Abstract
Effects of maternal cytoplasmic environment (MCE) on development rate in rainbow trout were evaluated within a quantitative trait loci (QTL) analysis framework. Previous research had identified QTL for development rate in doubled haploid (DH) progeny produced from a cross between the Oregon State University (OSU) and the Swanson (SW) River rainbow trout clonal lines. In this study, progeny for QTL mapping were produced from a cross between the OSU and Clearwater (CW) River clonal lines. Doubled haploids were produced from the OSU × CW F1 by androgenesis using eggs from different females (or MCEs); with androgenesis, the maternal nuclear genome was destroyed by irradiation and diploidy was restored by blocking the first embryonic cleavage by heat shock. All embryos were incubated at the same temperature and development rate quantified as time to hatch. Using a linkage map constructed primarily with AFLP markers, QTL mapping was performed, including MCE covariates and QTL × MCE effects in models for testing. The major QTL for development rate in the OSU×SW cross overlaps with the major QTL found in this OSU × CW cross; effects at this locus were the same across MCEs. Both MCE and QTL × MCE effects contribute to variability in development rate, but QTL × MCE were minor and detected only at small-effect QTL.
MATERNAL effects, or the influence of maternal resources and behavior on the environment and phenotype of developing embryos and young, can have significant implications for the evolution of developmental phenotypes in all taxa (for review, see Mousseau and Fox 1998). Female mate choice, maternal care, maternal allocation of resources, maternal transfer of cytoplasmic factors to eggs and young, and the influence of environment on female reproduction all have important implications for the survival of developing offspring. Maternal effects play an important role in the early development of salmonid fishes (Einum and Fleming 2000, 2004; Brown et al. 2006; Pakkasmaa et al. 2006; Wang et al. 2006), contributing both environmental and additive genetic effects to variability in offspring phenotype. Most studies in salmonids have focused on the maternal effects of egg size on the viability, survival, growth, and size of embryos and postemergence fry (Bams 1967; Hutchings 1991; Einum and Fleming 1999, 2000). Maternal egg size appears to have the greatest effects on size and growth during early embryonic development (Heath et al. 1999), but investigators have failed to find a correlation between egg size and development rate (Ferguson et al. 1985; Einum and Fleming 1999). Furthermore, egg size alone does not encompass many maternally derived egg components that may significantly affect development rate. More recently, using the same rainbow trout clonal line system described herein, variation in mitochondrial haplotype in a constant nuclear background has shown significant association with variation in development rate, suggesting that the mitochondrial haplotype alone can influence the rate of development (Brown et al. 2006). Here, we aim to determine if the egg source, or maternal cytoplasmic environment (MCE) in total, significantly affects the expression of quantitative trait loci (QTL) associated with development rate in Oncorhynchus mykiss. Maternal cytoplasmic effects could include, but are not limited to, maternal hormones, mitochondrial DNA, and maternally derived RNA deposited in the egg yolk.
The rate of development in the early life history of salmonids has important implications for fitness with respect to emergence timing and optimal conditions for foraging, predator avoidance, and migration (Einum and Fleming 2000; Sundstrom et al. 2004, 2005). Salmonid fishes suffer severe mortality when they emerge from the gravel nests in which they are fertilized (Elliot 1989). Emergence timing is also influenced by spawning date in salmonids, but embryonic development rate, thought to be adapted to local thermal environments (Tallman 1986; Beacham 1988; Hebert et al. 1998), also has significant effects on emergence size and timing. Fish that hatch earlier have a size advantage over fish that hatch later (Mason and Chapman 1965), but timing of emergence must be carefully balanced with conditions for food availability (Sundstrom et al. 2004, 2005). In addition to correlation of development rate and early life history traits, investigators have observed a correlation among faster development rate, growth, and age at sexual maturity (Allendorf et al. 1983).
Previous research has shown significant variation in the development rates among strains of rainbow trout (Ferguson et al. 1985; Robison et al. 1999). Considerable additive genetic variance for development rate has been revealed through quantitative genetic studies (Smoker 1986; Beacham 1988; Hebert et al. 1998), and heritability has been reported as high as 0.23 in steelhead trout, O. mykiss (McIntyre and Blanc 1972). Many studies have shown that variation in development rate and survival prior to hatch is also significantly influenced by maternal effects (McIntyre and Blanc 1972; Blanc and Poisson 1983; Smoker 1986; Brown et al. 2006). Although a few experiments have revealed the role of single regulatory loci in development rate in some populations of fishes (Allendorf et al. 1983; DiMichele and Powers 1991), the number and nature of genetic loci influencing variation in embryonic development rate in other populations has only recently been investigated. More recently, a major QTL segregating for development rate, contributing up to 25% of the phenotypic variation, has been identified in a two different crosses between clonal lines of rainbow trout (O. mykiss) (Robison et al. 2001; Martinez et al. 2005; Sundin et al. 2005). Alleles in this genome region from the male grandparent in both studies significantly increase the rate of development, shortening the time from fertilization to hatch.
The external fertilization and ease with which salmonid gametes can be handled to manipulate chromosome set numbers provide an excellent system for studying the effects of maternal cytoplasmic environment on developmental phenotype, independent of the direct transfer of maternal nuclear genes. Androgenetically derived isogenic strains of rainbow trout have been produced from outbred populations of interest in two generations (Young et al. 1996). These lines have been crossed to produce segregating doubled haploid individuals by androgenesis in the second generation from F1 hybrids and have provided the resources for linkage mapping and quantitative trait locus analysis of developmental phenotypes (Young et al. 1998; Robison et al. 2001; Nichols et al. 2003). With androgenesis, the maternal nuclear genome is destroyed by irradiation (May and Grewe 1993), irradiated eggs are fertilized and activated with sperm, and diploidy is restored by blocking the first embryonic cleavage with a heat or pressure shock (Arai et al. 1979; Parsons and Thorgaard 1984; Bongers et al. 1994; Young et al. 1996). Androgenesis with sperm from hybrids between lines with divergent phenotypes and different egg sources makes it possible to investigate the contribution of maternal cytoplasmic effects on the expression of QTL in doubled haploid families, independent of the direct transmission of the maternal nuclear genome.
In this study, we investigated whether development rate QTL exhibit significantly different additive effects in different MCEs. Robison et al. (2001) revealed a single QTL of large effect and a few QTL of small effect segregating for development rate in doubled haploid progeny produced from a cross between a hatchery-derived Shasta-type rainbow trout obtained from Oregon State University (OSU) and a semiwild strain of rainbow trout from the Swanson River, Alaska (SW). A difference in development rate also exists between the OSU clonal line and another clonal line derived from a Clearwater River steelhead trout (CW); introgression of this region using advanced backrosses of OSU × CW have shown the same region contributing to a significant proportion of variation in development rate (Sundin et al. 2005). Using doubled haploids produced from different egg sources, we have conducted a QTL analysis of development rate in a cross between OSU and CW to test the hypothesis that QTL by maternal cytoplasmic environment interaction (QTL × MCE) significantly affect variation in development rate.
MATERIALS AND METHODS
Crosses and culture:
Clonal line production:
O. mykiss clonal lines have been developed from outbred populations of interest in two generations by gyno- and androgenesis (Young et al. 1996) and are maintained at the Washington State University Trout Hatchery. O. mykiss clonal families and F1 hybrids between these families were further propagated from existing clonal lines to characterize differences in development rate between two clonal lines and within isogenic families of clonal hybrids. The OSU clonal line is a Shasta-type rainbow trout line originating from an outbred stock maintained at a research hatchery at Oregon State University (Corvallis, OR). The OSU line was developed from genetic females by two rounds of gynogenesis, and thus all individuals in this line are genetic females (XX). The CW line originated from a hatchery stock of steelhead trout from the Clearwater River at the Dworshak National Fish Hatchery (Ahsahka, ID). The CW line was developed by androgenesis in two generations and all individuals are YY genetic males. For this study, OSU and CW clonal rainbow trout families were propagated by androgenesis at Washington State University in December 1999, as described by Young et al. (1996). Briefly, eggs from three outbred females obtained from a commercial rainbow trout egg source (Troutlodge, Sumner, WA) were irradiated with gamma radiation to destroy maternal nuclear DNA. Sperm from a sex-reversed clonal OSU male (XX) and from a normal clonal CW male were used to fertilize irradiated eggs on December 8, 1999. For each of these groups, the first embryonic cleavage was blocked by a heat shock to restore diploidy with all-paternal nuclear DNA inheritance, and embryonic development proceeded as in normal diploid embryos. One additional group of OSU clones (O × O) was made on December 7, 1999, by fertilizing clonal OSU eggs (from one female) with sperm from a sex-reversed clonal OSU male to evaluate differences in development rate between clones propagated by androgenesis and traditional fertilization. F1 hybrids were produced from the same eggs as O × O clones on December 7, 1999.
Doubled haploid production:
Doubled haploid progeny used for linkage and QTL analysis were produced from hybrids from a cross between the OSU and CW clonal lines. Eggs from one OSU female were fertilized with sperm from one CW male to produce a family of F1 progeny. Sperm from the resulting F1 male hybrid clones was used to produce doubled haploid progeny by androgenesis, as described by Young et al. (1998). Outbred eggs were obtained from Troutlodge in December 1999 and from the Spokane State Fish Hatchery (Washington Department of Fish and Wildlife, Spokane, WA) in January 2000. Eggs from each of three outbred females from Troutlodge (TL) and from each of six outbred females from the Spokane (SP) hatchery were irradiated with gamma irradiation to destroy the maternal nuclear DNA. Irradiated eggs were fertilized with sperm from the same F1 hybrid male on December 8, 1999, and on January 18, 2000, respectively. During embryonic development, the first embryonic cleavage was blocked by heat shock to restore diploidy. Families of doubled haploids are named according to the population (TL or SP) followed by a number indicating the female within populations used to produce the families (1–3 for TL and 1–6 for SP); unique names correspond to individual MCEs.
Culture:
Fertilized eggs for clonal lines, hybrids, and doubled haploid families were maintained in recirculating stack egg incubators in a climate-controlled chamber. Water temperature was maintained at 11° and was monitored either by StowAway XTI temperature loggers every 9 min or manually by thermometer at each check on the embryos. Each cross and family was separately maintained in multiple enclosed boxes in the incubator within the same incubator tray. Previous evaluations of embryonic development rate among divergent clonal lines have not exhibited statistically significant box effects on the time to hatch (B. Robison, personal communication). Embryos were maintained in constant darkness, except during periodic removal of inviable embryos.
Phenotyping:
Embryonic development rate was quantified as time to hatch, which correlates with the relative rate of appearance of important development stages (Ferguson et al. 1985). Once eye pigmentation was visible, embryos were transferred to individual wells in an 80-well box within the stack incubators (Robison et al. 1999). Both clonal line and doubled haploid embryos were checked at 4-hr intervals once hatching began, as described by Robison et al. (1999). At each interval, hatched individuals were counted and moved to separate enclosed boxes so that female family (or MCE), cross, and hatch time were identical for all individuals in each box. Time to hatch (tth) was calculated for each fish as accumulated temperature units (ATU), where ATU (degree days) = (incubation temperature) × (days from fertilization to hatch).
Notes on gross skeletal and yolk-sac deformities were made on each fish at swim-up at the time fin clips were taken for genotyping. Deformities noted included abnormal curvature of the spine, craniofacial abnormalities, and yolk sac abnormalities.
Genotyping:
Fin clips were taken nonlethally from swim-up fry when yolk-sac absorption was complete. Clips were preserved in 95% ethanol. DNA was extracted with a Puregene DNA extraction kit (Gentra Systems, Minneapolis). Amplified fragment length polymorphic (AFLP) markers were screened on all individuals according to the methods of Vos et al. (1995) as modified by Robison et al. (2001). Fourteen EcoRI and MseI primer sets were used. These primer sets contained syntenic markers mapped in our other crosses in the laboratory (Young et al. 1998; Robison et al. 2001; Nichols et al. 2003), which all share the OSU maternal parent. AFLPs are named according to the +3 selective bases used for the EcoRI and MseI adapter primers. The number at the end of the marker name indicates either the AFLP size for those markers mapped in other crosses or an arbitrary number given to the band on the AFLP fingerprints. The final characters for syntenic loci indicate the parent in which the band was present (for those markers syntenic with those in other crosses): “o” for OSU, “a” for Arlee, “c” for Clearwater, and “s” for Swanson. Markers that are codominant end with an “o*.” “E” at the beginning of some of the names indicated the EcoRI six-cutter enzyme, following the convention of Young et al. (1998). In addition to AFLP markers, three other markers were genotyped. Two microsatellite markers, OMM1009 (Rexroad et al. 2001) and OmyFGT12TUF (Nichols et al. 2003), were genotyped by size separation on a 1.5% agarose gel. These markers were added to AFLP markers on the basis of inferred synteny from other rainbow trout maps in the region of the major development rate QTL. In addition, one known gene, growth hormone 1 (GH1; (Oakley and Phillips 1999), identified as a potential candidate gene for development rate, was genotyped by the identity of a single nucleotide polymorphism (SNP) in intron C. GH1 SNP genotypes were detected by the ABI SNaPshot technique (Applied Biosystems, Foster City, CA).
Statistical analyses:
Summary statistics:
Difference in time to hatch among clones was tested by one-way analysis of variance, and pairwise differences among clones was tested with a Tukey–Kramer test (PROC GLM, SAS Institute, Cary, NC). For the doubled haploid progeny, analysis of variance was conducted to test the significance of maternal population (TL and SP), MCE nested within population, and deformity effects on time to hatch. Tests for significance of population effects were made within a mixed model (PROC MIXED, SAS Institute), treating MCE as random effects nested within populations and estimating the degrees of freedom for the denominator of the test statistic using Satterthwaite's method. Given no fixed differences between populations, least-squares means were used to test significance among all pairs of female families or MCEs in the doubled haploid (DH) analysis using a Tukey–Kramer test (PROC GLM, SAS Institute). Difference in the incidence of deformity among MCEs was tested using logistic regression (PROC LOGISTIC, SAS Institute). Type I error rate was set at α = 0.05.
Genetic map construction:
Prior to map construction, segregation distortion or deviation from expected 1:1 Mendelian segregation ratios was tested for each marker with a χ2 test (α = 0.05). Markers with significant segregation distortion were checked for scoring errors and reliability and were corrected or, if unreliable, were removed from the analysis. For the combined data set of all families, markers for which <80% of individuals were scored were removed prior to linkage analysis. Markers were grouped and then ordered into linkage groups using the Kosambi map function for the doubled haploid design in Mapmaker for Mac (v. 2.0, Scott Tingey, Dupont Experimental Station, Wilmington, DE) and Mapmaker/EXP (Lander et al. 1987). Linkage groups were formed by estimating the recombination fractions among all pairs of markers; markers were grouped at a minimum log of the odds (LOD) score of 3.0 and a maximum recombination fraction of 0.40. Marker order within each linkage group was evaluated by maximum likelihood. For groups that contained eight or fewer markers, all possible marker orders were compared. For groups with greater than eight markers, an initial set of eight markers was chosen, and a framework map was formed by comparing all possible orders of those eight markers. Additional markers were then added to this framework, one at a time, placing each marker in the position giving the maximum likelihood, taking the framework markers and all previously placed markers as fixed. Orders of the larger groups were then checked further by considering all possible orders of a sliding window of seven markers, keeping markers outside the window in a fixed order. The final map was drawn with Mapchart 2.1 (Voorips 2002).
QTL analyses:
QTL analyses for development rate were performed combining the analysis for all MCEs, using interval mapping and multiple QTL mapping within R/qtl (Broman et al. 2003). Because R/qtl does not include a model for doubled haploids, a backcross model coding for the doubled haploids was used since models for both cross types utilize two genotypic classes for tests of significant QTL effects. Standard interval mapping (scanone) was performed testing for significant single-locus effects, with MCE effects included as additive covariates in the model. Analysis was performed at the markers and on a 1-cM grid along the genome. Significant loci were identified as those with LOD scores exceeding the 95% upper tail of distributions generated by 1000 permutations of the data (Churchill and Doerge 1994). The MCE effects were preserved in this permutation test: the MCE to which a particular phenotype was assigned was retained, and the genotype data were shuffled across all individuals.
We further performed a form of composite interval mapping (Zeng 1994): highly statistically significant loci were included as marker cofactors in a scan for additional QTL on chromosomes that did not show significant QTL in standard interval mapping. Controlling for large-effect QTL in this way can reduce the residual variation and allow loci of more modest effect to be more clearly seen.
A two-dimensional, two-QTL scan was also performed. This allows the identification of interacting QTL with limited main effects, which would remain hidden in the single-QTL analysis, and also an assessment of evidence for multiple linked QTL that appear as a single peak in the single-QTL analysis.
Once single and interacting QTL were identified, QTL positions were refined on the linkage groups using the approach used in multiple interval mapping (Kao et al. 1999), iteratively refining the position of each QTL, one at a time, in the context of a multiple QTL model (with all other significant QTL, QTL × QTL interactions, and MCE covariates in the model) until the QTL positions converged. Positions with the greatest LOD score (and lowest significant P-value) on each linkage group with a significant QTL (from composite interval mapping) were chosen as the starting positions for the refinement of QTL positions. To increase our power for the detection of locus by MCE (QTL × MCE) for very small-effect loci, tests for QTL × MCE effects were made only at significant QTL positions, including MCE covariates and large-effect QTL as covariates. To do this, the full model containing QTL, MCE, and QTL × MCE effects were compared to the reduced model containing only QTL and MCE effects, subtracting the LOD scores. Significance of QTL × MCE interactions were determined by permutation tests conducted only for the positions tested. The MCE effects were preserved in the permutations, but the QTL effects were eliminated, and so we use as the reference the distribution of the LOD score for the QTL × MCE interaction in the absence of either the interaction or the marginal QTL effect. The fact that the main and interaction effects are orthogonal suggests that the null distribution of the QTL × MCE interaction LOD score will be approximately the same, whether or not a marginal QTL effect is present, and so this permutation test is appropriate.
To estimate the effects of multiple quantitative trait loci detected in our prior analyses, we utilized a drop-one-term multiple imputation analysis (Sen and Churchill 2001), simultaneously estimating all significant epistatic, MCE, and QTL × MCE effects. To do this, genome positions selected from our refined multiple interval mapping analysis, MCE covariates, and significant QTL × MCE effects were chosen for model testing. Multiple QTL models were used to test for the significance of the main effects of QTL and MCE, for interaction effects between MCE and QTL, and for all possible epistatic interactions among QTL, evaluating the full-effects model in comparison with the reduced model with each term (and all higher-order terms including that term) dropped. Significant terms were evaluated in a reduced model and additive effects (± standard errors) and percentage of variation explained were estimated. QTL analyses for the incidence of deformity, coded as a binary trait, were also performed as described above, utilizing the binary model for analyses.
To test whether or not QTL identified in this cross (OC) overlapped with those found by Robison et al. (2001) in the OSU × Swanson (OS) cross, 2-LOD support intervals for QTL position in OS were constructed and evaluated for overlap with 2-LOD support intervals for QTL position in this OC cross. Two-LOD support intervals have been shown to be >95% C.I. (Liu 1998) and asymptotically represent 99.8% C.I. (Visscher et al. 1996).
RESULTS
Phenotypic differences among clonal lines:
Analysis of variance for differences among clonal lines revealed a highly significant difference in the development rate between the OSU and CW lines. OSU clonal families developed at a much slower rate, hatching later relative to the CW clonal families; F1 hybrids were intermediate in the time to hatch (Table 1). No difference was observed between CW clonal families produced from the eggs of TL1 or TL3. OSU embryos produced from TL1 had significantly slower development rates than those produced from TL3 eggs. However, the sample size for TL1 was very small for the OSU family (n = 13). No difference was observed between the average development rate of androgenetic and intercrossed OSU (O × O) clones (Table 1). TL2 female eggs were inviable for both the clonal lines and the doubled haploids.
TABLE 1.
Development rate means for clonal lines and hybrids
| Family | N | Time to hatch (ATU) (mean ± SEM) |
|---|---|---|
| CW clones (TL1) | 70 | 291.3 ± 1.0 (a) |
| CW clones (TL3) | 35 | 292.4 ± 1.3 (a) |
| OSU clones (TL1) | 13 | 352.4 ± 3.1 (b) |
| OSU clones (TL3) | 54 | 339.1 ± 1.5 (c) |
| OSU × OSU clones | 47 | 334.9 ± 1.0 (c) |
| F1 hybrid clones | 32 | 304.3 ± 1.0 (d) |
Tukey groupings of significantly different means are indicated by lowercase letters in parentheses in “Time to hatch” column. TL, eggs from Troutlodge; SP, eggs from Spokane Hatchery.
Phenotypic differences among DH families:
Eight doubled haploid families produced from different outbred egg sources were intermediate in development rate compared to OSU and CW clonal lines (Table 2). Population differences observed in the overall model were attributed to differences among individual MCE within the populations; when the hypothesis for population main effect was tested using nested MCE as the error term, population-level effects were not observed (F = 2.77, d.f. = 1, P = 0.1454). Given no fixed effect of population, MCE effects were tested. MCE, nested within population, was a significant source of variation in development rate (F = 10.64, d.f. = 6, P < 0.0001). The presence or absence of deformity did not significantly contribute to the variability in time to hatch (F = 1.21, d.f. = 1, P = 0.2722). Pairwise comparisons among MCEs revealed that progeny produced from the TL1 female consistently had a significantly greater mean time to hatch than progeny from TL3, SP1, SP2, SP3, SP5, and SP6 (Table 1). This observation is consistent with that observed between OSU clones produced with TL1 and TL3 eggs (above). A significant difference in the incidence of deformities was observed among MCE (Table 2; χ2 = 18.07, d.f. = 7, P = 0.0117).
TABLE 2.
Embryonic development rate means for doubled haploids produced from different outbred female egg sources or MCEs
| MCE | N | Time to hatch (ATU) (mean ± SEM) | % deformed |
|---|---|---|---|
| TL1 | 53 | 331.1 ± 2.3 (a) | 28.3 |
| TL3 | 109 | 318.2 ± 1.2 (bd) | 18.4 |
| SP1 | 82 | 321.1 ± 1.0 (bc) | 15.8 |
| SP2 | 83 | 316.1 ± 1.3 (bcd) | 19.3 |
| SP3 | 29 | 316.1 ± 2.0 (bcd) | 13.8 |
| SP4 | 8 | 322.8 ± 2.5 (abcd) | 12.5 |
| SP5 | 22 | 316.2 ± 2.2 (bcd) | 36.4 |
| SP6 | 168 | 314.3 ± 0.8 (d) | 8.9 |
Tukey groupings for significant differences between means are indicated by lowercase letters within parentheses; shared letters indicate a nonsignificant difference between means. Development rate means are expressed as ATUs from fertilization to hatch. TL, Troutlodge females; SP, Spokane Hatchery females.
Genetic map construction:
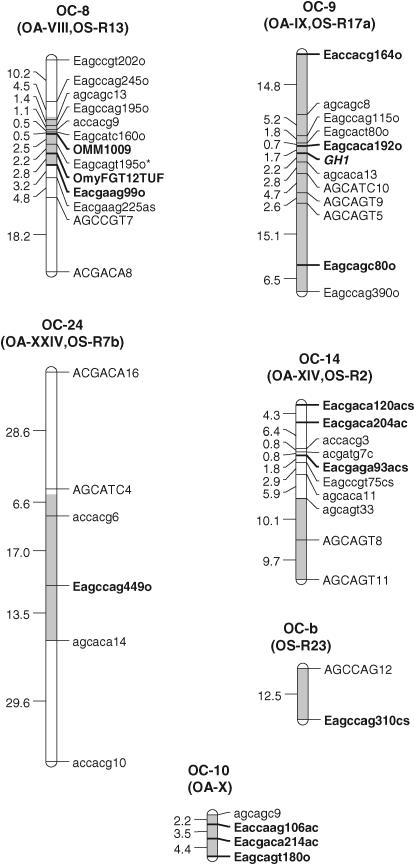
With 184 AFLP, two microsatellites, and one known gene in the linkage analysis, 28 linkage groups were identified (Figure 1). On the basis of the karyotypes of the OSU and CW lines, we would expect 29 linkage groups to result with linkage mapping. OSU has 60 chromosomes and CW has 58, and it is expected that 2 chromosomes from OSU would pair with the homologous fused pair of chromosomes from CW during meiosis (Ristow et al. 1998; Phillips et al. 2005). This fusion results in the joining of markers from OA linkage groups IV and XXV into a single linkage group OC-4-25 in the OSU × CW cross. Of the total number of markers genotyped, 172 were mapped on the linkage groups identified and 15 remained unlinked. The average intermarker distance was 9.7 cM. Of the 28 linkage groups, 26 match to linkage groups delineated from our OSU × Arlee (OA) doubled haploid mapping panel (Young et al. 1998; Nichols et al. 2003) on the basis of shared AFLP and microsatellite markers. All but one linkage group upon which significant QTL were identified have been matched to the OA map. The mapping of the sex phenotype in a small subset of these individuals that were grown to sexual maturity placed sex on the linkage group OC1 and shared AFLP markers in other mapping families of OSU × CW corroborate the synteny of the sex linkage groups of OA and OC (Felip et al. 2005). In addition, markers mapped from additional AFLP primer sets in additional studies with OC progeny indicated synteny of OC3, OC5, OC11, OC12, OC16, OC17, OC24, OC29, and OC31 with their homologous OA linkage groups (K. M. Nichols, unpublished data). Markers with synteny between this OC map and the OSU × Swanson (OS) map (Robison et al. 2001) have also been matched, including markers on all linkage groups for which OS progeny showed significant development rate QTL. Of the 27 linkage groups from the OS cross (Robison et al. 2001), 18 matched to linkage groups in this cross on the basis of syntenic AFLP markers. One additional linkage group, OC11, matched to the OS linkage group R22 on the basis of an additional marker mapped in another study with OC progeny (K. Nichols, unpublished data). Only two linkage groups in this study, OC-a and OC-b, remained unmatched to linkage groups in the OA (Young et al. 1998; Nichols et al. 2003). Linkage group OC8, upon which the largest QTL was identified in this cross, matched to linkage group OA-VIII and to the OS linkage group R13 (Figure 2). This linkage group is syntenic with linkage groups J and N in an unrelated outcrossed rainbow trout mapping panel on the basis of the syntenic microsatellite markers OMM1009 and OmyFGT12TUF (Nichols et al. 2003). Neither of the other linkage groups identified with significant QTL in OS, R6, and R9 (corresponding to OC23 and OC-a in this cross, respectively), were syntenic with OC linkage groups for which significant QTL were identified; however, a reanalysis of this data accounting for temperature environmental effects in the mapping families (Martinez et al. 2005) show QTL for time to hatch on OSU × SW linkage groups R6, R13, and R17a. R17a is syntenic with linkage group OC-9 in this article.
Figure 1.—
OSU × CW doubled haploid linkage groups with significant QTL for development rate. Linkage groups are named according to the cross (OC for OSU × CW, and an Arabic number corresponding to the Roman numeral of the OA linkage group with which it is syntenic). Names in parentheses correspond to the OSU × Arlee (OA) and the OSU × Swanson (OS) linkage groups to which OC groups are syntenic. Significant main-effect QTL, with 2-LOD support intervals, are indicated with shading within linkage groups. Markers in boldface type are syntenic with markers from the OA and OS linkage maps.
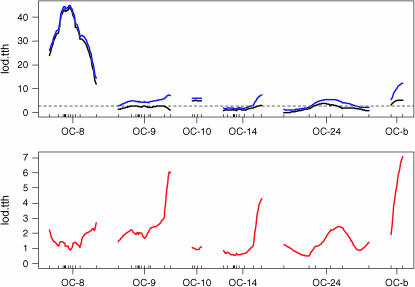
Figure 2.—
Composite interval mapping LOD profiles for linkage groups with significant QTL. (Top) LOD scans for models including MCE covariates (black) and both MCE covariates and QTL × MCE interaction effects (blue). Dashed line indicates 95% permutation threshold for MCE covariate model. (Bottom) LOD difference (red) between the two models in the top, evaluating LOD scores for an overall QTL × MCE effect at detected QTL.
QTL analyses:
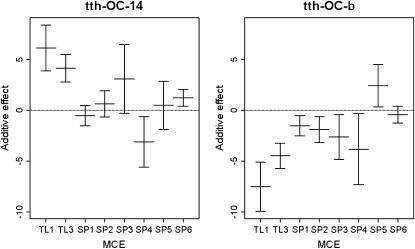
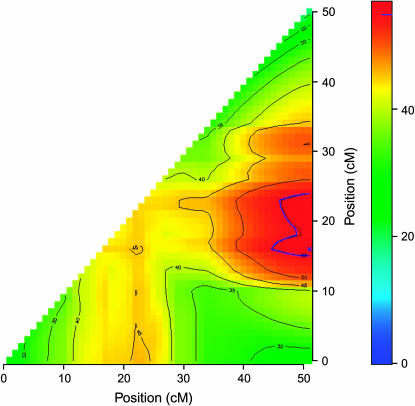
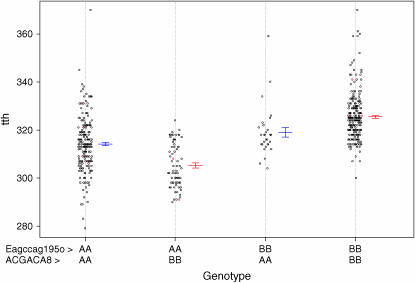
In the genomewide single-QTL scan, six significant QTL were identified. Simple interval mapping with MCE covariates revealed a major QTL on OC-8, called tth-OC-8a (Figures 1 and 2). Composite interval mapping, including both major QTL, tth-OC-8a, and MCE effects as covariates, revealed additional small-effect QTL on OC-9, OC-10, OC-14, and OC-24 (Figures 1 and 2). The same QTL were identified in both the full data set, and a pruned data set that included only MCEs with a sample size of n > 50 (data not shown). No QTL were detected for presence/absence of deformity (data not shown). Tests for QTL × MCE at significant main-effect QTL revealed that two loci (tth-OC-14 and tth-OC-b) exhibit minor, but significant, differences in genotypic effects among MCE (Figure 2) on the basis of permutation tests conducted at each significant locus. Although QTL × MCE appears significant on OC-9 (Figure 2), the position showing this trend is not at the peak of the QTL for this linkage group. To increase the power to detect QTL × MCE interactions, we tested for such interactions only at the position with the greatest QTL effect, and so the QTL × MCE interaction involving tth-OC-9 was not included in the final model. Genotypic effects at the significant QTL × MCE loci (tth-OC-14 and tth-OC-b) indicate that significant QTL × MCE arise from QTL of no effect in some MCEs and of small effects in others (Figure 3). The MCE TL1 was the greatest contributor to evidence for a QTL × MCE interaction. TL1 have a much larger effect on QTL tth-OC-14 and tth-OC-b than all other MCEs, which have little or no additive effects at these loci (Figure 3). Two-dimensional scans for linked QTL on linkage groups for which significant QTL were detected revealed a significant epistatic interaction (nonadditive gene action) between tth-OC-8a, and another QTL, tth-OC-8b, with only epistatic effects also on OC-8 (Figure 4); the joint QTL–QTL LOD score was 56.98 and the epistasis LOD score was 12.47. Gene–gene combinations of the CW genotype at tth-OC-8a and the OSU genotype at tth-OC-8b on the end of the chromosome are associated with significantly earlier time to hatch relative to all other gene combinations (Figure 5). Two-dimensional scans on other linkage groups indicated that only one QTL existed on each of the other linkage groups and that no significant interactions existed among detected QTL.
Figure 3.—
Additive effect means (± standard errors of the mean) for QTL exhibiting significant QTL × MCE interaction effects.
Figure 4.—
Two-dimensional scan for significant linked QTL on linkage group OC-8, showing the joint LOD scores for every pair of positions on a 1-cM grid. LOD contours are shown in black. The 2-LOD support interval for a significant joint LOD score is outlined in blue, with the LOD peak indicated with a blue “x.”
Figure 5.—
Joint genotypic or epistatic effects of two loci on linkage group OC-8. Each point is an individual; red symbols were calculated from imputed genotypes in missing individuals. Means and standard errors of the means are plotted next to each cloud of points.
When all six QTL, identified from one- and two-dimensional scans, were included in a single model, each was seen to contribute to variation in the phenotype (Table 3). Furthermore, QTL × QTL interactions, MCE, and QTL × MCE interactions were also shown to play a role in variation in time to hatch, as seen in Table 3, which contains the results of an analysis in which each term in the model was omitted, one at a time. (Note that when a particular effect was omitted, all interactions containing the effect were also omitted; the term TL1 has 3 d.f., as the LOD score for TL1 concerns a comparison of the full model to the reduced model in which TL1 and its interaction with each of the OC-14 and OC-b loci were omitted.) All six-way and lesser epistatic interactions among QTL were tested, but only one two-way interaction among loci was identified. The P-values in Table 3 are pointwise and so do not include an adjustment for the selection of the model; a complete account of the model selection in the assessment of multiple QTL models is difficult, but P-values <0.0001 indicate good evidence for the respective terms. As seen above, and further corroborated by these multiple QTL models, environmental effects from the female egg source (MCEs) were a significant source of variation in time to hatch, explaining up to 13.0% of the variation in time to hatch in the case of female TL1 (Table 3). QTL × MCE effects were a significant, but minor, source of variation in time to hatch, explaining up to 1.45% of the variation in time to hatch (Table 3). The major QTL, tth-OC-8a, was responsible for the single largest effect on time to hatch, accounting for 23.2% of the variation in phenotype. Differences in the genotypic means at this locus [OSU (BB)–CW (AA)] are 11.65 ± 1.07 degree days; individuals with the CW genotype at this locus have a mean hatching time ∼1 day earlier (at 11°) than individuals with the OSU genotype. The final reduced model, with only significant terms including QTL, MCE, QTL × QTL, and QTL × MCE, explained 53.1% of the variation in time to hatch and was highly significant (LOD = 91.1, d.f. = 12, P < 0.0001). These results suggest that the maternal cytoplasmic environment can differentially affect the expression of minor-effect QTL for development rate and that MCE alone has a significant influence on development rate.
TABLE 3.
Multiple QTL modeling analysis of variance for significant model terms
| Model terms
|
Closest marker | Genotypic effects
|
||||||
|---|---|---|---|---|---|---|---|---|
| Term | Position (cM) | d.f.a | LOD | % variation | χ2P-value | (ATU; degree days) | SE | |
| tth-OC-8a | 16 | Eagccag195o | 2 | 48.41 | 23.2 | <0.0001 | 11.65 | 1.07 |
| tth-OC-8b | 51.9 | ACGACA8 | 2 | 12.91 | 5.31 | <0.0001 | −0.6686 | 1.0674 |
| tth-OC-9 | 18 | Eagccag115o | 1 | 1.84 | 0.722 | 0.004 | 2.158 | 0.782 |
| tth-OC-10 | 10.1 | Eagcagt180o | 1 | 1.67 | 0.656 | 0.006 | 2.603 | 0.891 |
| tth-OC-14 | 39 | AGCAGT11 | 2 | 2.90 | 1.14 | 0.001 | 1.07 | 0.81 |
| tth-OC-24 | 47 | Eagccag449o | 1 | 3.19 | 1.26 | 0.00012 | 2.876 | 0.813 |
| tth-OC-b | 11 | Eagccag310cs | 2 | 4.92 | 1.96 | <0.0001 | −1.337 | 0.976 |
| tth-OC-8a × tth-OC-8b | 16 × 51.9 | 1 | 10.46 | 4.26 | <0.0001 | 15.02 | 2.15 | |
| TL1 | 3 | 29.46 | 13 | <0.0001 | 13.35 | 1.56 | ||
| SP1 | 1 | 5.43 | 2.17 | <0.0001 | 5.271 | 1.057 | ||
| tth-OC-14 × TL1 | 1 | 1.69 | 0.665 | 0.005 | 7.061 | 2.946 | ||
| tth-OC-b × TL1 | 1 | 3.65 | 1.45 | <0.0001 | −11.18 | 3.47 | ||
SE, standard error of genotypic effects; ATU, genotypic effects expressed as accumulated temperature units; % variation, percentage of variation explained by model term and all higher-order terms containing that term.
One term was omitted from the model at a time; when a main effect was omitted, all interactions containing it were also omitted.
Synteny with the major development rate QTL previously identified:
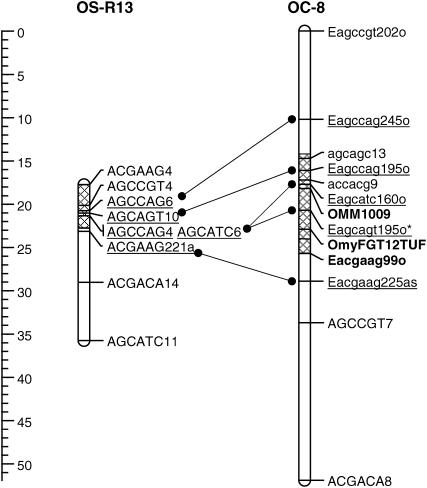
The major QTL on OC-8 is in the same QTL region as that identified for time to hatch in the doubled haploid mapping population derived from a cross between OSU and Swanson clonal lines (Robison et al. 2001) on the basis of the overlap of the 2-LOD support interval for the OS cross with the 2-LOD support interval for tth-OC-8a in this cross (Figure 6). QTL mapping in the OSU × CW cross was conducted with a greater number of progeny (n = 554) than the OSU × SW study (n = 170), and this increase in power shows a more precise localization of the QTL for time to hatch in this study. Additionally, minor-effect QTL for development rate were detected in the same region on linkage groups OC-9 in this cross and R17a in the OSU × SW cross (data not shown) (Martinez et al. 2005).
Figure 6.—
Comparison of OSU × CW linkage group OC8 with syntenic OSU × SW linkage group R13, with localized time-to-hatch QTL. The OSU × SW linkage group was redrawn from Robison et al. (2001). Cross-hatching indicates the QTL regions (2-LOD support interval) for the most significant QTL in each study. Lines drawn between the linkage groups indicate specific syntenic markers. Underlined markers are those that are syntenic between the maps.
DISCUSSION
This QTL analysis of development rate has revealed that the major QTL segregating in the cross between the OSU line and a clonal line derived from the SW line (Robison et al. 2001) is likely also segregating in this cross between OSU and a clonal line derived from the CW line (Sundin et al. 2005). Fine-scale mapping and identification of genes are needed to determine if indeed the same genes are responsible for developmental timing in these two crosses. This major QTL alone is responsible for 12° days, or ∼1 day shift for earlier development rate in individuals with the CW genotype relative to individuals with the OSU genotype at this locus. The fact that the same major QTL may be segregating in these two line crosses is not completely surprising, however, since the same female line (OSU) was used in both analyses. The most parsimonious explanation for the similar results in the two studies is that the OSU line has a constant effect at this QTL that causes a slower development rate relative to the paternal lines CW and SW, which have originated from geographically distinct populations. Across the salmonid genome, great differences exist in male and female recombination rates. Males usually have much smaller recombination rates in the centromere regions, and greater recombination rates than females toward the telomeres (Sakamoto et al. 2000; Danzmann et al. 2005). However, the major QTL region for development rate is in an area of greatly reduced recombination in both males and females (Danzmann et al. 2005). Furthermore, tth-OC-8a colocalizes to the same region as spawn-timing QTL in rainbow trout on the basis of synteny among rainbow trout linkage maps (O'Malley et al. 2002; Danzmann et al. 2005; Leder et al. 2006). The greatly reduced recombination rate in this QTL region, together with colocalization of QTL for an important reproductive life-history trait suggests the possible existence of a large gene complex in this region, responsible for the expression of multiple, correlated life-history traits in this species. Further detailed mapping and sequencing in this region is needed to address this hypothesis.
With a growing number of genes and ESTs mapped in rainbow trout, synteny among all O. mykiss linkage maps (Young et al. 1998; Sakamoto et al. 2000; Nichols et al. 2003; Danzmann et al. 2005) may reveal possible candidate genes for development rate in this region. One known gene, Clock, is currently mapped to this region (Danzmann et al. 2005; Leder et al. 2006). Clock plays a well-known role in circadian rhythms in animals, and expression of this gene is likely entrained by oscillations in daily light cycles. Photoperiod is tightly associated with maturation, spawning, and developmental metamorphosis in many organisms, including the salmonid fishes, but the role of this gene in the rate of development needs further exploration. Robison et al. (2001) ruled out the possibility that liver PGM1, an allozyme variant associated with faster development rate in a hatchery strain of rainbow trout (Allendorf et al. 1983), was associated with development rate divergence between the OSU and SW clonal lines. Expression of liver PGM1 was not evaluated in this cross. Aside from the single major QTL identified in most maternal environments in this study, five other QTL of much smaller effects were identified, and some of these show significantly different effects among maternal cytoplasmic environments. Only one QTL of smaller effect was found on a linkage group syntenic with small-effect QTL identified in more recent analyses of the Robison et al. (2001) data (Martinez et al. 2005). It is possible that the two crosses have different genes of small-to-moderate effect modulating embryonic development rate or that maternal or other environmental factors play a significant role in the expression of small-effect loci.
Most studies that have tested the formal hypothesis of QTL by environment interaction by testing equal QTL effects across environments have utilized recombinant inbred lines (Fry et al. 1998; Leips and Mackay 2000; Vieira et al. 2000; Borevitz et al. 2002). These lines afford replication of individuals with identical genotypes (at all loci) in different environments for formal tests of equal additive effects among environments within the maximum-likelihood models developed for composite interval mapping (Jiang and Zeng 1995). Other studies have evaluated QTL by environment interactions by performing QTL analyses in each single environment, overlaying the results on a common map to determine whether QTL identified in single environments are shared across environments. In this study, we have generated doubled haploid individuals for our QTL analysis. The generation time and space required for the propagation of each of these doubled haploid individuals into doubled haploid lines is a monumental task for rainbow trout; thus we must test initial hypotheses for significant QTL in the first generation of doubled haploids. Among the types of crosses that have been utilized for QTL mapping, the doubled haploid design is the most powerful (Beavis 1998; Martinez et al. 2002). Rather than overlay results from multiple independent models, we formally tested MCE and QTL × MCE effects within a single model. With this formal modeling approach, we have identified small-effect loci that show differential genotypic effects among maternal cytoplasmic environments.
Our QTL × MCE results suggest that minor QTL are significant in some, but not all, maternal cytoplasmic environments. The differences observed in MCEs could be due to the disparity in sample sizes (and thus power). QTL effects have been shown to be distributed in a negative exponential fashion, such that QTL of large effect are relatively few, and QTL of very small effect relatively numerous (Lynch and Walsh 1998 and references therein). The power of QTL studies can significantly affect the ability to detect QTL of small-to-moderate effect and can lead to an upward bias in estimates of QTL effect and percentage of variance explained for the loci that are detected (Beavis 1998). This may have been the case for MCEs with small sample sizes relative to SP6, but there are a few QTL of small effect identified in MCEs with smaller sample sizes that were not detected in SP6, suggesting significant environmental effects on the expression of QTL in the different maternal cytoplasmic environments. The maternal cytoplasmic effects in this study indicate differences in the magnitude, rather than in the direction, of additive effects among mothers. These effects are not nearly as striking as the opposite effects of QTL in different environments observed in Drosophila (Leips and Mackay 2000; Vieira et al. 2000), but antagonistic pleiotropy is not common in studies detecting significant QTL × environment interaction (Fry et al. 1998; Borevitz et al. 2002).
Previous research on the maternal influence on offspring in salmonids has focused primarily on differences in progeny survival, growth, and size with differences in female egg size. Differences observed in these characters have primarily been attributed to egg size or the amount of resources available to the developing embryo (Smoker 1986; Hutchings 1991; Einum and Fleming 1999; Heath et al. 1999; Vandeputte et al. 2002), but studies in brown trout (Einum and Fleming 1999) and brook trout (Hutchings 1991) have not found significant correlations between egg size and development rate. While these studies may address maternal effects as a function of resource quantity in eggs, quality and quantity of other maternal cytoplasmic factors that may be independent of egg size also play a large role in embryonic development.
Research in many organisms has shown that mitochondrial haplotype (Gerber et al. 2001; Brown et al. 2006), maternal hormones (Eising et al. 2001), and maternal mRNA in eggs (Nagler 2000) can influence embryonic development. Variation in mitochondrial haplotypes can affect rate of development, oxygen consumption, growth, and fitness independent of nuclear genetic background as well as by interaction with the nuclear genome (Gerber et al. 2001; Brown et al. 2006; and references therein). Androgens, corticosteriods, and thyroid hormones are among the maternally derived hormones with significant roles during embryonic development in vertebrates. Maternal androgens in bird eggs have been shown to influence hatching time, survival, and behavior (Eising et al. 2001). Maternal corticosteriods have been shown to affect growth rate pulses in damselfish prior to hatching (McCormick and Nechaev 2002), but this study and a study in coho salmon (Stratholt et al. 1997) did not find differences in hatching time or development rate as a result of maternal corticosteroids. Maternal thyroid hormones in eggs have long been known for their role in growth and metamorphosis in teleosts and other vertebrate animals (Dickhoff et al. 1990 and references therein). In rainbow trout, maternal mRNA has been shown to direct the progression of embryonic development until gastrulation when transcription of the embryonic nuclear genome begins (Nagler 2000). Genomic imprinting or the differential methylation of the genome depending on maternal or paternal inheritance has been shown to be necessary for normal development in mammals, but genomic imprinting has not been demonstrated in fish species. In fact, on the basis of results from this study and numerous other fish species, the successful production of viable offspring from all-paternal (androgenesis) or all-maternal (gynogenesis) inheritance clearly shows that both genomes and the associated epigenetic differences are not necessary for normal development in fish species in which it has been studied (McGowan and Martin 1997).
Despite the number of studies investigating the effects of these maternal effects on the fitness and physiology of developing organisms, few studies have examined these effects in a constant nuclear genetic background. Among the studies in fishes evaluating maternal effects on embryonic development rate, only two have tested maternal differences within a constant nuclear genetic background. Bongers et al. (1995) found differences in the incidence of deformity and survival among embryos of different egg sources used to produce androgenetic clones of carp (Cyprinus carpio), as was observed in this study. These authors suggest the possibility that maternal RNAs in batches of eggs from different females might be differentially damaged by the irradiation used to destroy maternal nuclear DNA in the process of androgenesis. However, their experiment did not ascertain if differences observed among females in deformities and survival were due to reduced homeostasis due to the homozygous nature of the embryos, differences in mitochondrial–nuclear interactions, differences in egg nutritional quality, or differences in the quantity and nature of maternal factors deposited in the eggs. In our laboratory, Parsons and Thorgaard (1984) found no statistical differences in the survival of rainbow trout produced by gyno- or androgenesis, but sublethal effect differences on the time to hatch have not been evaluated (although we observed no significant difference in development rate between O × O and OSU androgenetic clones in this study). Separating components of maternal cytoplasmic factors into environmental and mitochondrial genetic effects (either direct effects of the mitochondrial genome or mitochondrial–nuclear interaction) is difficult. However, using our rainbow trout clonal lines, Brown et al. (2006) were able to evaluate differences in developmental rate and physiology in a constant nuclear and cytoplasmic (extramitochondrial) background with variable mitochondrial haplotypes. Variable oxygen consumption and rate of development were significantly associated with mitochondrial haplotype, even in eggs produced from the same second-generation female clonal line (OSU line) possessing different mitochondrial genomes; in these females, mitochondria are different, but a constant nuclear background minimizes maternal genetic effects, either by mtDNA–nuclear interaction or in the deposition of maternal mRNA in the eggs. Each of these studies suggests that cytoplasmic factors may play a significant role in modulating nuclear genes associated with the rate of embryonic development rate. Further studies examining the effects of mitochondrial haplotypes, hormones, and mRNA within a constant nuclear genetic background may elucidate the role and significance of each factor in modulating embryonic gene expression.
Although tth-OC-8a, uninfluenced by QTL × MCE interaction, has the largest contribution to the variance in the phenotype, this study suggests that maternal cytoplasmic components can also affect the time to hatch, independent of the direct transmission of the maternal nuclear genome. The production of doubled haploid families from different females by androgenesis has provided a unique opportunity to assess the significance of maternal factors in the expression of QTL. Our results suggest that future QTL analyses should be carefully designed to consider or account for the effects of both maternal environmental factors alone and maternal × QTL interaction on phenotypic variability. It is not known whether the maternal effects observed here are the results of nongenetic maternal influences (age or nutritional status), whether the mother's genome has directly and significantly affected the environment of the egg by differential deposition of mRNA or other cytoplasmic factors such as hormones, or whether maternal mitochondrial–nuclear interactions exist. If the maternal effects observed in this study have been derived indirectly from nuclear or directly from mitochondrial genes of the female, maternal influence may prove to be a significant selective agent in the survival of developing embryos and fry.
Acknowledgments
The authors thank Sara Johnson and Steve Patton for laboratory and hatchery assistance. Barrie Robison provided useful discussions on the quantitative genetic analysis of development rate as well as comments on drafts of this manuscript. Thanks to Marc Evans and Julia Sharp for discussions on statistical analyses. Caird Rexroad and Ruth Phillips generously provided aliquots of primers for OMM1009 and GH1, respectively. Comments of anonymous reviewers significantly improved the manuscript. This research was funded by the National Science Foundation to G.H.T. (IBN-0082773), in part by National Research Initiative grant no. 2006-35205-16728 from the United States Department of Agriculture Cooperative State Research, Education, and Extension Service to G.H.T., and in part by the National Institutes of Health to K.W.B. (R01, GM074244).
References
- Allendorf, F. W., K. L. Knudsen and R. F. Leary, 1983. Adaptive significance of differences in the tissue-specific expression of a phosphoglucomutase gene in rainbow trout. Proc. Natl. Acad. Sci. 80: 1397–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai, K., H. Onozato and F. Yamakazi, 1979. Artificial androgenesis induced with gamma irradiation in masu salmon, Oncorhynchus masou. Bull. Fac. Fish. Hokkaido Univ. 30: 181–186. [Google Scholar]
- Bams, R. A., 1967. Differences in performance of naturally and aritifically propagated sockeye salmon migrant fry, as measured with swimming and predation tests. J. Fish. Res. Board Canada 24: 1117–1153. [Google Scholar]
- Beacham, T. D., 1988. A genetic analysis of early development in pink (Oncorhynchus gorbuscha) and chum salmon (Oncorhynchus keta) at 3 different temperatures. Genome 30: 89–96. [DOI] [PubMed] [Google Scholar]
- Beavis, W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by A. H. Paterson. CRC Press, New York.
- Blanc, J. M., and H. Poisson, 1983. Parental sources of variation in hatching and early survival rates of Salmo trutta × Salvelinus fontinalis hybrid. Aquaculture 32: 115–122. [Google Scholar]
- Bongers, A. B. J., E. P. C. In't Veld, K. Abo-Hashema, I. M. Bremmer, E. H. Eding et al., 1994. Androgenesis in common carp (Cyprinus carpio) using UV irradiation in a synthetic ovarian fluid and heat shocks. Aquaculture 122: 119–132. [Google Scholar]
- Bongers, A. B. J., J. B. Abarca, B. Z. Coulabi, E. H. Eding, J. Komen et al., 1995. Maternal influence on development of androgenetic clones of common carp, Cyprinus carpio L. Aquaculture 137: 139–147. [Google Scholar]
- Borevitz, J. O., J. N. Maloof, J. Lutes, T. Dabi, J. L. Redfern et al., 2002. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman, K. W., H. Wu, S. Sen and G. A. Churchill, 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown, K. H., R. W. Lee and G. H. Thorgaard, 2006. Use of androgenesis for estimating maternal and mitochondrial genome effects on development and oxygen consumption in rainbow trout, Oncorhynchus mykiss. Comp. Biochem. Physiol. 143B: 415–421. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzmann, R. G., M. Cairney, W. S. Davidson, M. M. Ferguson, K. Gharbi et al., 2005. A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae). Genome 48: 1037–1051. [DOI] [PubMed] [Google Scholar]
- Dickhoff, W. W., C. L. Brown, C. V. Sullivan and H. A. Bern, 1990. Fish and amphibian models for developmental endocrinology. J. Exp. Zool. 4: 90–97. [Google Scholar]
- DiMichele, L., and D. A. Powers, 1991. Allozyme variation, developmental rate, and differential mortality in the teleost Fundulus heteroclitus. Physiol. Zool. 64: 1426–1443. [Google Scholar]
- Einum, S., and I. A. Fleming, 1999. Maternal effects of egg size in brown trout (Salmo trutta): norms of reaction to environmental quality. Proc. R. Soc. Lond. B Biol. Sci. 266: 2095–2100. [Google Scholar]
- Einum, S., and I. A. Fleming, 2000. Selection against late emergence and small offspring in Atlantic salmon (Salmo salar). Evolution 54: 628–639. [DOI] [PubMed] [Google Scholar]
- Einum, S., and I. A. Fleming, 2004. Environmental unpredictability and offspring size: conservative versus diversified bet-hedging. Evol. Ecol. Res. 6: 443–455. [Google Scholar]
- Eising, C. M., C. Eikenaar, H. Schwabl and T. G. G. Groothuis, 2001. Maternal androgens in black-headed gull (Larus ridibundus) eggs: consequences for chick development. Proc. R. Soc. Lond. B Biol. Sci. 268: 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot, J. M., 1989. Mechanisms responsible for population regulation in young migratory trout, Salmo trutta. I. The critical time for survival. J. Anim. Ecol. 58: 987–1002. [Google Scholar]
- Felip, A., W. P. Young, P. A. Wheeler and G. H. Thorgaard, 2005. An AFLP-based approach for the identification of sex-linked markers in rainbow trout (Oncorhynchus mykiss). Aquaculture 247: 35–43. [Google Scholar]
- Ferguson, M. M., R. G. Danzmann and F. W. Allendorf, 1985. Developmental divergence among hatchery strains of rainbow trout (Salmo gairdneri). I. Pure strains. Can. J. Genet. Cytol. 27: 289–297. [Google Scholar]
- Fry, J. D., S. V. Nuzhdin, E. G. Pasyukova and T. F. C. Mackay, 1998. QTL mapping for genotype-environment interaction for fitness in Drosophila melanogaster. Genet. Res. 71: 133–141. [DOI] [PubMed] [Google Scholar]
- Gerber, A. S., R. Loggins, S. Kumar and T. E. Dowling, 2001. Does nonneutral evolution shape observed patterns of DNA variation in animal mitochondrial genomes? Annu. Rev. Genet. 35: 539–566. [DOI] [PubMed] [Google Scholar]
- Heath, D. D., C. W. Fox and J. W. Heath, 1999. Maternal effects on offspring size: variation through early development of chinook salmon. Evolution 53: 1605–1611. [DOI] [PubMed] [Google Scholar]
- Hebert, K. P., P. L. Goddard, W. W. Smoker and A. J. Gharrett, 1998. Quantitative genetic variation and genotype by environment interaction of embryo development rate in pink salmon (Oncorhynchus gorbuscha). Can. J. Fish. Aquat. Sci. 55: 2048–2057. [Google Scholar]
- Hutchings, J. A., 1991. Fitness consequences of variation in egg size and food abundance in brook trout Salvelinus-Fontinalis. Evolution 45: 1162–1168. [DOI] [PubMed] [Google Scholar]
- Jiang, C. J., and Z. B. Zeng, 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.-H., Z. B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E., J. Abrahamson, A. Barlow, M. Daly, S. Lincoln et al., 1987 Mapmaker: a computer package for constructing genetic-linkage maps. Cytogenet. Cell Genet. 46: 642. [DOI] [PubMed]
- Leder, E. H., R. G. Danzmann and M. M. Ferguson, 2006. The candidate gene, Clock, localizes to a strong spawning time quantitative trait locus region in rainbow trout. J. Hered. 97: 74–80. [DOI] [PubMed] [Google Scholar]
- Leips, J., and T. F. C. Mackay, 2000. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155: 1773–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. H., 1998. Statistical Genomics: Linkage, Mapping and QTL Analysis. CRC Press, Boca Raton, FL.
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Martinez, V. A., W. G. Hill and S. A. Knott, 2002. On the use of doubled haploids for detecting QTL in outbred populations. Heredity 88: 423–431. [DOI] [PubMed] [Google Scholar]
- Martinez, V., G. Thorgaard, B. Robison and M. J. Sillanpaa, 2005. An application of Bayesian QTL mapping to early development in double haploid lines of rainbow trout including environmental effects. Genet. Res. 86: 209–221. [DOI] [PubMed] [Google Scholar]
- Mason, J. C., and D. W. Chapman, 1965. Significance of early emergence, environmental rearing capacity, and behavioral ecology of juvenile coho salmon in stream channels. J. Fish. Res. Board Can. 22: 173–190. [Google Scholar]
- May, B., and P. M. Grewe, 1993. Fate of maternal mtDNA following 60Co inactivation of maternal nuclear DNA in unfertilized salmonid eggs. Genome 36: 725–730. [DOI] [PubMed] [Google Scholar]
- McCormick, M. I., and I. V. Nechaev, 2002. Influence of cortisol on developmental rhythms during embryogenesis in a tropical damselfish. J. Exp. Zool. 293: 456–466. [DOI] [PubMed] [Google Scholar]
- McGowan, R. A., and C. C. Martin, 1997. DNA methylation and genome imprinting in the zebrafish, Danio rerio: some evolutionary ramifications. Biochem. Cell Biol. 75: 499–506. [PubMed] [Google Scholar]
- McIntyre, J. D., and J. M. Blanc, 1972. A genetic analysis of hatching time in steelhead trout (Salmo gairdneri). J. Fish. Res. Board Can. 30: 137–139. [Google Scholar]
- Mousseau, T. A., and C. W. Fox, 1998. The adaptive significance of maternal effects. Trends Ecol. Evol. 13: 403–407. [DOI] [PubMed] [Google Scholar]
- Nagler, J. J., 2000. In vivo treatment with cycloheximide or actinomycin D inhibits early embryonic development in rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem. 22: 61–66. [Google Scholar]
- Nichols, K. M., W. P. Young, R. G. Danzmann, B. D. Robison, C. Rexroad et al., 2003. A consolidated linkage map for rainbow trout (Oncorhynchus mykiss). Anim. Genet. 34: 102–115. [DOI] [PubMed] [Google Scholar]
- Oakley, T. H., and R. B. Phillips, 1999. Phylogeny of salmonine fishes based on growth hormone introns: Atlantic (Salmo) and Pacific (Oncorhynchus) salmon are not sister taxa. Mol. Phylogenet. Evol. 11: 381–393. [DOI] [PubMed] [Google Scholar]
- O'Malley, K. G., T. Sakamoto, R. G. Danzmann and M. M. Ferguson, 2002. Quantitative trait loci for spawning date and body weight in rainbow trout: testing for conserved effects across ancestrally duplicated chromosomes. J. Hered. 94: 273–284. [DOI] [PubMed] [Google Scholar]
- Pakkasmaa, S., O. P. Penttinen and J. Piironen, 2006. Metabolic rate of Arctic charr eggs depends on their parentage. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 176: 387–391. [DOI] [PubMed] [Google Scholar]
- Parsons, J. E., and G. H. Thorgaard, 1984. Induced androgenesis in rainbow trout. J. Exp. Zool. 231: 407–412. [Google Scholar]
- Phillips, R. B., M. R. Morasch, P. A. Wheeler and G. H. Thorgaard, 2005. Rainbow trout (Oncorhynchus mykiss) of Idaho and Alaskan origin (2n=58) share a chromosome fusion relative to trout of California origin (2n=60). Copeia 3: 661–664. [Google Scholar]
- Rexroad, C. E., III, R. L. Coleman, A. M. Martin, W. K. Hershberger and J. Killefer, 2001. Thirty-five polymorphic microsatellite markers for rainbow trout (Oncorhynchus mykiss). Anim. Genet. 32: 316–331. [DOI] [PubMed] [Google Scholar]
- Ristow, S. S., L. D. Grabowski, C. Ostberg, B. Robison and G. H. Thorgaard, 1998. Development of long-term cell lines from homozygous clones of rainbow trout. J. Aquat. Anim. Health 10: 75–82. [Google Scholar]
- Robison, B. D., P. A. Wheeler and G. H. Thorgaard, 1999. Variation in development rate among clonal lines of rainbow trout (Oncorhynchus mykiss). Aquaculture 173: 131–141. [Google Scholar]
- Robison, B. D., P. A. Wheeler, K. Sundin, P. Sikka and G. H. Thorgaard, 2001. Composite interval mapping reveals a major locus influencing embryonic development rate in rainbow trout (Oncorhynchus mykiss). J. Hered. 92: 16–22. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoker, W. W., 1986. Variability of embryo development rate, fry growth, and disease susceptibility in hatchery stocks of chum salmon. Aquaculture 57: 219–226. [Google Scholar]
- Stratholt, M. L., E. M. Donaldson and N. R. Liley, 1997. Stress induced elevation of plasma cortisol in adult female coho salmon (Oncorhynchus kisutch) is reflected in egg cortisol content, but does not appear to affect early development. Aquaculture 158: 141–153. [Google Scholar]
- Sundin, K., K. H. Brown, R. E. Drew, K. M. Nichols, P. A. Wheeler et al., 2005. Genetic analysis of a development rate QTL in backcrosses of clonal rainbow trout, Oncorhynchus mykiss. Aquaculture 247: 75–83. [Google Scholar]
- Sundstrom, L. F., M. Lohmus, J. I. Johnsson and R. H. Devlin, 2004. Growth hormone transgenic salmon pay for growth potential with increased predation mortality. Proc. R. Soc. Lond. B Biol. Sci. 271: S350–S352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom, L. F., M. Lohmus and R. H. Devlin, 2005. Selection on increased intrinsic growth rates in coho salmon, Oncorhynchus kisutch. Evolution 59: 1560–1569. [PubMed] [Google Scholar]
- Tallman, R. F., 1986. Genetic differentiation among seasonally distinct spawning populations of chum salmon, Oncorhynchus keta. Aquaculture 57: 211–217. [Google Scholar]
- Vandeputte, M., E. Quillet and B. Chevassus, 2002. Early development and survival in brown trout (Salmo trutta fario L.): indirect effects of selection for growth rate and estimation of genetic parameters. Aquaculture 204: 435–445. [Google Scholar]
- Vieira, C., E. G. Pasyukova, Z. B. Zeng, J. B. Hackett, R. F. Lyman et al., 2000. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher, P. M., R. Thompson and C. S. Haley, 1996. Confidence intervals in QTL mapping by bootstrapping. Genetics 143: 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTL. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I. A., E. H. Leder, W. W. Smoker and A. J. Gharrett, 2006. Timing of development during epiboly in embryos of second-generation crosses and backcrosses between odd- and even-broodyear pink salmon, Oncorhynchus gorbuscha. Environ. Biol. Fishes 75: 325–332. [Google Scholar]
- Young, W. P., P. A. Wheeler, R. D. Fields and G. H. Thorgaard, 1996. DNA fingerprinting confirms isogenicity of androgenetically derived rainbow trout lines. J. Hered. 87: 77–81. [DOI] [PubMed] [Google Scholar]
- Young, W. P., P. A. Wheeler, V. H. Coryell, P. Keim and G. H. Thorgaard, 1998. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]