes from both testes and ovaries. Reciprocal crosses were performed in which each trichimera functioned as first sperm donor and then egg donor.
RESULTS
Experimental Design.
B. schlosseri colonies selected for this experiment came from laboratory stocks that all shared at least one allele at the Fu/HC locus (29, 30) and, therefore, would fuse via their vascular ampullae on contact. Each colony had asexually reproduced for several generations, and at the time of the experiment consisted of many asexually derived blastozooids. Eleven colonies, chosen from the three pedigrees shown in Fig. 2 and designated by the letters A–K, were genetically typed for microsatellite alleles and assigned to one of five experimental sets consisting of three colonies each (Table 1). Four of the colonies (A–C and H) were assigned to more than one experimental set. Within an experimental set, subclones (systems of genetically identical blastozooids) from each colony were paired with equal sized subclones from each of the other colonies and placed on glass slides to form bichimeras and combined all together to form trichimeras. Except for set II, each bichimera and trichimera was replicated three times. Replicates were separated in time from each other by several asexual cycles. Once the chimeras were created, they were housed in aquaria and fed as described (34), and weekly observations were made with regard to position and size of chimeric partners.
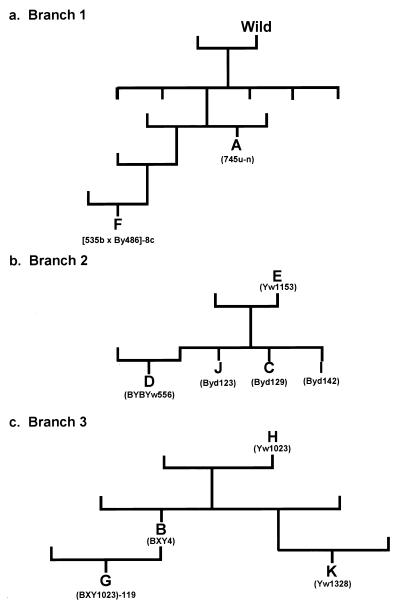
Figure 2.
Pedigree of laboratory colonies. (A) The branch of the pedigree showing the two genetic relationship among the two most dominant germ-cell competitors. (B) Another branch of the pedigree showing the genetic relationship among five other good germ-cell competitors. (C) A third branch of the pedigree showing the genetic relationship among the 4 poorest germ cell competitors. All of the colonies within the three branches, with perhaps the exception of colony A, have at least one parent who was a descendant from an initial pair of colonies that founded our laboratory stocks. Within this larger pedigree, the three branches are all distantly related to each other. Letters indicate colony identity. Listed below the identifier is the actual name of the colony in the pedigree (available on request).
Table 1.
Directionality of colony resorption and somatic and gametic identification of phenotypic winner in five experimental sets 2 months after fusion
| Line | Experimental set | Fusion partners | Replicate | Resorptive winner | Genotype of the survivors
|
|
|---|---|---|---|---|---|---|
| Buds | Sperm | |||||
| 1 | I | A = B | 1 | A | A | A |
| 2 | 2 | A | A | A | ||
| 3 | 3 | A | A | A | ||
| 4 | A = C | 1 | C | C | C | |
| 5 | 2 | C | C | A | ||
| 6 | 3 | A | A | A | ||
| 7 | B = C | 1 | C/B | C/B | C > B/C | |
| 8 | 2 | C | C | C | ||
| 9 | 3 | C | C | C | ||
| 10 | A = B = C | 1 | A | A | A | |
| 11 | 2 | A | A | A | ||
| 12 | 3 | C | C > A | A | ||
| 13 | II | A = D | 1 | A | A | A |
| 14 | 2 | A | A | A | ||
| 15 | A = E | 1 | A | A | A | |
| 16 | 2 | A | A | A | ||
| 17 | D = E | 1 | E | E | E | |
| 18 | 2 | E | E | E | ||
| 19 | A = D = E | 1 | A | A | A | |
| 20 | 2 | E | E > A | A > E | ||
| 21 | III | B = F | 1 | B/F | B/F | F/F |
| 22 | 2 | B | BF | F | ||
| 23 | 3 | B/F | B/F | F/F | ||
| 24 | B = G | 1 | B | G | G | |
| 25 | 2 | B | G | G | ||
| 26 | 3 | B | G | G | ||
| 27 | F = G | 1 | G | G | F | |
| 28 | 2 | G | F | F | ||
| 29 | 3 | F | F | F | ||
| 30 | B = F = G | 1 | G | G > F | F | |
| 31 | 2 | B | G | F | ||
| 32 | 3 | B | G | F | ||
| 33 | IV | C = H | 1 | C | C > H | C |
| 34 | 2 | C | C > H | C > H | ||
| 35 | 3 | C | C > H | C | ||
| 36 | C = I | 1 | C | C | I | |
| 37 | 2 | C | C | I | ||
| 38 | 3 | C | C | I > C | ||
| 39 | H = I | 1 | H | I > H | I | |
| 40 | 2 | H | I > H | I | ||
| 41 | 3 | H | I > H | I | ||
| 42 | C = H = I | 1 | C | C > H | I > C | |
| 43 | 2 | C | C > H | I > C | ||
| 44 | 3 | C | C > H | I > C | ||
| 45 | V | H = J | 1 | H | H | J |
| 46 | 2 | H | H | J | ||
| 47 | 3 | H | H > J | J | ||
| 48 | H = K | 1 | H | H | K | |
| 49 | 2 | H | H | K | ||
| 50 | 3 | H | H | K | ||
| 51 | J = K | 1 | J | JK | J | |
| 52 | 2 | J | J | JK | ||
| 53 | 3 | J | — | JK | ||
| 54 | H = J = K | 1 | K | J > H | J | |
| 55 | 2 | H | J > H | J | ||
| 56 | 3 | H | J > H | J | ||
Listed are the genotypes of the colonies that either physically resorbed the other colony(ies) in the bi(tri)chimeras or were expressed within the somatic and gametic tissues of the chimera 2 months after fusion. /, both members of the chimera were still present 2 months after fusion. Two letters together indicate that the tissues expressed both genotypes. >, the genotype to the right of the symbol represents a minor component of a tissue.
Bichimeras.
In 39 of 42 bichimeras, one of the two paired colonies ceased budding and was macroscopically resorbed, as determined by visual inspection (35) within the first 2 months postfusion (Table 1). As described (35), the colony resorption results were consistent across replicates. In only 4 of 15 pairings was the resorptive winner also the somatic and germ cell winner in all replicates (Table 1, lines 1–3, 13–14, 15–16, and 17–18). Although evidence of complete replacement of the somatic tissues of one colony by the tissues of the resorbed colony was found in only 2 of 15 pairings (Table 1, lines 24–26 and 28), partial somatic replacement (presence of both genotypes in the bud) occurred in 5 of 15 pairings (Table 1, lines 22, 33–35, 39–41, 47, and 51). In 5 of 15 pairings, there was complete germ-cell replacement of the colony resorption winner’s testis by germ cells of the other in all three replicates (Table 1, lines 21–23, 24–26, 39–41, 45–47, and 48–50). In two pairings, complete gametic replacement of the resorption winner was found in at least two of three replicates (Table 1, lines 27, 28, 36, and 37), and almost complete replacement was found in the third replicate (Table 1, line 38). Partial gamete replacement was observed in another 2 pairings (Table 1, lines 34 and 52–53). Both complete or partial somatic and germ-cell replacement of the resorption winner could occur within the same chimera (Table 1, lines 24–26, 28, and 39–41), but more often the gametic and somatic replacement of resorption winner tissues were independent (Table 1, lines 5, 21–23, 27, 36–38, 45–47, and 48–50 and possibly 51 and 52).
A linear hierarchy was a good descriptor of the competitive relationships among psc (Table 1, Fig. 3). The relative ranking of somatic winner genotypes was not identical to the resorption rankings, although there was a strong bias for the resorptive winner to retain its somatic genotype (Table 1 and Fig. 3). Of the 15 bichimeric replicates tested, 9 of the resorptive winners were the somatic winner in all replicates. Five of the 15 replicates were either derived completely from the fused partner’s genotype or were mixtures of both somatic genotypes in at least one of the replicates. In two cases (lines 24–26 and lines 39–41 of Table 1), the resorptive winner in all 3 replicates was wholly or mostly derived from the loser’s somatic tissues.
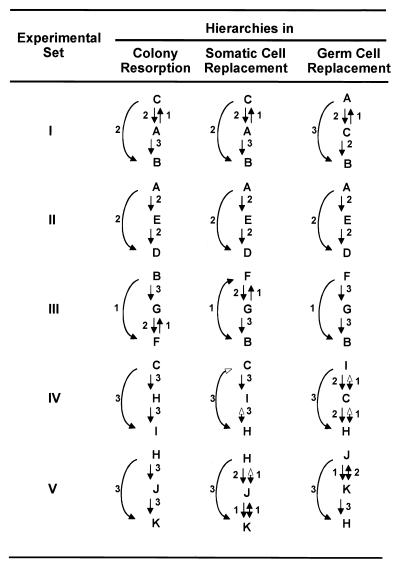
Figure 3.
Linear hierarchies for resorption, somatic replacement, and gametic replacement in B. schlosseri. Arrows point to the competitive losers in five experimental sets (Table 1). Only cases where complete colony resorption occurred within 2 months of fusion are included. Arrows possessing two arrowheads indicate reciprocal outcomes. Open arrowheads indicate cases where only minute PCR amplifications of one of the two genotypes were recorded. Numbers alongside the arrows show the number of replicates for which that outcome was observed.
A linear competitive hierarchy also appears to be a good descriptor of the competitive relationships among pgc (Table 1, Fig. 3). There was little correlation between the gametic hierarchy and the resorption hierarchy because the gametic tissues frequently expressed the germ line of the resorptive loser. For example, it would appear that the germ line of genotype A is a good competitor for access to reproductive sites regardless of whether it is physically resorbed. On the other hand, the germ line of genotype B seems to be an invariably poor competitor (sets I and III). The results of the gametic competition were consistent among temporally separated clonal replicates. The fact that genotypes could be placed into a competitive hierarchy suggests that the ability of pgc to compete for reproductive sites is probably heritable.
Trichimeras.
Trichimeras were useful for determining the competitive abilities of pgc and psc where three genotypes fused and had access to germ line and somatic niches. In all five trichimeras, the winner of the gametic competition in all replicates was predictable from the bichimera results, and in most cases, a single pgc genotype completely populated all gonads. For example, as in B = G bichimeras, in the B = F = G trichimeras, colony B appeared to be the resorption winner but in fact was the somatic loser to colony G, whereas both lost the gametic competition to F (lines 30–32). Somatic chimeras could be found in at least one replicate in all five sets of trichimeras (Table 1, lines 12, 20, 30, 42–44, and 54–56).
The somatic hierarchy for the trichimeras also roughly paralleled the resorption hierarchy, although to a lesser degree than in the bichimeras (Table 1, Fig. 3). In fact, in most (9 of 14) trichimeras, no single genotype dominated the competition for access to somatic sites. These results suggest that competitive interactions among genotypes for access to somatic sites can be modified by the presence of additional genotypes. As was also the case for the bichimeras, among the trichimeras there was little correlation between the gametic hierachy and either the resorption or somatic hierarchies (Table 1, Fig. 3).
Comparing the results of the genetic analyses performed at 5 months with the genetic analyses performed at 2 months, there were no changes in either the genetic identities of the somatic and gametic tissues or in the relative contribution of genotypes in those cases where there was partial replacement (Tables 1 and 2).
Table 2.
Resorptive, somatic and gametic winners of seven chimeric combinations after 5 months
| Line | Experimental set | Fusion partners | Replicate | Resorptive winner | Genotype of the Survivors
|
|
|---|---|---|---|---|---|---|
| Buds | Sperm | |||||
| 1 | I | A = B | 3 | A | A | A |
| 5 | A = C | 2 | C | C | A | |
| 8 | B = C | 3 | C | C | C | |
| 12 | A = B = C | 3 | C | C > A | A | |
| 32 | III | B = F = G | 2 | B | G | F |
| 43 | IV | C = H = I | 3 | C | C > H | I > C |
| 54 | V | H = J = K | 1 | K | J > H | J |
Listed are the genotypes of the colonies that either physically resorbed the other colony(ies) in the bi(tri)chimeras or were expressed within the somatic and gametic tissues of the chimera 5 months after fusion. Two letters together indicate that the tissues expressed both genotypes. >, the genotype to the right of the symbol represents a minor component of a tissue. Lines refers to corresponding lines of same fusion partners in Table 1.
We tested whether the spermatic winners represented pgc that gave rise to both functional sperm and eggs by breeding these trichimeras to a wild colony, wherein each trichimera served successively as an egg and then as a sperm donor. We expected ∼50% of the offspring to bear the microsatellite markers of the pgc winner. Of 81 progeny assayed from four crosses, 44–48% of the progeny were derived from the pgc winner, as identified by the unique band derived from the testicular winner. Only 1 of the 81 progeny was identified as having been sired by an undetected genotype (Table 3). G tests showed that within all four crosses, the proportion of progeny in each genotype class were as would be expected if the testicular winner was the gamete donor (Table 3). These results were obtained whether the trichimeras served as sperm or egg donor. Thus, we assert, as we did previously (18, 19), that the sperm output of each genotype also predicts male and female reproductive output. Because the trichimera crosses shown in Table 3 indicate that the ability to capture testes germ-line sites correlates with the ability to capture ovary germ-line sites in the same colony, we also suspect that the competitive elements are pgc rather than testis or ovary committed protogametes.
Table 3.
Progeny testing of two trichimeras from experimental sets I and III
| Trichimera (Set) | Microsatellite bands of component colonies | Microsatellite bands of test colony | Crosses
|
n | Microsatellite bands of progeny | Expected results from cross if testicular winner was gamete donor, n | Observed results from cross, n | G test | |
|---|---|---|---|---|---|---|---|---|---|
| Egg donor | Sperm donor | ||||||||
| A = B = C | *A = 1,3 | 0,1 | TC | TRI | 30 | 1,3 | 7.5 | 8 | P > 0.1 |
| (I) | B = 1 | 1,2 | 0 | 1 | |||||
| C = 1,2 | 0,3 | 7.5 | 6 | ||||||
| 0,2 | 0 | 0 | |||||||
| 0,1 + 1,1 | 15.0 | 15 | |||||||
| 0,1 | TRI | TC | 9 | 1,3 | 2.25 | 0 | P > 0.1 | ||
| 1,2 | 0 | 0 | |||||||
| 0,3 | 2.25 | 4 | |||||||
| 0,2 | 0 | 0 | |||||||
| 0,1 + 1,1 | 4.50 | 5 | |||||||
| B = F = G | B = 1 | 2,2 | TC | TRI | 29 | 2,3 | 14.5 | 14 | P > 0.5 |
| (III) | *F = 1,3 | 2,1 | 14.5 | 15 | |||||
| G = 1 | |||||||||
| 0,1 | TRI | TC | 13 | 1,3 | 3.25 | 4 | P > 0.5 | ||
| 0,3 | 3.25 | 3 | |||||||
| 0,1 + 1,1 | 6.50 | 6 | |||||||
Reciprocal crosses were performed in which the trichimera acted as either functional male or functional female. Microsatellite bands of component colonies and progeny are the bands observed on an autoradiograph when microsatellite locus PB41 is amplified. The banding patterns of test colonies were inferred from microsatellite analysis of tissues from the test colonies using locus PB41 and the results from the crosses. For example, microsatellite analysis of the third test colony suggested that its genotype could have been either 0,2 or 2,2. But given the genotypes of the progeny from the cross in which it participated, we inferred that its genotype was most likely 2,2. The last column gives results from a G test of the hypothesis that the testicular winner from each of the trichimeras was the gametic donor. ∗, testicular winner identified by the microsatellite analysis of sperm from the trichimera. TC, test colony; TRI, trichimera.
Pedigree of Experimental Colonies.
One method for determining whether a phenotypic trait has a genetic basis is pedigree analysis. Heritable traits would be expected to be transmitted by sexual reproduction to progeny in a pedigree. We have raised the B. schlosseri used in the experiments at Hopkins Marine Station for 10–12 generations and have generated a pedigree for most of the colonies produced during this time period. The colonies used in this experiment are a small subset of this larger pedigree, and all shared at least one allele at the Fu/HC locus. An analysis of this pedigree showed a strong correlation between the genealogical tree (Fig. 2) and the winners in the germ cell competition (Table 1, Fig. 3). The best gametic competitors were either F2s (colony A) or F4s (colony F) of branch 1 or F1s (colonies C, I, and J) of colony E in branch 2. In gametic competitions involving just these two branches, colonies from branch 1 always defeated colonies from branch 2. A survey of the rank order of colonies within the five experimental sets revealed that colonies from branches 1 and 2 of the pedigree were often also the second best gametic competitor within an experimental set. In each case where one of these colonies came in second, there was always more than one colony from these two branches present within an experimental set (Fig. 2, Table 1). The poorest gametic competitors were usually F1s (colony B) and F2s (colonies G and K) of colony H as well as colony H itself from branch 3. This branch of the pedigree contained four of five losers and two of five of the second place colonies (Fig. 2, Table 1). Interestingly, when members of this branch were in second place (colony G), another member of this branch always occupied last place. It is of interest that both branches 1 and 2 contain independent ancestral wild colonies within the previous five generations (the full pedigree chart is available on request).
What is the probability that these results occurred by chance? For each bichimera replica in the pedigree analysis, the probability that the gametic winner was from a particular colony is 0.5 and for each trichimera is 0.33. In Table 4, we present the observed vs expected outcomes of the bichimera and trichimera germ-line competitions when interpedigree partners were involved. In every interpedigree interaction, the probability that the germ-line winners occurred by chance in bichimeras or trichimeras between pedigree 1 vs. pedigree 2 or pedigree 3 and pedigree 2 vs. pedigree 3 is <0.05 and in most cases <0.001. It is fair to conclude that there is a nonrandom distribution of pgc winners and losers when colonies between pedigrees are tested and therefore that the pgc competition outcomes are heritable. It is beyond the scope of this study to demonstrate the mode of inheritance.
Table 4.
Interpedigree win–loss record with respect to germ cell competitions
| Pedigrees Compared | Observed wins per pedigree
|
Expected wins per pedigree
|
df | G | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | ||||
| Bichimeras | |||||||||
| 1 vs. 2 | 6 | 1 | — | 3.50 | 3.50 | — | 1 | 3.9624 | <0.05 |
| 1 vs. 3 | 9 | — | 0 | 4.50 | — | 4.50 | 1 | 12.4766 | <0.001 |
| 2 vs. 3 | — | 13* | 1** | — | 7.00 | 7.00 | 1 | 12.2032 | <0.001 |
| Trichimeras | |||||||||
| 1 vs. 2 vs. 3 | 3 | 0 | 0 | 1.00 | 1.00 | 1.00 | |||
| 1 vs. 2 vs. 2 | 2 | 0 | — | 0.66 | 1.34 | — | |||
| 1 vs. 3 vs. 3 | 3 | — | 0 | 1.00 | — | 2.00 | |||
| 2 vs. 2 vs. 3 | — | 3 | 0 | — | 2.00 | 1.00 | |||
| 2 vs. 3 vs. 3 | — | 3 | 0 | — | 1.00 | 2.00 | |||
| Trichimeras-Totals | |||||||||
| 1 vs. non-1 | 8 | 0 | 0 | 2.66 | 2.34 | 3.00 | 1 | 17.6178 | <0.001 |
| 2 vs. 3 | — | 6 | 0 | — | 3.00 | 3.00 | 1 | 8.3178 | <0.005 |
A goodness-of-fit test is used to verify the hypothesis that there is no competitive difference among colonies from each of the three pedigrees used in our study. Assuming equal competitive ability, colonies from each pedigree are expected to be the competitive winner 50% of the time within bichimeras and 33% of the time within trichimeras.
, Includes replicate where both genotypes were present within the gonads, but one genotype was dominant.
, Does not include two replicates in which there was no clear winner.
There was a weaker correlation between these pedigrees and the somatic hierarchy. Four of the five somatic winners came from the same two branches (branches 1 and 2, Fig. 2) of the pedigree that contained the gametic winners. However, the somatic winner in set V, colony H, came from a branch that contained all the gametic losers (branch 3 in Fig. 2), and in experimental set I there was no one clear winner.
CONCLUSIONS AND SPECULATIONS
We show here that pgc and psc lineages may have traits that make them likely units of natural selection. Specifically, we found with respect to both the pgc and psc competitions in chimeric B. schlosseri that the order of winning and losing was consistent among clonal replicates, was stable over tens of asexual generations, could be arranged into a linear hierarchy (Fig. 3), and was heritable via sexual reproduction over 3–4 generations of crosses (Fig. 2, Table 4). The lack of correlation in the rank order of somatic and gametic winners suggests that the outcomes of the somatic vs. gametic competitions are independent. Because the reproductive differential between the winners and losers of gametic cell competition was essentially all or none, it would appear that cell lineage selection could powerfully impact the fitness of B. schlosseri. Technically, cell lineage selection is still selection of a distinct genome and is therefore a form of individual selection. However, selection of a genome within a composite soma will differ from most instances of individual selection in that under cell lineage selection, fitness will not necessarily be linked to the survival of the soma.
Buss (6, 7, 16) and Buss and Green (15) proposed that in colonial invertebrates such as B. schlosseri the cells that pass from one asexual generation to the next are totipotent cells, with lineage commitments to somatic and gametic differentiation occurring as late but regulated events. It seems most likely that the relative independence of successful gonadal vs. somatic competitions in the same chimeras is inconsistent with this model. We propose alternatively that there is a separation of pgc and psc in the fetal oozooid and that these psc and pgc probably represent populations of germ-line stem cells and somatic stem cells. Stem cells are cells that are capable, at the clonal level, of self-renewal as well as multilineage differentiation (36, 37). At each round of asexual reproduction of B. schlosseri the nascent bud is made up of morphologically undifferentiated cells that proliferate and differentiate to generate fully developed organ systems; hence, it is likely that these undifferentiated bud cells are in fact collections of stem cells. It is also likely that asexual reproduction and colony size expansion provide a requirement for the expansion of these putative stem cell pools, i.e., by self-renewal. If so, the competitive nature of these putative stem cell populations could depend on their ratio of self-renewing to differentiating cell divisions. Clearly, it shall be important to identify and isolate the cells that are responsible for somatic and gonadal invasion and to test whether their totipotent common progenitors exist only in the embryonic oozooid or continue to exist during asexual expansion of blastozooids.
It is evident that one level of selection occurs via the winner of germ-line competition. But the survivability of the somatic host is a selectable trait as well, and perhaps it will turn out that there are cooperating elements between the genes that encode the body and genes that are expressed in the germ cells that use the body. For example, insofar as the Fu/HC system limits the opportunity of fusion to kin (29), kin-shared elements would be passengers along with the trait or traits that encode successful pgc replacement of host gonads. Successful pgc clones that carry genes which promote fitter somatic features should eventually overcome equally competitive pgc clones that carry less successful somatotype genes.
The preceding discussion might give the impression that the fitness of the host colony depends solely on the genotype of the host. However, genetic analyses of the somatic tissues of fused colonies shows that they can be a composite of both genotypes (Tables 1 and 2). It is conceivable that the fitness of a fused colony may actually depend on the joint fitness of the genomes that contribute cells to somatic function. If two psc genotypes have different environmental tolerances, one would expect that as environmental conditions change, so would the composition of the chimera, a process that is enabled by the dynamic nature of somatic cell lineage competitions in each asexual generation (a new round of asexually derived blastozooids takes over each week in Monterey B. schlosseri). Thus, the existence of psc chimerism may provide the “chimeric entity” with a wider range of responses than it would the individuals.
The finding that there are some genotypes that are always better pgc than others suggests that there could arise supercompetitor lineages within a population that spread by fusing with other members of the population. How far such pgc supercompetitor lineages will spread will be limited primarily to kin by the high polymorphism of the Fu/HC locus (29). The spread of the mobile stage tadpole larva of B. schlosseri is nonrandom; laboratory experiments performed by Grosberg and Quinn (21) demonstrate that sibling larvae of B. schlosseri preferentially settle next to histocompatible kin (including their parents).
These results have several potential implications. First, the finding that pgc competition is a heritable trait that might lead to the emergence of specialist genotypes suggests that at least within chimeric B. schlosseri, selection is acting on cell lineages as well as on individuals. These findings imply that in order to understand the evolution of organisms with life histories like B. schlosseri such as sponges (38, 39), hydroids (40), corals (41, 42), and bryozoans (43, 44), one must accept the possibility that their evolution occurred as the result of the action of selection acting on several levels of organization (6–10). Also, when surveying natural populations of organisms that can form genetic chimeras, one should not rely solely on morphological observation to make inferences about population structure (literally, you cannot believe your eyes) in that the phenotype that is represented by the body may not represent the genotype that is transmitted by the gonads. Finally, these findings confirm the hypotheses of Buss (6, 7), Grosberg and Quinn (21), Grosberg (22), and Rinkevich and Weissman (45) that polymorphic histocompatibility loci may have arisen or been maintained to limit supercompetitor lineages to histocompatible kin. This provides an alternative (but not exclusive) hypothesis to account for the widespread development of polymorphic histocompatibility loci within the metazoa.
The experiments described here show that there is probably a genetic basis to pgc and psc competition within B. schlosseri chimeras, presumably related to the self-renewal and/or the migratory properties of these cell lineages. In the future, we hope to use genomic markers (46) to test the Mendelian inheritance of the pgc competitive ability, as well as identifying, isolating, and transplanting candidate pgc and psc cells.
Acknowledgments
We thank K. Ishizuka and K. Palmeri for their excellent work in raising and maintaining the animals and animal crosses and D. Baltimore, A. DeTomaso, M. Fagan, L. Hood, D. Laird, and B. Magor for discussions and/or comments on earlier versions of this manuscript. This study was supported by the U.S.–Israel Binational Science Foundation (B.R. and I.L.W.), by the International Human Frontier Science Program (B.R.), by the Minerva Center for Marine Invertebrate Immunology and Developmental Biology (B.R.), and primarily by Systemix/Sandoz grants (I.L.W.). We also are grateful for support from the Israeli Academy of Sciences.
ABBREVIATIONS
- pgc
primordial germ cells
- psc
primitive somatic cells
Footnotes
A Commentary on this article begins on page 8801.
References
- 1.Lewontin R C. Annu Rev Ecol Syst. 1970;1:1–18. [Google Scholar]
- 2.Mayr E. Proc Natl Acad Sci USA. 1997;94:2091–2094. doi: 10.1073/pnas.94.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher R A. The Genetical Theory of Natural Selection. Oxford: Clarendon; 1930. [Google Scholar]
- 4.Wright S. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haldane J B S. The Causes of Evolution. New York: Longmans; 1932. [Google Scholar]
- 6.Buss L W. Proc Natl Acad Sci USA. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buss L W. The Evolution of Individuality. Princeton: Princeton Univ. Press; 1987. [Google Scholar]
- 8.Dawkins R. The Selfish Gene. Oxford: Oxford Univ. Press; 1976. [Google Scholar]
- 9.Gould S J. Science. 1982;216:380–387. doi: 10.1126/science.7041256. [DOI] [PubMed] [Google Scholar]
- 10.Michod R B. Am Nat. 1997;149:607–645. [Google Scholar]
- 11.Weismann A. Das Keimplasma: Eine Theorie der Verebung. Jena, Germany: Fisher; 1892. [Google Scholar]
- 12.Nussbaum M. Arch Mikrobiol Anat. 1880;18:1–121. [Google Scholar]
- 13.Nieuwkoop P D, Sutasurya L A. Primordial Germ Cells in the Invertebrates. New York: Cambridge Univ. Press; 1981. [Google Scholar]
- 14.Nieuwkoop P D, Sutasurya L A. Primordial Germ Cells in the Chordates. New York: Cambridge Univ. Press; 1979. [Google Scholar]
- 15.Buss L W, Green D R. Dev Comp Immunol. 1985;9:191–201. doi: 10.1016/0145-305x(85)90110-7. [DOI] [PubMed] [Google Scholar]
- 16.Buss L W. Paleobiology. 1988;14:313–321. [Google Scholar]
- 17.Steinbeck J. The Log from the Sea of Cortez. New York: Penguin; 1951. pp. 136–137. [Google Scholar]
- 18.Pancer Z, Rinkevich B. Biol Bull. 1995;189:106–112. doi: 10.2307/1542460. [DOI] [PubMed] [Google Scholar]
- 19.Stoner D S, Weissman I L. Proc Natl Acad Sci USA. 1996;93:15254–15259. doi: 10.1073/pnas.93.26.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabbadin A, Zaniolo B J. J Exp Zool. 1979;207:289–304. [Google Scholar]
- 21.Grosberg R K, Quinn J F. Nature. 1986;322:456–459. [Google Scholar]
- 22.Grosberg R K. Q. Rev Biol. 1988;63:377–412. [Google Scholar]
- 23.Feldgarden M, Yund P O. Biol Bull. 1992;185:155–158. doi: 10.2307/1542190. [DOI] [PubMed] [Google Scholar]
- 24.Nauta M J, Hoekstra R F. Evolution. 1994;48:979–995. doi: 10.1111/j.1558-5646.1994.tb05287.x. [DOI] [PubMed] [Google Scholar]
- 25.Den Boer R J. Mol Biol Evol. 1995;12:494–502. [Google Scholar]
- 26.Berrill N J. The Tunicata. London: Ray Society; 1950. [Google Scholar]
- 27.Oka H, Watanabe H. Proc Jpn Acad Sci. 1957;33:657–659. [Google Scholar]
- 28.Sabbadin A. Am Zool. 1982;22:765–773. [Google Scholar]
- 29.Scofield V L, Schlumpberger J, West L A, Weissman I L. Nature (London) 1982;295:499–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- 30.Weissman I L, Saito Y, Rinkevich B. Immunol Rev. 1990;113:227–241. doi: 10.1111/j.1600-065x.1990.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 31.Burnet F M. Nature (London) 1971;232:230–235. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- 32.Stoner D S, Quattro J M, Weissman I L. Mol Mar Biol Biotechnol. 1997;6:163–171. [PubMed] [Google Scholar]
- 33.Pancer Z, Gershon Z, Rinkevich B. Biochem Biophys Res Commun. 1994;203:646–651. doi: 10.1006/bbrc.1994.2231. [DOI] [PubMed] [Google Scholar]
- 34.Boyd H C, Brown S K, Harp J A, Weissman I L. Biol Bull. 1986;170:91–109. [Google Scholar]
- 35.Rinkevich B, Weissman I L. J Zool London. 1987;213:717–733. [Google Scholar]
- 36.Spangrude G J, Heimfeld S, Weissman I L. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 37.Morrison S J, Uchida N, Weissman I L. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 38.Fell P E. In: Reproduction of Marine Invertebrates. Giese A C, Pearse J S, editors. Vol. 1. New York: Academic; 1974. pp. 51–132. [Google Scholar]
- 39.Ilan M, Loya Y. Biol Bull. 1990;179:279–286. doi: 10.2307/1542319. [DOI] [PubMed] [Google Scholar]
- 40.Shenk M A, Buss L W. J Exp Zool. 1991;257:80–86. [Google Scholar]
- 41.Fadlallah Y D. Coral Reefs. 1983;2:129–150. [Google Scholar]
- 42.Hidaka M. Coral Reefs. 1985;4:111–116. [Google Scholar]
- 43.Winston J E, Jackson J B C. J Exp Mar Biol Ecol. 1984;76:1–21. [Google Scholar]
- 44.Craig S F. In: Biology and Paleobiology of Bryozoans. Hayward P J, Ryland J S, Taylor P D, editors. Fredensborg, Denmark: Olsen and Olsen; 1994. pp. 51–54. [Google Scholar]
- 45.Rinkevich B, Weissman I L. Symbiosis. 1987;4:117–134. [Google Scholar]
- 46.De Tomaso A W, Saito Y, Ishizuka K J, Palmeri K J, Weissman I L. Genetics. 1998;149:277–287. doi: 10.1093/genetics/149.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]