Abstract
Background and aim: The importance of the hepatocyte ploidisation pattern to the control of cell proliferation and differentiation has been well established. However, there are no data that have characterised hepatocyte ploidy at various stages of chronic liver inflammation and fibrosis in vivo.
Methods: We therefore investigated hepatocyte ploidy/binuclearity patterns in 57 patients with chronic hepatitis, using a recently developed methodology which allows simultaneous hepatocyte ploidy and binuclearity analyses on the same liver section.
Results: The percentage of mononuclear diploid hepatocytes was significantly reduced in patients with high hepatitis activity and marked fibrosis (low activity: 75.1 (18.8)% v high activity: 61.8 (21.6)%, p = 0.0111, and low fibrosis: 77.3 (13.8)% v high fibrosis: 57.4 (23.3)%, p = 0.0002). Accordingly, the percentage of mononuclear polyploid hepatocytes increased in patients with high hepatitis activity and marked fibrosis (low activity: 11.9 (15.5)% v high activity: 22.2 (20.1)%, p = 0.0166, and low fibrosis: 9.4 (10.7)% v high fibrosis: 26.4 (21.6)%, p = 0.0001). In addition, the fraction of binuclear hepatocytes was significantly higher in patients with hepatitis B virus (HBV) than in those with hepatitis C virus (HCV) infections (HBV: 18.2 (7.6)% v HCV: 12.0 (4.8)%; p = 0.0020). Under multivariate analysis, HBV infection was an independent factor accounting for the larger binuclear hepatocyte fraction (p = 0.0294).
Conclusion: Our results revealed an increase in the polyploid hepatocyte fraction which correlates with the severity of chronic hepatitis; moreover, we demonstrated that HBV and HCV related chronic hepatitis exhibited distinctive hepatocyte ploidy patterns, thus allowing the suggestion that these two viral infections may modulate liver ploidy through different mechanisms.
Keywords: chronic viral hepatitis, DNA ploidy, binuclearity, hepatocyte proliferation
Hepatocyte polyploidisation and binucleation are important features of liver growth and physiology. This process starts in the liver after birth, with the successive appearance of polyploid cell classes. In the liver of the newborn rat, hepatocytes are exclusively diploid (2n) whereas in the liver of the adult rat, about 10% of hepatocytes are diploid, 70% are tetraploid, and 20% octoploid. If we consider the polyploid fraction, 20–30% of hepatocytes are binuclear (either 2×2n or 2×4n).1,2 The degree of polyploidisation varies among mammals,3 and particularly in humans where the number of polyploid cells averages 20–45% in adults.4–7 There is evidence for the importance of the control of hepatocyte polyploidisation in both the normal and pathological liver. Polyploidy in the liver is generally considered to indicate terminal differentiation and senescence8,9,10 and to lead to both progressive loss of cell pluripotency and markedly decreased replication capacity.11 Within the liver nodule, differences in the ploidy of hepatocytes are recognised, with periportal cells exhibiting less ploidy and perivenous cells greater ploidy.12 Liver polyploidisation is differentially regulated on loss of liver mass and liver damage. Partial hepatectomy induces marked cell proliferation followed by an increase in liver ploidy, with a reduction in the proportion of binuclear cells and a rise in the proportion of tetraploid mononuclear cells and, to a lesser extent, octoploid cells.1,11,13
In contrast, liver lesions induced in the rat by antioxidant enzymes (copper-zinc-superoxide dismutase, glutathione peroxidase, or aminoguanidine)14,15 or chemical carcinogens (diethylnitrosamine and 2-acetyl-aminofluorene)2,16 lead to an overall reduction in liver ploidy and expansion of the diploid cell population. Studies in humans have also shown a shift towards diploid cell growth during hepatocarcinogenesis. Increases in diploid mononuclear hepatocytes with a decrease in polyploid hepatocytes (including binuclear hepatocytes) have been reported in human euploid hepatocellular carcinoma (HCC).6,7,17,18 Diploid cells are also frequently detected in preneoplastic nodules, suggesting an early shift to diploid cell expansion during hepatocarcinogenesis.2,19
However, only a few studies have analysed hepatocyte ploidy/binuclearity in human chronic hepatitis liver samples.6,17 Therefore, it is still unclear at which stage of chronic hepatitis a shift in the hepatocyte ploidy/binuclearity pattern occurs.
We have recently developed a method based on quantitative fluorescence imaging to determine in vivo the profile of liver ploidy.20 Our investigations have shown that this method is suitable for determining the proportion of mononuclear 2n, binuclear 2×2n, and mononuclear 4n hepatocytes in liver samples. Thus we performed detailed analyses of patients with chronic hepatitis without HCC who displayed different activity and fibrosis scores (including cirrhosis) to clarify the hepatocyte ploidy pattern during chronic inflammation and the early stages of liver carcinogenesis.
PATIENTS AND METHODS
Patients
Fifty seven patients with histologically and virologically confirmed chronic hepatitis B or C and various grades of activity and fibrosis were enrolled in the study (41 males and 16 females; mean age 39.1 years). An ultrasonography guided needle liver biopsy had been performed in all patients between March 1996 and March 2001. Twenty four patients were chronically infected with hepatitis B virus (HBV) while the remaining 33 patients were infected with hepatitis C virus (HCV). HBV patients had detectable hepatitis B surface antigen. At the time of liver biopsy, 11 patients had negative HBV DNA and 13 had positive serum HBV DNA (eight were infected by pre-C mutant virus and five by wild-type virus) using a hybridisation test (cut off value 700 000 copies/ml). There was no evidence of occult HBV infection in HCV patients.
All patients included in this study had no evidence of alcohol related liver disease. Based on histological findings, patients were grouped as a function of the degree of activity of hepatitis and degree of fibrosis, according to the METAVIR scoring system.21
The study was approved by the institutional review board for Hopital Necker Enfants Malades (Paris, France) and was carried out in accordance with the Helsinki declaration. Written informed consent was obtained from all patients before liver biopsy and prior to the study.
Immunofluorescence microscopy: definition of liver DNA ploidy
Liver tissues fixed in 10% phosphate buffered formalin were embedded in paraffin. Tissue sections (3 μm thick) were obtained using a conventional microtome. Sections were deparaffinised in xylene and placed in 100% ethanol. Sections were rehydrated in a descending gradient of ethanol-water and then boiled for 3×5 minutes in 10 mmol/l citrate buffer, pH 6.2, in a microwave oven. All subsequent antibody incubations were carried out in an antibody diluent (Dako, Trappes, France). Incubation with the primary antibody (anticytokeratin 1:50; KL1, Immunotech, SA, Marseille, France) was carried out for one hour at room temperature, followed by three washes with phosphate buffered saline (PBS). A secondary reagent was used to detect the primary antibody: biotinylated swine antigoat, mouse, rabbit immunoglobulin solution (1:200; Dako) (30 minutes’ incubation) followed by FITC conjugated streptavidin (1:200; Dako) (30 minutes’ incubation). Hoechst 33342 (1 μg/ml) was included in the last incubation to stain DNA (20 minutes’ incubation). Following three final washes with PBS, the slides were mounted in 100% glycerol.
Tissue sections were then examined under a Zeiss (Axiovert 35; Carl Zeiss, Gottingen, Germany) inverted microscope equipped for epi-illuminations. Zeiss Plan Neafluar objectives 40× (Na = 0.75) were chosen, enabling the collection of light from the entire thickness of the nucleus, these conditions being essential for adequate determination of DNA content.22 Images were captured using a cooled charged coupled device camera (KAF 1400-G2, class 2; Photometrics, Tucson, Arizona, USA) on 4056 grey levels. Automatic quantitative image analysis was performed in 12 bits using the IPLab Spectrum version 3.1 software. For measurement of nuclear DNA content on the image, we carefully selected only nuclei with a clear edge in order to avoid the effect of diffraction on the evaluation. Nuclei were termed mono- or binucleated hepatocytes by comparing fluorescent and membrane labelling images. Other liver cell types (defined by their morphological characteristics) overlapping nuclei or debris were eliminated interactively by inspection on the screen, using zoom and contrast enhancement where necessary. Parameters such as integrated fluorescence intensity or area of nucleus were stored in computer files for analysis using the Kaleidagraph and Imastat software (Imastat as developed by P Debey).20,22 A minimum of 300 hepatocytes was studied, observed on 8–12 separate fields (analyse of at least two sections). Fields were randomly selected but always contained the same number of periportal, centrolobular, and mediolobular area from one patient to another. In order to eliminate fluorescence intensity value due to nuclear truncature, a control slide consisting on a pure 2n hepatocyte population was include in each experiments. This slide allowed us to determine the fluorescence intensity values for 2n DNA content; lower values (corresponding to nuclear truncature) were not taken into account. To ensure that the same cells were not counted twice, slides were read by two observers in a systematic manner, moving from the right to the left of the slide and then on successive descending lines. All analyses were performed blind.
Statistics
Results are expressed as mean (SD). Differences in proportions were tested by the χ2 test. Mean quantitative values were compared using the non-parametric Mann-Whitney U test. Correlations were evaluated using Spearman’s test. Multivariate analysis was performed using logistic regression; the variables analysed were age at biopsy (⩽40 years v >40 years), sex, infective virus (HBV v HCV), activity of liver inflammation (A0/A1 v A3), and degree of liver fibrosis (F0-2 v F3/F4). Data analyses were performed with the SAS statistical package version 6 (SAS Institute, Cary, North Carolina, USA, 1990). All p values were two tailed and a level of <0.05 was accepted as statistically significant.
RESULTS
Ploidy/binuclearity analyses by in vivo fluorescence imaging
Using a recently developed approach,20 we analysed the modification of ploidy during chronic hepatitis. By simultaneous nuclear and plasma membrane labelling of liver tissue sections, we were able to determine the number of nuclei per cell and thus calculate the binuclear hepatocyte fraction (fig 1A ▶). Moreover, the use of a stoichiometric DNA labelling agent, Hoechst 33342, allowed determination of the DNA content in all cells by measuring either the area of each nucleus or the intensity of fluorescence. For each measurement, a typical histogram of the data obtained per section is shown in fig 1B ▶ (area: left panel, intensity: right panel). The first peak is representative of a hepatocyte population with 2n DNA content. The second peak is positioned around twice the value of the first peak and is representative of hepatocyte populations with 4n DNA content. Statistical analysis was performed to calculate the distribution of each hepatocyte population. For each sample analysed there was a significant correlation between the proportion of 2n and 4n nuclei determined by measuring the intensity of fluorescence and that obtained by measuring the area of nuclei (Spearman’s rank correlation 0.85; p<0.001 for all patients); an example in one patient is shown in fig 1C ▶. Using this approach, we then determined hepatocyte ploidy in vivo during HBV and HCV associated chronic hepatitis.
Figure 1.
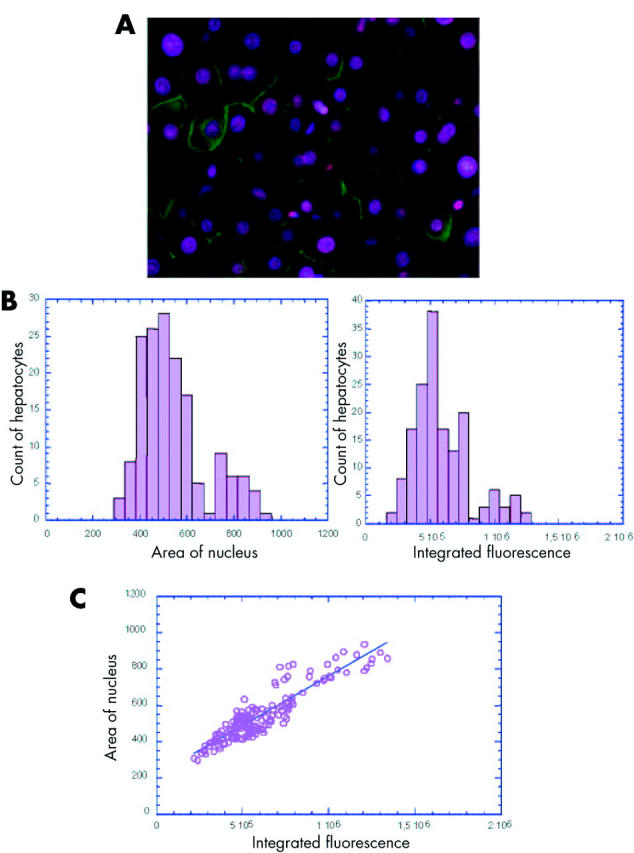
In vivo analysis of hepatocyte ploidy/binuclearity in patients with chronic hepatitis. (A) Image of a liver section after double staining with Hoechst 33342 (nuclear labelling, pink) and cytokeratin (plasma membrane labelling, green) which enabled a distinction between mononuclear and binuclear hepatocytes. (B) Representative histogram of the DNA content distribution of mononuclear hepatocytes for one patient. DNA content was evaluated by recording either the area of each nucleus (left panel) or the intensity of Hoechst integrated fluorescence (right panel). The first peak is representative of hepatocytes with 2n DNA content. The second peak is positioned around twice the value of the first peak and is representative of hepatocytes with 4n DNA content. (C) Integrated fluorescence was correlated with the area of nucleus in each specimen analysed (example for one patient, p<0.0005). A significant correlation was observed between the two parameters (p<0.001 for the 57 patients, Spearman’s rank correlation 0.85).
We selected 57 patients with chronic hepatitis caused by HBV or HCV infection (24 with HBV and 33 with HCV). Based on their histological findings, we grouped these patients in relation to the degree of activity of hepatitis and the degree of fibrosis, according to the METAVIR scoring system (table 1 ▶).21 It is important to note that hepatocyte ploidy/binuclearity is reported to change according to age in normal human liver.5 Therefore, we selected age matched patients (39.1 (5.0) years) in order to reduce the influence of this parameter. Firstly, we analysed hepatocyte ploidy/binuclearity in age matched normal human liver samples (n = 3). The fractions of mononuclear diploid, mononuclear polyploid, and binuclear hepatocytes were 76.1 (2.7)%, 9.6 (2.2)%, and 14.3 (3.6)%, respectively. Only a very few studies have analysed normal human livers but our values showed a proportion of the different fractions of cells which were “intermediate” between those found in two independent studies.5,7
Table 1.
Patient characteristics
| Total | Low activity (A0, A1) Low fibrosis (F0–F2) | High activity (A3) Low fibrosis (F0–F2) | Low activity (A0, A1) High fibrosis (F3, F4) | High activity (A3) High fibrosis (F3, F4) | |
| No of patients | 57 | 15 | 15 | 11 | 16 |
| Sex (M/F) | 41/16 | 7/8 | 10/5 | 10/1 | 14/2 |
| Age (y) (mean (SD)) | 39.1 (5.0) | 37.5 (4.6) | 39.8 (5.5) | 41.8 (4.6) | 39.1 (5.1) |
| Hepatitis virus (HBV/HCV) | 23/33 | 5/10 | 6/9 | 5/6 | 8/8 |
Activity and fibrosis were scored according to the METAVIR scoring system.21
HBV, hepatitis B virus; HCV, hepatitis C virus.
Ploidy/binuclearity distribution according to the severity scores of the disease
Given the very low fraction of mononuclear 8n and binuclear 2×4n in our studied cell population, the results were expressed globally as mononuclear polyploid and binuclear hepatocytes. Thus proportions of mononuclear diploid hepatocytes, mononuclear polyploid hepatocytes, and binuclear hepatocytes were compared according to activity and fibrosis scores, respectively (fig 2 ▶). The percentage of mononuclear diploid hepatocytes was significantly reduced in both patients with high hepatitis activity and marked fibrosis (low activity: 75.1 (18.8)% v high activity: 61.8 (21.6)%, p = 0.0111 (fig 2A ▶); and low fibrosis: 77.3 (13.8)% v high fibrosis: 57.4 (23.3)%, p = 0.0002 (fig 2B ▶)). Accordingly, the percentage of mononuclear polyploid hepatocytes increased in both patients with high hepatitis activity and marked fibrosis (low activity: 11.9 (15.5)% v high activity: 22.1 (20.1)%, p = 0.0166 (fig 2C ▶); and low fibrosis: 9.4 (10.7)% v high fibrosis: 26.4 (21.6)%, p = 0.0001 (fig 2D ▶)). In contrast, we did not find any difference in binuclear hepatocyte percentages between either patients with low and high activity, or between patients with low and high fibrosis (fig 2E ▶, F). Multivariate analysis showed that fibrosis was an independent factor associated with increased mononuclear polyploid (p = 0.035) and decreased mononuclear diploid (p = 0.04) cell fractions.
Figure 2.
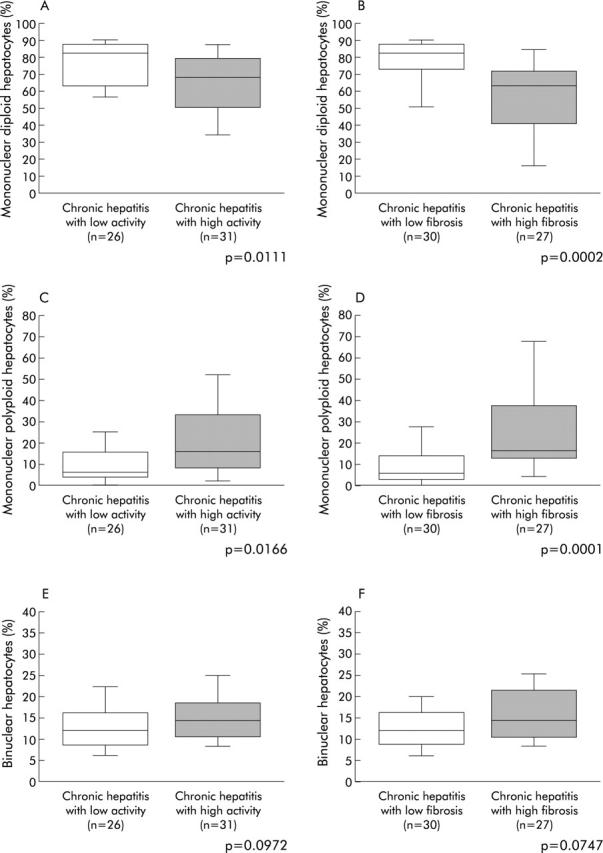
Ploidy/binuclearity distributions, according to the progression of chronic hepatitis. (A, B) Fraction of mononuclear diploid hepatocyte; (C, D) fraction of mononuclear polyploid hepatocytes; and (E, F) fraction of binuclear hepatocytes. (A, C, E) Comparison between low and high activity of inflammation; and (B, D, F) comparison between low and high degrees of fibrosis. Box plots for the percentage of hepatocytes in each population (mononuclear diploid hepatocytes, mononuclear polyploid hepatocytes, and binuclear hepatocytes) out of the total number of hepatocytes analysed. The ends of each box represent the quartiles, and the median of distribution is indicated by a line within the box; n corresponds to the number of patients in each group. All p values were calculated using the Mann-Whitney U test.
These results demonstrated a shift in the distribution of hepatocyte ploidy towards an increase in mononuclear polyploid hepatocytes associated with activity and, more strongly, with fibrosis (that is, severity of chronic hepatitis).
Distribution of the fraction of mononuclear cells in the population of polyploidy hepatocytes (FMP) according to progression of hepatitis
In order to investigate changes in the balance between mononuclear (4n, 8n) and binuclear (2×2n, 2×4n) hepatocytes within the polyploid hepatocyte population, the fraction of mononuclear hepatocytes in the latter (FMP) was calculated for each patient.
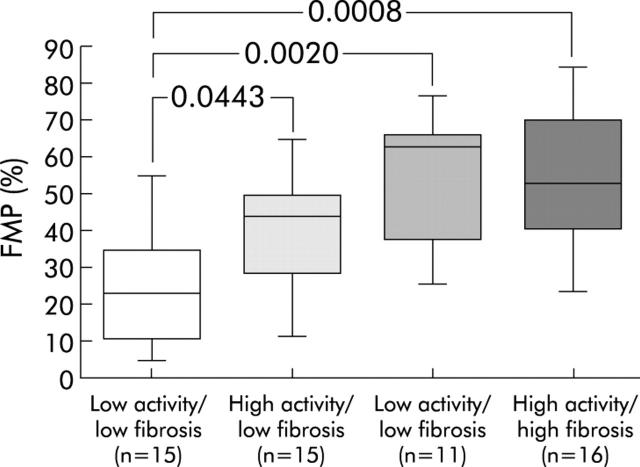
FMP levels increased in patients with high hepatitis activity (low activity: 0.36 (0.24) v high activity: 0.47 (0.22); p = 0.0807) and in those with marked fibrosis (low fibrosis: 0.32 (0.20) v high fibrosis: 0.54 (0.21); p = 0.0005). When the analysis was based on a combination of activity and fibrosis, we observed a gradual increase in FMP levels, together with an increase in the activity and fibrosis scores (fig 3 ▶).
Figure 3.
Comparison of the fraction of mononuclear cells in the population of polyploid hepatocytes (FMP) in patients with chronic hepatitis. Four groups of patients were defined: low activity and low fibrosis; high activity and low fibrosis; low activity and high fibrosis; and high activity and high fibrosis. FMP percentages were calculated as follows: FMP = number of mononuclear polyploid hepatocytes/number of all polyploid hepatocytes (mononuclear polyploid hepatocytes+binuclear hepatocytes). The ends of each box represent the quartiles, and the median of distribution is indicated by a line within the box; n corresponds to the number of patients in each group. All p values were calculated using the Mann-Whitney U test.
Comparison of ploidy/binuclearity profiles during HBV and HCV infections
The distribution of hepatocyte ploidy/binuclearity was compared between patients with chronic hepatitis B and C to test for a direct effect of HBV and HCV on the distribution of hepatocyte ploidy/binuclearity.
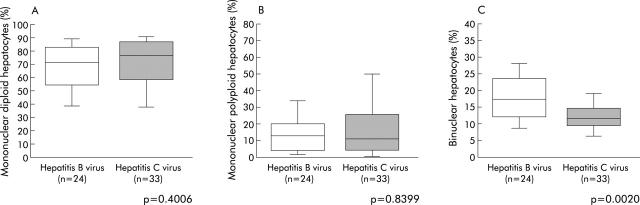
There were no significant differences in the distribution of mononuclear diploid and mononuclear polyploid hepatocyte fractions between patients with HBV and HCV infections although there was a trend towards a lower mononuclear diploid hepatocyte fraction in HBV infected patients (fig 4A, 4B ▶ ▶). In contrast, the fraction of binuclear hepatocytes was significantly higher in patients with HBV than in those with HCV (HBV: 18.2 (7.6)% v HCV: 12.8 (4.8)%; p = 0.0020) (fig 4C ▶). In addition, multivariate analysis showed that HBV infection was an independent factor accounting for the higher percentage of binuclear hepatocytes (p = 0.0294, table 2 ▶). Our study therefore demonstrated a difference in the pattern of hepatocyte binucleation between patients with HBV and HCV related chronic hepatitis.
Figure 4.
Comparison of ploidy/binuclearity distributions between patients with chronic hepatitis B and C. (A) Fraction of mononuclear diploid hepatocyte; (B) fraction of mononuclear polyploid hepatocyte; and (C) fraction of binuclear hepatocyte levels. Box plots of the percentage of hepatocytes in each population (mononuclear diploid hepatocytes, mononuclear polyploid hepatocytes and binuclear hepatocytes) out of the total number of hepatocytes analysed. The ends of each box represent the quartiles, and the median of distribution is indicated by a line within the box; n corresponds to the number of patients in each group. All p values were calculated using the Mann-Whitney U test.
Table 2.
Multivariate analysis of factors affecting the increase in the fraction of binuclear hepatocytes
| Total | Odds ratio (95% CI) | p Value | |
| Age | ⩽40 y v >40 y | 1.5 (0.4–5.8) | 0.5371 |
| Sex | Males v females | 0.7 (0.1–3.6) | 0.6368 |
| Infected virus | HBV v HCV | 0.2 (0.05–0.9) | 0.0294 |
| Activity of inflammation | Low v high | 2.4 (0.6–9.3) | 0.2020 |
| Degree of fibrosis | Low v high | 1.0 (0.3–3.9) | 0.9866 |
Multivariate analysis was performed using logistic regression analysis.
Low activity of inflammation = A0, A1; high activity of inflammation = A3, according to the METAVIR scoring system.21
Low degree of fibrosis = F0–F2; high degree of fibrosis = F3, F4, according to the METAVIR scoring system.21
HBV, hepatitis B virus; HCV, hepatitis C virus; 95% CI, 95% confidence interval.
DISCUSSION
In this study, we assessed the evolution of liver cell ploidy status during progression of chronic hepatitis. We developed a methodology that allowed direct and simultaneous evaluation of cellular DNA content in liver cells identified in tissue sections. Using this approach, we demonstrated that chronic inflammation and fibrosis are associated with a marked increase in polyploid hepatocytes. In addition, our results showed distinct ploidy profiles in liver cells from patients with HBV and HCV related chronic hepatitis, suggesting different molecular pathways for the two viral infections.
In previous studies, human liver cell nucleus DNA content was evaluated in vitro by flow cytometry7,17,18 or Feulgen spectrocytophotometry.4–6,19 In some studies, DNA quantification was combined with a morphological evaluation of hepatocyte binuclearity, performed on serial sections from the same sample.5–7,17 In the present study, we validated a technique which allowed us to simultaneously identify on one tissue section mono- and binuclear liver cells and determine their DNA content in vivo.
Although several studies in humans have investigated patients with HCC only a few made a detailed analysis of liver sections from subjects with chronic hepatitis. In HCC, mononuclear diploid hepatocytes were mostly evidenced in precancerous nodules, early stage HCC, and well differentiated HCC.18,19 An increase in the mononuclear diploid hepatocyte fraction was also noted in the liver of patients with chronic hepatitis or cirrhosis.6,17 However, these studies did not compare patients with differing severities of inflammation and fibrosis, the two principal parameters indicating worsening of chronic hepatitis and progression to cirrhosis and HCC. In fact, we observed a decrease in mononuclear diploid hepatocyte fractions associated with an increase in mononuclear polyploid hepatocyte fractions in patients with high hepatitis activity and marked liver fibrosis. The contrast between our results and those of previous studies is likely to be due to the comparison we made between patients at different stages of liver disease. Furthermore, we selected age matched patients (57 patients, aged 39 (5) years (range 32–49)); this important issue was not taken into account for the 10 patients analysed by Melchiorri and colleagues.6 In addition, for the 18 patients analysed by Anti and colleagues,17 there was a marked distribution for patient age (58 (15) years (range 23–81)). Also, one study reported a similar distribution of ploidy/binuclearity in non-tumoral cirrhotic liver samples adjacent to HCC.7
Another important finding in our study was the increased proportion of mononuclear versus binuclear fractions of polyploid hepatocytes (as estimated by FMP determination) in patients with high fibrosis and activity scores. High FMP levels have been reported in HCC in comparison with chronic hepatitis liver samples, and the suggestion was made that increased FMP levels may be of predictive value regarding the development of HCC.6 The mechanisms of liver cell ploidisation are still largely unknown. The role of binuclear cell formation as an important step towards liver cell ploidisation has been suggested. In fact, we have recently provided direct evidence for a pivotal role of such binuclear cells in the formation of 4n mononuclear cells and suggested the importance of centrosome homeostasis in this process.20 Our results may therefore be consistent with stimulation of binuclear cell DNA replication in the context of chronic inflammation and fibrosis. In addition, they do not exclude the possibility of a direct shift from mononuclear 2n to mononuclear 4n cells, in addition to intermediate binuclear cells.
Overall, our results in human chronic hepatitis liver samples did not demonstrate a shift of hepatocyte ploidy/binuclearity towards a predominance of mononuclear diploid hepatocytes, as was previously reported in human and animal experimentally induced HCC samples. One may hypothesise that the decrease in mononuclear diploid, and increase in polyploid, hepatocytes observed in our patients may reflect compensatory proliferation of hepatocytes in response to continuous loss of hepatocytes caused by chronic hepatitis, as has been observed after partial hepatectomy.13 It is generally accepted that continuous liver injury, unlike partial hepatectomy, may promote the proliferation of mononuclear 2n cells which would then become predominant in pre-tumorous livers. However, in contrast with the situation observed during chemically induced liver carcinogenesis in rodents, our data suggest that in humans, chronic liver inflammation mainly promotes increased hepatocyte ploidisation despite ongoing liver injury. It is plausible that increased ploidisation may represent a protective mechanism as polyploid hepatocytes exhibit advancement towards terminal differentiation and nuclear senescence (apoptosis).9–11 One cannot exclude the possibility of genetic changes in such cells which would favour the emergence of precancerous cell clones.9 Alternatively, however, our results may suggest the expansion of a small proportion of mononuclear 2n liver cells, undetected by our methodology, which would constitute the pre-tumoral cell reservoir.
Another intriguing finding was the different hepatocyte binuclearity profiles in patients infected by HBV and HCV. Thus the proportion of binuclear hepatocytes was significantly increased in patients with HBV compared with HCV related chronic hepatitis. This increase was independent of other factors such as age, sex, activity of inflammation, and degree of fibrosis by multivariate analysis. In contrast, there was no significant difference in overall ploidy status between HBV and HCV infected subjects. The difference in the balance between mononuclear polyploid and binuclear hepatocytes indicated distinct patterns of hepatocyte polyploidisation/binucleation in HBV and HCV viral infections. HBV and HCV both trigger chronic liver inflammation and cirrhosis, a major risk factor for HCC. However, there is now strong evidence that these two viruses also act in liver carcinogenesis through direct and distinct molecular pathways (see reviews by Brechot and colleagues23 and Hildt and Hofschneider24). Our in vivo findings may pave the way towards mechanistic investigations; in particular, it is important to test whether HBV and HCV proteins may trigger some of the molecular signals which control the mononuclear/binuclear balance.
Acknowledgments
We would like to thank Jacques Emmanuel Guidotti, France Demaugre, and Dina Kremsdorf for helpful discussions. We would also like to thank Pascal Debey for the imaging facility at INRA UC806/EA2703 Muséum National d’Histoire Naturelle.
Abbreviations
HBV, hepatitis B virus
HCV, hepatitis C virus
FMP, fraction of mononuclear cells in the population of polyploidy cells
HCC, hepatocellular carcinoma
PBS, phosphate buffered saline
Conflict of interest: None declared.
REFERENCES
- 1.Nadal C , Zajdela F. Hepatic polyploidy in the rat. IV. Experimental changes in the nucleolar volume of liver cells and their mechanisms of regulation. Exp Cell Res 1967;48:518–28. [DOI] [PubMed] [Google Scholar]
- 2.Saeter G , Schwarze E, Seglen O. Shift from polyploidizing to nonpolyploidizing growth in carcinogen-treated rat liver. J Natl Cancer Inst 1988;80:950–8. [DOI] [PubMed] [Google Scholar]
- 3.Wheatley DN. Binucleation in mammalian liver. Studies on the control of cytokinesis in vivo. Exp Cell Res 1972;74:455–65. [DOI] [PubMed] [Google Scholar]
- 4.Koike Y , Suzuki Y, Nagata A, et al. Studies on DNA content of hepatocytes in cirrhosis and hepatoma by means of microspectrophotometry and radioautography. Histochemistry 1982;75:549–62. [DOI] [PubMed] [Google Scholar]
- 5.Kudryavtsev BN, Kudryavtseva MV, Sakuta GA, et al. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch B Cell Pathol Incl Mol Pathol 1993;64:387–93. [DOI] [PubMed] [Google Scholar]
- 6.Melchiorri C , Bolondi L, Chieco P, et al. Diagnostic and prognostic value of DNA ploidy and cell nuclearity in ultrasound-guided liver biopsies. Cancer 1994;74:1713–19. [DOI] [PubMed] [Google Scholar]
- 7.Saeter G , Lee CZ, Schwarze PE, et al. Changes in ploidy distributions in human liver carcinogenesis. J Natl Cancer Inst 1988;80:1480–5. [DOI] [PubMed] [Google Scholar]
- 8.Brodsky WY, Uryvaeva IV. Cell polyploidy: its relation to tissue growth and function. Int Rev Cytol 1977;50:275–332. [DOI] [PubMed] [Google Scholar]
- 9.Gupta S . Hepatic polyploidy and liver growth control. Semin Cancer Biol 2000;10:161–71. [DOI] [PubMed] [Google Scholar]
- 10.Sigal SH, Gupta S, Gebhard DF, et al. Evidence for a terminal differentiation process in the rat liver. Differentiation 1995;59:35–42. [DOI] [PubMed] [Google Scholar]
- 11.Gorla GR, Malhi H, Gupta S. Polyploidy associated with oxidative injury attenuates proliferative potential of cells. J Cell Sci 2001;114:2943–51. [DOI] [PubMed] [Google Scholar]
- 12.Schmucker DL. Hepatocyte fine structure during maturation and senescence. J Electron Microsc Tech 1990;14:106–25. [DOI] [PubMed] [Google Scholar]
- 13.Sigal SH, Rajvanshi P, Gorla GR, et al. Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. Am J Physiol 1999;276:G1260–72. [DOI] [PubMed] [Google Scholar]
- 14.Diez-Fernandez C , Sanz N, Alvarez AM, et al. Influence of aminoguanidine on parameters of liver injury and regeneration induced in rats by a necrogenic dose of thioacetamide. Br J Pharmacol 1998;125:102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakatani T , Inouye M, Mirochnitchenko O. Overexpression of antioxidant enzymes in transgenic mice decreases cellular ploidy during liver regeneration. Exp Cell Res 1997;236:137–46. [DOI] [PubMed] [Google Scholar]
- 16.Schwarze PE, Pettersen EO, Shoaib MC, et al. Emergence of a population of small, diploid hepatocytes during hepatocarcinogenesis. Carcinogenesis 1984;5:1267–75. [DOI] [PubMed] [Google Scholar]
- 17.Anti M , Marra G, Rapaccini GL, et al. DNA ploidy pattern in human chronic liver diseases and hepatic nodular lesions. Flow cytometric analysis on echo-guided needle liver biopsy. Cancer 1994;73:281–8. [DOI] [PubMed] [Google Scholar]
- 18.Oriyama T , Yamanaka N, Fujimoto J, et al. Progression of hepatocellular carcinoma as reflected by nuclear DNA ploidy and cellular differentiation. J Hepatol 1998;28:142–9. [DOI] [PubMed] [Google Scholar]
- 19.Hoso M , Nakanuma Y. Cytophotometric DNA analysis of adenomatous hyperplasia in cirrhotic livers. Virchows Arch A Pathol Anat Histopathol 1991;418:401–4. [DOI] [PubMed] [Google Scholar]
- 20.Guidotti JE, Bregerie O, Robert A, et al. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem 2003;278:19095–101. [DOI] [PubMed] [Google Scholar]
- 21.Bedossa P , Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996;24:289–93. [DOI] [PubMed] [Google Scholar]
- 22.Chassoux D , Franchi J, Cao T, et al. DNA content by in situ fluorescence imaging and S-phase detection, with chromatin structure preserved. Anal Quant Cytol Histol 1999;21:489–97. [PubMed] [Google Scholar]
- 23.Brechot C , Gozuacik D, Murakami Y, et al. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol 2000;10:211–31. [DOI] [PubMed] [Google Scholar]
- 24.Hildt E , Hofschneider PH. The PreS2 activators of the hepatitis B virus: activators of tumour promoter pathways. Recent Results Cancer Res 1998;154:315–29. [DOI] [PubMed] [Google Scholar]