Abstract
Background: Hereditary non-polyposis colorectal cancer (HNPCC) is clinically defined by familial clustering of colorectal cancer and other associated tumours.
Methods: By thorough molecular and clinical evaluation of 41 families, two different groups were characterised: group 1, 25 families with truncating mutations in MLH1 or MSH2 (12 novel mutations); and group 2, 16 Amsterdam positive families without mutations in these genes and without microsatellite instability in their corresponding tumours.
Results: Significant clinical differences between these two groups were found. Firstly, earlier age of onset for all colorectal cancers (median 41 v 55 years; p<0.001) and all tumours (median 43 v 56 years; p = 0.022) was observed, comparing groups 1 and 2. Secondly, 68% of the index colorectal cancers were localised proximally of the splenic flexure in group 1 compared with 14% in group 2 (p<0.010). Thirdly, more synchronous and metachronous colorectal (p = 0.017) and extracolorectal tumours (p<0.001) were found in group 1. Fourthly, a higher colorectal adenoma/carcinoma ratio (p = 0.030) and a tendency towards more synchronous or metachronous adenomas in group 2 (p = 0.084) was observed, indicating a slower progression of adenomas to carcinomas. As three mutation negative tumours revealed chromosomal instability after comparative genomic hybridisation, these tumours may be caused by one or more highly penetrant disease alleles from the chromosomal instability pathway.
Conclusion: These data show that HNPCC includes at least two entities with clinical and molecular differences. This will have implications for surveillance programmes and for cancer research.
Keywords: colorectal cancer, hereditary non-polyposis colorectal cancer, microsatellite instability, mutation
Originally, hereditary non-polyposis colorectal cancer (HNPCC) described a clinical entity of familial clustering of colorectal cancer, according to the Amsterdam criteria. Early onset colorectal cancer with predominantly right sided tumours, marked lymphocyte infiltration, and a high synchronous and metachronous tumour risk have been demonstrated.1–3 Identification of microsatellite instability (MSI) in a major subset of tumours and later the detection of germline mutations in DNA mismatch repair genes gave the impression of HNPCC as a molecularly defined hereditary tumour entity.4–6 After the introduction of experience based surveillance programmes, it became obvious that there was a significant number of families fulfilling the Amsterdam criteria of HNPCC with neither MSI in their corresponding tumours nor germline mutations in DNA mismatch repair genes. Surveillance strategies in these families cannot be limited to mutation carriers but must be offered to at least all first degree relatives. Regarding these families with the clinical HNPCC syndrome but with no molecular features of high microsatellite instability (MSI-H), it is not yet clear whether they should be managed in the same way as those families harbouring a germline mutation in one of their mismatch repair genes. Amsterdam positive but mutation negative families very often have been classified as clinical HNPCC without indicating that this is probably a molecular and perhaps also a clinically distinct entity harbouring at least two subgroups: microsatellite stable and microsatellite instable tumours.
In this study, we present the clinical features of MSI-H mutation positive compared with mutation negative families with microsatellite stable tumours meeting the Amsterdam criteria in order to better describe this subgroup and make suggestions regarding clinical management and genetic counselling in these families.
MATERIAL AND METHODS
Patients
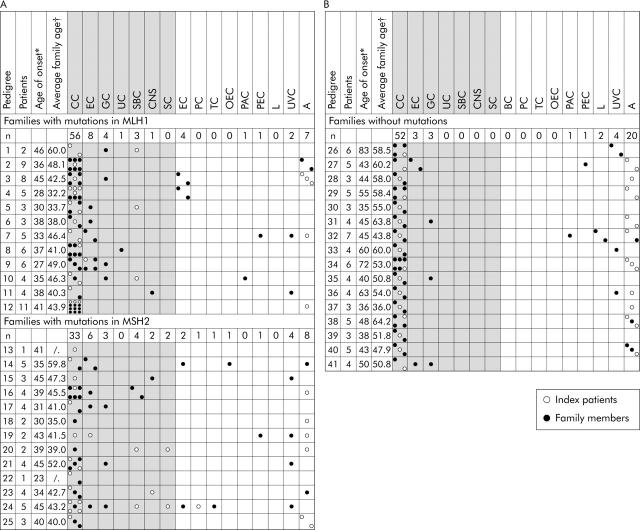
Forty one consecutively registered index patients and their families were included, all fulfilling the Bethesda criteria or the Muir Torre phenotype, with complete data obtained on first degree relatives. Twenty five index patients and their families (106 patients with tumours or colorectal adenomas; 50 females, 56 males) were included with truncating mutations in MLH1 or MSH2, excluding missense mutations (fig 1 ▶).
Figure 1.
Tumour spectrum of families with truncating mutations in MLH1 or MSH2 (A) and without mutations (B). Tumour diseases of the index patients and those of family members are shown. Hereditary non-polyposis colorectal cancer associated tumour diseases are shown in the shaded areas. *Age of onset, age of the index patient at the time of the first tumour disease; †average family age, sum of the age of onset of all tumour patients in a family divided by the number of patients; A, adenomatous polyp; BC, breast cancer; CC, colorectal cancer; CNS, central nervous system cancer; EC, endometrial cancer; GC, gastric cancer; L, leukaemia; OEC, oesophageal cancer; PAC, pancreatic carcinoma; PC, prostate cancer; PEC, pelvic carcinoma; SBC, small bowel cancer; SC, sebaceous cancer; TC, thyroid cancer; UC, urothelial cancer; UVC, unverified cancer.
Sixteen index patients and their families (71 patients with tumours or colorectal adenomas; 33 females, 38 males) were included who fulfilled the Amsterdam I (n = 14) or Amsterdam II criteria (n = 2) with proven microsatellite stability, immunohistochemical positivity for mismatch repair gene expression of MLH1 and MSH2, or negative mutation screening (denaturing high performance liquid chromatography (DHPLC)/exon deletion) for MLH1 and MSH2, and partly MSH6 (fig 2 ▶). Patients were recruited by genetic counselling or from the departments of surgery and gastroenterology of the Universities of Munich. All patients gave written informed consent to take part in this study funded by the German Cancer Aid and approved by the ethics committees.
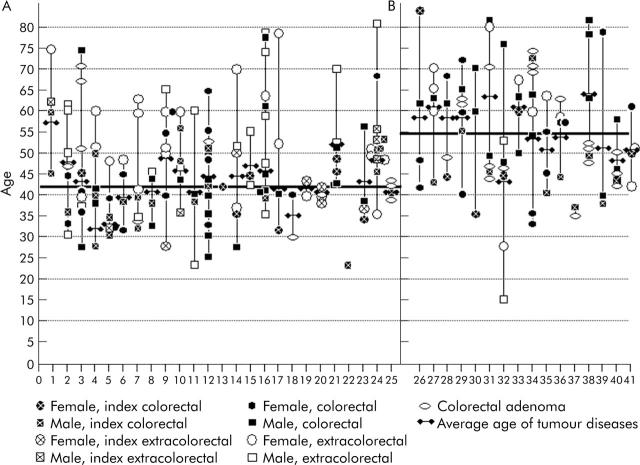
Figure 2.
Mutation positive (A) and mutation negative (B) families with age of onset and tumour type.
DHPLC analysis of MLH1, MSH2, and MSH6
All exons of the MLH1, MSH2, and MSH6 gene were amplified using primers previously published.7–9 MLH1 and MSH2 were analysed for all patients whereas MSH6 was analysed additionally for all patients without mutations in MLH1 or MSH2.
DHPLC analysis was carried out on an automated DHPLC device equipped with a DNA separation column (WAVE; Transgenomic, San Jose, California, USA), as published previously.7
Deletion screening
Genomic DNA of patients without truncating point mutations was analysed for exon deletions for all exons of MLH1 and MSH2 using the MLPA kit according to the manufacturer’s instructions (MRC Holland). Polymerase chain reaction (PCR) products were run on an ABI 3100 genetic analyser.
Microsatellite analysis
After microdissection and extraction, mono- and dinucleotide repeat sequences were used for microsatellite analysis following standard procedures.10,11 The polymorphic markers BAT25, BAT26, D2S123, D5S346, and D17S250 were amplified from tumour DNA and from genomic DNA isolated from peripheral blood leucocytes of the respective patients. If the tumours did not reveal a MSI-H phenotype, an extended marker panel including p53, D7S1824, D18S58, TGFβRII, MYCL1, and D10S197 was analysed.12 PCR products were run on an Applied Biosystems 377 DNA sequencer. Samples were judged as MSI-H showing MSI for at least two of the five or four out of 10 loci, as MSI-low with MSI at one out of four or 1–3 out of 10 loci, and as microsatellite stable without MSI.
Immunohistochemistry for MLH1, MSH2, and MSH6
Monoclonal mouse antihuman MSH2 antibody (Calbiochem Oncogene Research Products, Heidelberg, Germany) and MSH6 antibody (Becton Dickinson, Heidelberg, Germany) were used at a dilution of 1:200 or 1:400, respectively. Monoclonal mouse antihuman MLH1 antibody (Zymed, Berlin, Germany) was used at a dilution of 1:80.
Immunohistochemical stainings on microdissected paraffin embedded tissue blocks were performed following standard procedures.13–15
Staining pattern for the antibodies was nuclear. A tumour was defined as positive if more than 10% of the tumour cells showed a nuclear staining pattern. A negative staining reaction in tumour cells was only regarded as loss of protein expression in the presence of a positive staining reaction in stromal cells and lymphocytes serving as controls. Specificity of staining was proven by replacement of the primary antibody with the mouse IgG isotype (Southern Biotechnology Associates INC., Birmingham, UK) resulting in a negative staining reaction.
Comparative genomic hybridisation (CGH)
Tumorous and non-tumorous DNA was extracted from paraffin embedded tissue by manual microdissection. Dissected DNA and reference DNA of a healthy donor were labelled by nick translation using fluorescent labelled nucleotides. Equal amounts of both were hybridised with human metaphase chromosomes. A computer based analytical system has been used to evaluate chromosomal gains and losses. The CGH technique was applied as described by Aust and colleagues.16
Statistics
Differences in localisation and frequencies of tumour diseases for the MLH1 and MSH2 group as well as for the mutation negative group were analysed using the Mann-Whitney U test. A χ2 test was used to compare the size of the tumour subgroups and age of onset (SPSS software package; SPSS Inc., Chicago, Illinois, USA). Statistical significance was assumed at p<0.05.
RESULTS
Families
We analysed the clinical differences between families with and without mutations in MLH1 or MSH2. We compared 25 families with truncating mutations in MLH1 or MSH2 who were regarded as mutation positive families with 16 Amsterdam positive families without mutations in DNA mismatch repair genes and without MSI (mutation negative families).
Among the 16 Amsterdam positive families without mutations in DNA mismatch repair genes and without MSI, family No 41 showed one gastric and one endometrial cancer apart from two colorectal cancers. Intensive mutation screening with DHPLC for MLH1, MSH2, and MSH6 and deletion screening for MLH1 and MSH2 did not reveal a disease causing mutation in this special case, so the family was judged to be mutation negative. MSH6 was also screened in family No 27 where two colorectal and one endometrial cancer had occurred.
Mutation in MLH1 and MSH2
Twenty different mutations in 25 families were identified. The precise genetic changes in the MLH1 and MSH2 genes of the families are shown in table 1 ▶.
Table 1.
Results of microsatellite analysis and immunohistochemistry for patients with truncating mutations in MLH1 or MSH2
| Pedigree | Exon | Codon/ amino acid change | Nucleotide change | Consequence | Criteria | MSI quotient* | MLH1 EXP | MSH2 EXP | MSH6 EXP | Reference |
| 1 | H1 3 | R100X | c.298 C>T | Nonsense | B† | 4/5 | + | + | ND | Samowitz17 |
| 2 | H1 9 | c.790+1G>A | Splice defect | A I‡ | 4/5 | − | + | + | Cunningham18 | |
| 3 | H1 9 | c.790+1del G | Splice defect | A I | 5/5 | + | + | Novel | ||
| 4 | H1 9 | c.790+1delG | Splice defect | A I | 6/7 | − | + | + | Novel | |
| 5 | H1 10 | 295 | c.884+2T>C | Splice defect | A II§ | 5/5 | ND | ND | ND | Novel |
| 6 | H1 13 | 488 | c.1463delA | Frameshift | A I | 4/5 | − | + | + | Novel |
| 7 | H1 13 | 497 | c.1490insC | Frameshift | B | 4/7 | − | + | ND | Möslein 19 |
| 8 | H1 13 | 497 | c.1490insC | Frameshift | A II | 5/8 | − | + | + | Möslein 19 |
| 9 | H1 13 | 497 | c.1490insC | Frameshift | A II | 5/5 | − | − | ND | Möslein 19 |
| 10 | H1 13 | 497 | c.1490insC | Frameshift | B | 5/7 | − | + | ND | Möslein 19 |
| 11 | H1 15 | Y561X | c.1683C>G | Nonsense | B | 5/5 | − | + | + | Novel |
| 12 | H1 16 | c.1732-1G>A | Splice defect | A I | 4/5 | − | + | + | Novel | |
| 13 | H2 4 | 239 | c.717_721delGGACCinsTTA | Frameshift | B | 7/7 | + | − | − | Novel |
| 14 | H2 6 | c.1076+1G>A | Splice defect | B | 0/0 | ND | ND | ND | Novel | |
| 15 | H2 7 | R383X | c.1147C>T | Nonsense | B | 0/0 | ND | ND | ND | Buerstedde20 |
| 16 | H2 7 | 408 | c.1222insT | Frameshift | A I | 6/7 | + | − | ND | Novel |
| 17 | H2 7 | c.1226delAG | Frameshift | A II | 3/5 | ND | ND | ND | Möslein,19 Liu21 | |
| 18 | H2 7 | c.1226delAG | Frameshift | B | 5/5 | + | − | − | Möslein,19 Liu21 | |
| 19 | H2 7 | c.1077- 2A>G | Splice defect | B | 8/8 | + | − | − | Novel | |
| 20 | H2 8 | c.1386+1G>A | Splice defect | MT¶ | 5/7 | + | − | ND | Novel | |
| 21 | H2 8 | Q429X | c.1285C>T | Nonsense | A I | 5/5 | + | − | ND | Wijnen 22 |
| 22 | H2 9 | 469 | c.1408delG | Frameshift | B | 8/8 | + | − | ND | Novel |
| 23 | H2 9 | c.1497delA | Frameshift | AI | 5/5 | + | − | ND | Novel | |
| 24 | H2 11 | c.1720delC | Frameshift | B/MT | 4/5 | + | − | ND | Novel | |
| 25 | H2 12 | c.2005 + 2T>C | Splice defect | A I | 5/5 | + | − | ND | Novel |
*MSI quotient, number of unstable microsatellite markers to the number of markers tested.
†Bethesda positive; ‡Amsterdam I positive; §Amsterdam II positive; ¶Muir Torre. ND, not determined.
EXP, expression of protein in immunohistochemical staining of tumour tissue.
Of these 25 mutations, 12 were detected in MLH1 and 13 in MSH2, including 14 different new truncating mutations for MLH1 (six) and MSH2 (eight), seven affecting splice sites, six frameshift mutations, and one nonsense mutation. Remarkably, of the 12 truncating mutations found in MLH1, six (50%) affected only two different nucleotides in MLH1.
Microsatellite analysis
Within the group of 25 mutation positive families, we were able to analyse colorectal cancers from 23 families, all showing high grade MSI (for details see table 1 ▶).
Within the 16 mutation negative Amsterdam positive families, tumours of 10 families were available for analysis. From six families (Nos 27, 34, 38, 39, 40, 41) no tumour material was available for microsatellite analysis. Tumours showing loss of heterzygosity for one marker of the standard panel were analysed with the additional marker panel. None of the markers tested showed MSI.
Immunohistochemistry
Within the MSH2 mutation positive families, we were able to analyse tumours for 10 of 13 families. All tumours showed complete loss of MSH2 protein expression in tumour cells. Three tumours (Nos 13, 18, and 19) additionally analysed for MSH6 showed complete loss of protein expression. Within the MLH1 mutation positive families, we analysed tumour samples for 11 of 12 families. Ten tumours revealed complete loss of MLH1 expression while one tumour (No 1) showed decreased expression. Neither nucleotide exchanges nor splice mutations resulted in residual protein expression, detected by immunohistochemistry. Within the mutation negative group, all tumours analysed showed positive immunohistochemistry for the mismatch repair proteins tested (table 1 ▶).
Comparative genomic hybridisation
CGH experiments were done for tumours from three families (Nos 29, 30, 31) in the mutation negative group. Tumour tissue of the index patient of family No 29 showed losses for chromosomes 2q22, 4q, 6q12-q14, 11q13-q22, 18q, and gains for 13q, 16p, 20q, and 22q. Tumour tissue of the index patient of family No 30 showed losses for chromosome 18q and gains for 13q12-q14, 19q, and 20q. Tumour tissue of the index patient of family No 31 showed gains for chromosomes 7, 13q, 16p, 20q, and 20p11·2-qter, without clearly detectable losses.
Cancer occurrence
Colorectal cancers and adenomas
Colorectal cancer was the most frequently observed tumour, with 141 colorectal cancers in 41 families. The highest numbers of colorectal cancers were found in the mutation negative group (78%), followed by families with truncating mutations in MLH1 (70%), and then families with mutations in MSH2 (50%).
Three families (Nos 2, 12, 25) in the mutation positive group however showed 7, 12, and 3 colorectal cancers, respectively, without any extracolonic cancer occurrence. Age of onset for all documented cancers diagnosed in the families is shown in fig 2 ▶. The average age of onset for any tumour disease in the mutation positive families was 42 years compared with 54 years in mutation negative families (p<0.001) (table 2 ▶).
Table 2.
Clinical features of mutation positive and negative families
| Mutation positive | Mutation negative | p Value* | |
| Families | 25 | 16 | |
| Patients (with tumours and colorectal adenomas) | 106 | 71 | |
| Patients with tumours only | |||
| Index CRC | 21 | 16 | |
| Non-index CRC | 74 | 35 | |
| Non-CRC | 27 | 14 | |
| Overall | 101 | 65 | |
| Sex (all patients with tumours and CRC adenoma) | |||
| Female | 50 | 33 | |
| Male | 56 | 38 | |
| Sex tumour patients only | |||
| Female | 48 | 31 | |
| Male | 53 | 34 | |
| Overall | 101 | 65 | NS |
| CRC patients | |||
| Female | 25 | 20 | |
| Male | 40 | 32 | |
| Overall | 65 | 52 | NS |
| Non-CRC patients | |||
| Female | 23 | 11 | |
| Male | 13 | 2 | |
| Overall | 36 | 13 | NS† |
| Age of onset | |||
| Index CRC | 37.6 (39)‡ | 49.7 (45) | 0.0024 |
| All CRC | 43.5 (41) | 54.8 (55) | <0.001 |
| All tumours | 45.5 (43) | 54.8 (56) | 0.022 |
| Family age | 42.2 (42) | 54.1 (54·5) | <0.001 |
| Tumours | |||
| CRC | 89 | 52 | |
| Extracolorectal | 52 | 15 | 0.025 |
| Endometrial | 14 | 3 | |
| Gastric | 7 | 3 | |
| Small bowel | 7 | 0 | |
| Breast | 6 | 0 | |
| Other | 18 | 8 | NS |
| Gastrointestinal | 105 | 55 | |
| Non-gastrointestinal | 36 | 11 | NS |
| Overall | 141 | 66 | |
| Localisation index CRC | |||
| Proximal | 13 | 3 | |
| Splenic flexure | |||
| Distal | 8 | 13 | 0.010 |
| Splenic flexure | |||
| Syn-/metachronic (patients) | |||
| CRC | 12 | 1 | 0.017 |
| Extracolorectal tumours | 17 | 0 | <0.001 |
| Colorectal adenomas | 6 | 9 | (0.084) |
| Colorectal adenoma | |||
| Patients | 10 | 13 | (0.067) |
| Polyps | 15 | 20 | 0.030§ |
CRC, colorectal cancer; NS, not significant.
*Mann-Whitney U test/χ2 test.
†Tested with and without endometrial cancer.
‡Mean (median).
§Compared with number of CRC.
Fifty six colorectal cancers were diagnosed in the MLH1 mutation positive group and 33 were diagnosed in the MSH2 mutation positive group. The clinical features of the mutation positive families are shown in figs 1 ▶ and 3 ▶.
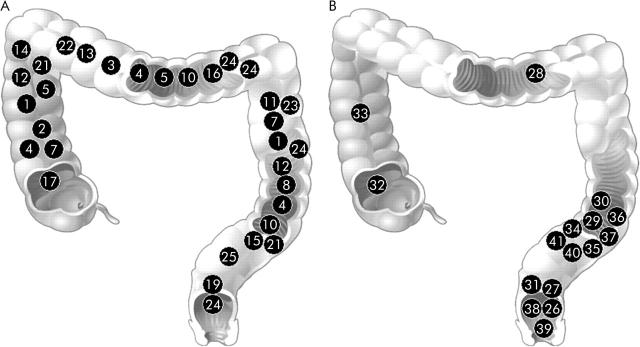
Figure 3.
Localisation of colorectal cancers of 25 families with truncating mutations in MLH1 or MSH2 (family Nos 1–25) (A) and 16 families without mutations (family Nos 26–41) (B).
The difference in age of onset within families increased with the number of tumours observed in families and reached a maximum of 50 years in one family. Mean age of onset for 89 colorectal cancers in the mutation positive group was 43.5 years.
Synchronous and metachronous colorectal cancers were found in 9% of mutation positive patients. Fifty three per cent of all index colorectal cancer tumours in the mutation positive group were diagnosed before the age of 40 years compared with only 13% in the mutation negative group, resulting in a significantly earlier age of onset for all colorectal cancers (median 41 v 55 years; p⩽0.001) when comparing groups 1 and 2. There were 12 syn- or metachronous colorectal cancers in the mutation positive group but just one synchronous colorectal cancer in the mutation negative group (p = 0.017).
Localisation of clearly documented colorectal cancer index cases showed a striking difference between the mutation positive and mutation negative families (fig 3 ▶). In the mutation positive families, 68% were found proximally and 32% distally of the splenic flexure. In the mutation negative families, apart from 3 (21%) index colorectal cancer tumours proximal to the splenic flexure, all clustered in the sigmoid colon or rectum (p = 0.010). Among 53 colorectal cancers, 20 adenomas were documented in the mutation negative group whereas only 15 adenomas but 89 colorectal cancers were documented in the mutation positive group (p = 0.030) (table 2 ▶).
Extracolonic cancers
The highest percentage of associated extracolorectal tumours was observed in the MSH2 mutation positive group: endometrial cancer (10%) followed by small bowel cancer (7%), gastric cancer (5%), CNS cancer (3%), breast cancer (3%), and sebaceous cancer (3%). Other tumours (oesophageal, skin, brain, urothelial, etc) were found in 19%. In the MLH1 mutation positive group, endometrial cancer (10%) was followed by gastric cancer (5%), breast cancer (5%), and small bowel cancer (4%). Other tumours were found in 6%. Comparing the occurrence of cancer types in the MLH1 and MSH2 mutation positive groups, the percentage of tumours was similar for endometrial, gastric, small bowel, and breast cancer but different for others, such as skin, brain, and urothelial tumours which were more frequent in the MSH2 mutation positive group. Comparing the MLH1 and MSH2 mutation positive groups, there was a trend regarding the occurrence of colorectal cancer compared with extracolorectal cancers (p = 0.087).
In the mutation negative group, 78% of the cancers diagnosed were colorectal cancer, followed by endometrial cancer (5%) and gastric cancer (5%). A striking difference was found for the occurrence of syn- or metachronous extracolorectal cancers, with 17 syn- or metachronous extracolorectal cancers in the mutation positive group compared with none in the mutation negative group (p<0.001).
DISCUSSION
High throughput and sensitivity techniques such as DHPLC make it easier to identify mutations in families suspected of having HNPCC.7 Nevertheless, it is important to have pre-selection based on family history.23,24 Selection of families on the basis of the Amsterdam criteria achieves a high rate (approximately 60%) of mutation detection. All of our families underwent genetic counselling with extensive evaluation of their family histories. We analysed MSI, immunohistochemistry, and the MLH1 and MSH2 genes for all index patients of families fulfilling the Amsterdam criteria, and the MSH6 gene if no mutation was found in MLH1 or MSH2. For families or patients fulfilling the Bethesda criteria, we first performed microsatellite analysis and immunohistochemistry followed by mutation analysis in case of MSI or loss of expression in the tumour tissue. In this way, we identified 25 families with disease causing truncating mutations in MLH1 or MSH2. We also defined 16 families fulfilling the Amsterdam criteria without truncating mutations in DNA mismatch repair genes and without MSI in their corresponding tumours in at least 10 of these families. These 41 families formed the basis for comparison of the clinical features of these two molecular defined entities. The major clinical differences found were age of onset, tumour spectrum, tumour localisation, and tumour progression.
Comparing the mutation positive and mutation negative groups, there was a significantly earlier age of onset for all colorectal tumours (median 41 v 55 years; p⩽0.001) and for all tumours (median 43 v 56 years; p = 0.022) in the mutation positive group. No difference in the age of onset for colorectal cancer was found between the MLH1 and MSH2 mutation positive groups.
The clinical features in the mutation positive group resembled those of known mutation positive HNPCC patients.25 The most frequent type of cancer diagnosed was colorectal cancer (63%) followed by endometrial cancer (10%), gastric cancer (5%), and cancer of the small bowel (5%). Other cancer types with typical mutations in DNA mismatch repair genes such as CNS and skin cancer were also observed. In the mutation negative group, we found 78% colorectal cancers, in addition to gastric (5%) and endometrial (5%) cancers. Other associated cancers typical of mutation positive HNPCC patients were not observed. A striking difference between the mutation positive and negative groups was the rate of syn- or metachronous extracolorectal cancer (p<0.001) where none was observed in the mutation negative group.
Wijinen et al described a method according to which a low average age of onset, presence of endometrial cancer, and fulfilment of the Amsterdam criteria increased the probability of mutation identification in HNPCC families.23 This is in agreement with our results with the exception of family Nos 27 and 41 which fulfilled the Amsterdam criteria and showed a tumour spectrum typical of mutation positive families.
Another striking difference between the mutation negative and positive groups was the localisation of colorectal cancers. Within the mutation positive group, 68% of the exactly documented colorectal cancers were found proximal to the splenic flexure, representing distribution over the whole colorectum. In the mutation negative group, 79% of cancers were located distal to the splenic flexure, all in the sigmoid colon and rectum. Hence, approximately 45% of the first colorectal cancers were proximal to the splenic flexure or 46% if metachronous cancers are taken into account. Other studies involving Amsterdam positive families with and without mutations in DNA mismatch repair genes showed 70% of colorectal cancers located proximal to the splenic flexure.25 The only explanation for this discrepancy is selection bias towards a higher number of Amsterdam positive families without mutations in DNA mismatch repair genes for the purposes of this special study.
Another important difference was the number of adenomas observed in the mutation negative and mutation positive groups. In the second group, apart from 89 colorectal cancers, 12 adenomas (14%) were documented, whereas in the mutation negative group 53 colorectal cancers were found and 20 adenomas (28%). Jass has advocated the “aggressive adenoma” theory that adenomas arise in HNPCC patients as often as in the general population; nevertheless, once formed, they progress to carcinoma faster or more frequently or both than their sporadic counterparts.26 Jass however did not discriminate between adenomas from mutation positive and mutation negative HNPCC families. The data presented here showed that adenomas in mutation positive patients were less frequent, but there was an almost equal number of colorectal cancers. Therefore, adenomas in mutation positive patients may proceed quickly to carcinomas.
Families with hereditary colorectal cancer without mutations in DNA repair genes, with a later age of onset, and a reduced frequency of associated cancers, seem to represent one subgroup of hereditary colorectal cancers with underlying highly penetrant germline mutations in as yet unknown genes, which overlap with sporadic cases that are caused mainly by exogenic factors and probably low penetrant mutations. Nevertheless, they seem to have the chromosomal instability (CIN) pathway in common. This was highlighted by the CGH data from three tumours in the mutation negative group which showed CIN and aneuploid DNA, typical of sporadic tumours following the CIN pathway.
Within the mutation positive group there was a tendency towards more extracolonic cancers observed in the MSH2 mutation positive group, with only 50% colorectal cancers versus 70% in the MLH1 mutation positive group. Including all gastrointestinal cancers such as gastric, small bowel, and oesophageal cancer, 68% of the cancers were within the gastrointestinal tract for the MSH2 and 79% for the MLH1 mutation positive group, which was not statistically significant.
Several studies have reported a higher risk of endometrial cancer in MSH2 mutation carriers.25,27,28 In our study, the most frequently diagnosed extracolonic cancer was endometrial cancer, with 10% in the MLH1 and 11% in the MSH2 mutation positive group, which suggests no difference in the risk of endometrial cancer for the two mutated genes.
Two publications describing the clinical differences and similarities between mutation positive and mutation negative families have been published. Bisgaard and colleagues29 found that families without a mutation fell into two clearly distinguishable subgroups. The major group without mutations had a significantly lower frequency of multiple colorectal cancers, as well as a lower frequency of HNPCC related cancers. Furthermore, rectal cancer was diagnosed more often and age of onset tended to be later than for mutation carriers. The minor group without mutations generally had the same characteristics as found in affected persons with identified mutations. Because tumours within this last group showed MSI, the authors assumed that deletions within the DNA mismatch repair genes might be causative. The clinical features for the major group of patients without mutations resembled the clinical features of our Amsterdam positive mutation negative group.
Data from Scott and colleagues30 concur with the results of our study cohort: a significantly later age of onset in the mutation negative group (+5 years; our study +10 years) and more endometrial and stomach cancers in the mutation positive group. However, in the study of Scott et al, statistical significance was reached only for the MLH2 mutation positive group.30 In this study, missense mutations of uncertain pathology were found in the MLH1 and MSH2 groups. No data concerning localisation of colorectal cancers, MSI, or immunohistochemistry were provided. Missense mutations may increase the average age of onset due to lower penetrance or missing pathology. The pathogenic relevance of these missense variants has yet to be revealed. Furthermore, genomic deletions are not detected by DGGE, which was used for mutation detection by Scott and colleagues.30
Our data show that a thorough evaluation of family history, including mutation and microsatellite analysis as well as immunohistochemistry in combination with clinical documentation, allows classification of Amsterdam positive families into two different clinical entities.
The major clinical differences were later age of onset, tumour spectrum, distal localisation of colorectal cancers, and slower tumour progression in mutation negative families. Firstly, this may have implications for surveillance programmes, especially in terms of intervals, as there was no evidence of accelerated tumour progression in the mutation negative group. Secondly, cancers in the mutation positive group followed the MIN pathway whereas tumours in the mutation negative group seemed to follow the CIN pathway. The search for the highly penetrant germline mutations causing the cancers in the mutation negative group should be performed for candidate genes in the CIN pathway.
Acknowledgments
This work was supported by the German Cancer Aid-Mildred Scheel Foundation.
Y Mueller-Koch and H Vogelsang substantiated the data collected equally, evaluated the data, and prepared the manuscript, including genetic and clinical counselling. R Kopp carried out patient counselling, data collection for the Department of Surgery, and participated in the discussion of the data. P Lohse was responsible for microsatellite analysis at the Ludwig Maximilian University,. G Keller was responsible for microsatellite and mutation analysis at the University of Polytechniques. M Muders and I Becker carried out tissue preparation for microsatellite analysis, immunohistochemistry, and histopathological evaluation of tumour and adenoma slides at the Ludwig Maximilian University and University of Polytechniques. D Aust was responsible for pathology reports as well as the CGH investigation. M Gross and U Schiemann were responsible for clinical counselling, endoscopic, diagnostic, and surveillance programmes. M Scholz was in charge of the internet platform and database necessary for multicentre documentation. B Kerker, J Daum, and M Grabowski were responsible for microsatellite and mutation analysis in two different laboratories (Ludwig Maximilian University). G Henke was in charge of data collection and data coordination. E Holinski-Feder was the laboratory supervisor at the Ludwig Maximilian University, in charge of the coordination of the whole working group, and together with Y Mueller-Koch and H. Vogelsang responsible for manuscript preparation.
Abbreviations
HNPCC, hereditary non-polyposis colorectal cancer
CGH, comparative genomic hybridisation
CIN, chromosomal instability
MSI, microsatellite instability
MSI-H, high microsatellite instability
DHPLC, denaturing high performance liquid chromatography
PCR, polymerase chain reaction
Published online first 14 June 2005
REFERENCES
- 1.Lynch HT, Watson P, Lanspa S, et al. Clinical nuances of Lynch syndromes I and II. Prog Clin Biol Res 1988;279:177–88. [PubMed] [Google Scholar]
- 2.Lynch HT, Lanspa S, Smyrk T, et al. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I & II). Genetics, pathology, natural history, and cancer control, Part I. Cancer Genet Cytogenet 1991;53:143–60. [DOI] [PubMed] [Google Scholar]
- 3.Lynch HT, Watson P, Lanspa SJ, et al. Natural history of colorectal cancer in hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II). Dis Colon Rectum 1988;31:439–44. [DOI] [PubMed] [Google Scholar]
- 4.Aaltonen LA, Peltomaki P, Mecklin JP. et al Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res 1994;54:1645–8. [PubMed] [Google Scholar]
- 5.Peltomaki P, Lothe RA, Aaltonen LA. et al Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res 1993;53:5853–5. [PubMed] [Google Scholar]
- 6.Kuska B. New diagnostic criteria for HNPCC are on the way. J Natl Cancer Inst 1997;89:11–12. [DOI] [PubMed] [Google Scholar]
- 7.Holinski-Feder E, Muller-Koch Y, Friedl W, et al. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J Biochem Biophys Methods 2001;47:21–32. [DOI] [PubMed] [Google Scholar]
- 8.Edelmann W, Yang K, Umar A, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell 1997;91:467–77. [DOI] [PubMed] [Google Scholar]
- 9.Hutter P, Couturier A, Scott RJ, et al. Complex genetic predisposition to cancer in an extended HNPCC family with an ancestral hMLH1 mutation. J Med Genet 1996;33:636–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res 1997;57:4749–56. [PubMed] [Google Scholar]
- 11.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–57. [PubMed] [Google Scholar]
- 12.Schiemann U, Muller-Koch Y, Gross M, et al. Extended microsatellite analysis in microsatellite stable, MSH2 and MLH1 mutation-negative HNPCC patients: genetic reclassification and correlation with clinical features. Digestion 2004;69:166–76. [DOI] [PubMed] [Google Scholar]
- 13.Hendriks Y, Franken P, Dierssen JW, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 2003;162:469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Leeuw WJ, Dierssen J, Vasen HF, et al. Prediction of a mismatch repair gene defect by microsatellite instability and immunohistochemical analysis in endometrial tumours from HNPCC patients. J Pathol 2000;192:328–35. [DOI] [PubMed] [Google Scholar]
- 15.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002;20:1043–8. [DOI] [PubMed] [Google Scholar]
- 16.Aust DE, Willenbucher RF, Terdiman JP, et al. Chromosomal alterations in ulcerative colitis-related and sporadic colorectal cancers by comparative genomic hybridization. Hum Pathol 2000;31:109–14. [DOI] [PubMed] [Google Scholar]
- 17.Samowitz WS, Curtin K, Lin HH, et al. The colon cancer burden of genetically defined hereditary nonpolyposis colon cancer. Gastroenterology 2001;121:830–8. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham JM, Kim CY, Christensen ER, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet 2001;69:780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Möslein G, Tester DJ, Lindor NM, et al. Microsatellite instability and mutation analysis of hMSH2 and hMLH1 in patients with sporadic, familial and hereditary colorectal cancer. Hum Mol Genet 1996;5:1245–52. [DOI] [PubMed] [Google Scholar]
- 20.Buerstedde JM, Alday P, Torhorst J, et al. Detection of new mutations in six out of 10 Swiss HNPCC families by genomic sequencing of the hMSH2 and hMLH1 genes. J Med Genet 1995;32:909–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Wahlberg S, Rubio C, et al. DGGE screening of mutations in mismatch repair genes (hMSH2 and hMLH1) in 34 Swedish families with colorectal cancer. Clin Genet 1998;53:131–5. [DOI] [PubMed] [Google Scholar]
- 22.Wijnen J, Vasen H, Khan PM, et al. Seven new mutations in hMSH2, an HNPCC gene, identified by denaturing gradient-gel electrophoresis. Am J Hum Genet 1995;56:1060–6. [PMC free article] [PubMed] [Google Scholar]
- 23.Wijnen J, Khan PM, Vasen H, et al. Hereditary nonpolyposis colorectal cancer families not complying with the Amsterdam criteria show extremely low frequency of mismatch-repair-gene mutations. Am J Hum Genet 1997;61:329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinimann K, Scott RJ, Buerstedde JM, et al. Influence of selection criteria on mutation detection in patients with hereditary nonpolyposis colorectal cancer. Cancer 1999;85:2512–18. [DOI] [PubMed] [Google Scholar]
- 25.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet 1999;36:801–18. [PMC free article] [PubMed] [Google Scholar]
- 26.Jass JR. Replication error phenotype in colorectal cancer. Gut 2000;47:597–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin KM, Shashidharan M, Thorson AG, et al. Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg 1998;2:67–71. [DOI] [PubMed] [Google Scholar]
- 28.Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999;116:1453–6. [DOI] [PubMed] [Google Scholar]
- 29.Bisgaard ML, Jager AC, Myrhoj T, et al. Hereditary non-polyposis colorectal cancer (HNPCC): phenotype-genotype correlation between patients with and without identified mutation. Hum Mutat 2002;20:20–7. [DOI] [PubMed] [Google Scholar]
- 30.Scott RJ, McPhillips M, Meldrum CJ, et al. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am J Hum Genet 2001;68:118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]