Abstract
Temperature-sensitive mutations in subunits of the Caenorhabditis elegans anaphase-promoting complex (APC) arrest at metaphase of meiosis I at the restrictive temperature. Embryos depleted of the APC co-activator FZY-1 by RNAi also arrest at this stage. To identify regulators and potential substrates of the APC, we performed a genetic suppressor screen with a weak allele of the APC subunit MAT-3/CDC23/APC8, whose defects are specific to meiosis. Twenty-seven suppressors that resulted in embryonic viability and larval development at the restrictive temperature were isolated. We have identified the molecular lesions in 18 of these suppressors, which correspond to five genes. In addition to a single intragenic suppressor, we found mutations in the APC co-activator fzy-1 and in three spindle assembly checkpoint genes, mdf-1, mdf-2, and mdf-3/san-1, orthologs of Mad1, Mad2, and Mad3, respectively. Reduction-of-function alleles of mdf-2 and mdf-3 suppress APC mutants and exhibit pleiotropic phenotypes in an otherwise wild-type background. Analysis of a single separation-of-function allele of mdf-1 suggests that MDF-1 has a dual role during development. These studies provide evidence that components of the spindle assembly checkpoint may regulate the metaphase-to-anaphase transition in the absence of spindle damage during C. elegans meiosis.
SUCCESSFUL cell division requires that a cell faithfully duplicate and segregate its chromosomes to two daughter cells. Errors in this process can lead to aneuploidy, in which insufficient or extra genetic material is incorporated into the new cells. Aneuploidy can result in cell abnormalities or death and has been associated with most types of human cancers. In meiosis, the process of producing haploid gametes from diploid germ cells, aneuploidy often results in embryonic lethality or birth defects.
The major regulator of eukaryotic chromosome segregation is the anaphase-promoting complex (APC), a highly conserved protein complex consisting of 10–15 subunits, depending on the species (Harper et al. 2002; Irniger 2006). The APC is an E3 ubiquitin ligase, which transfers multiple ubiquitin molecules from a ubiquitin-conjugating enzyme (E2) to various substrates. Once polyubiquitinated in this fashion, target proteins are degraded by the 26S proteasome.
The regulated destruction of proteins is an important component of progression through the cell cycle. During M-phase, the E3 ubiquitin ligase activity of the APC is required for the metaphase-to-anaphase transition, the hallmark of which is chromosome segregation. At metaphase, chromosomes are pulled toward the spindle poles by microtubules attached to the kinetochore of each sister chromatid. This force is counteracted by the cohesin complex, which holds the sister chromatids together. For chromosome segregation to occur, the cohesion between chromosomes must be dissolved. This is accomplished via the proteolytic cleavage of cohesin by the enzyme separase. APC-mediated ubiquitination and subsequent degradation of the inhibitory protein securin liberates active separase and initiates a cascade of events that typify anaphase. The pivotal role of the APC in this process is underscored by the observation that mutant subunits of the APC in several organisms result in metaphase arrest (Sikorski et al. 1993; Furuta et al. 2000; Golden et al. 2000; Wirth et al. 2004).
To prevent chromosome missegregation, the activity of APC is tightly regulated and multiple mechanisms of regulation have been described. First, the APC has a transiently associated subunit that acts as a co-activator of APC activity. To date, several co-activators have been identified: Cdh1, specific for APC activity required during G1, and Cdc20, the M-phase co-activator (Dawson et al. 1995; Fang et al. 1998; Kitagawa et al. 2002). In addition, meiosis-specific co-activators of the APC such as Ama1 in budding yeast and cortex in Drosophila have also been identified (Cooper et al. 2000; Chu et al. 2001). All other described APC regulatory mechanisms work by modifying co-activator–APC interactions. For example, the phosphorylation of several APC subunits occurs under certain conditions and this modifies the ability of Cdc20 to bind to and activate the APC, resulting in alterations of the ubiquitin ligase activity (Rudner and Murray 2000). Another point of APC regulation is by the spindle assembly checkpoint, which modulates APC activity by preventing the association of Cdc20 with the APC, thus inhibiting APC activity (Chan et al. 2005).
The spindle assembly checkpoint detects faulty or incomplete spindle assembly by monitoring the presence of unattached kinetochores or lack of tension in the spindle, which often results from monopolar chromosome attachment. When these defects occur, the spindle checkpoint is activated and transmits a “wait anaphase” signal, delaying the onset of anaphase by inhibiting APC activity through sequestration of Cdc20. This delay often is sufficient for a cell to correct the spindle problem, which satisfies the checkpoint and allows progression through the cell cycle. The spindle checkpoint was first characterized in budding yeast and includes six proteins (Mad1, Mad2, Mad3, Bub1, Bub3, Mps1), of which only Mps1 is essential (Hoyt et al. 1991; Li and Murray 1991; Poch et al. 1994). In metazoans, additional proteins such as Rod and Zw10, among others, also contribute to checkpoint activity (Basto et al. 2000; Encalada et al. 2005). A role for the checkpoint in meiosis has also been demonstrated (Shonn et al. 2000). Orthologs of checkpoint proteins in metazoans exhibit a conserved function in activating the spindle checkpoint in cells that have suffered spindle damage (Kitagawa and Rose 1999; Dobles et al. 2000). Importantly, it has been shown that the spindle checkpoint proteins monitor chromosome attachment and tension during an unperturbed cell cycle, permitting anaphase to occur only after all microtubule–chromosome attachments have been achieved (Taylor and McKeon 1997; Gorbsky et al. 1998). This role for the checkpoint in the wild-type cell cycle is consistent with the data that deletions of these genes are often lethal to the organism (Kitagawa and Rose 1999; Dobles et al. 2000; Baker et al. 2004, 2006). There is also a correlation of spindle assembly defects with genome instability in cancer (Bharadwaj and Yu 2004).
It is well established that the readout of the checkpoint is inhibition of the APC co-activator Cdc20 (Hwang et al. 1998). Both Mad2 and Mad3 are able to bind directly to Cdc20 and the Mad2/Cdc20 and Mad3/Cdc20 interactions reduce APC activity in vitro (Hwang et al. 1998; Kim et al. 1998). All other checkpoint proteins have roles upstream of these interactions with Cdc20. Bub1 and Bub3 are localized to kinetochores and may be involved in recruiting other checkpoint proteins to this site (Taylor and McKeon 1997; Millband and Hardwick 2002; Encalada et al. 2005). Mad1–Mad2 binding results in a conformational change in Mad2 in which the activated form can bind Cdc20 (Luo et al. 2002; Sironi et al. 2002).
The spindle checkpoint proteins are highly conserved across species, consistent with observations that these proteins have conserved functions. The Caenorhabditis elegans genome contains orthologs of five of the six canonical checkpoint proteins. mdf-1, mdf-2, and mdf-3/san-1 are the orthologs of budding yeast Mad1, Mad2, and Mad3, respectively (Kitagawa and Rose 1999; Nystul et al. 2003). Orthologs have also been identified for Bub1 (bub-1) (Oegema et al. 2001) and Bub3 (Y54G9A.6; our unpublished observations), but not for Mps1. Analysis of the function of these genes in C. elegans has thus far been restricted to RNA interference (RNAi) and an mdf-1 deletion mutant, mdf-1(gk2). To date, no alleles of these genes have been recovered in forward genetic screens. mdf-1 is essential in C. elegans, as strains homozygous for a deletion allele cannot be maintained for more than two or three generations (Kitagawa and Rose 1999). A reduction-of-function mutation in the APC subunit emb-30 restores viability to this strain (Furuta et al. 2000). Depletion of MDF proteins allows progression through the cell cycle upon nocodazole treatment in the mitotic germline and early embryos, reflecting a deficiency in the spindle checkpoint (Kitagawa and Rose 1999; Encalada et al. 2005). mdf-3/san-1 [suspended animation (anoxia-induced) defective] was characterized in an RNAi screen for genes that confer sensitivity to anoxia (Nystul et al. 2003). For clarity, and consistency within the cell cycle field, we use the gene name mdf-3/san-1 rather than san-1 throughout this report.
Previously, we and others identified nine orthologs of APC subunits in the nematode C. elegans (Furuta et al. 2000; Golden et al. 2000; Davis et al. 2002; Shakes et al. 2003). RNAi of seven of these subunits and of the Cdc20 ortholog fzy-1 results in a strong maternal-effect embryonic lethal phenotype: embryos arrest at the one-cell stage, specifically at metaphase of the first meiotic division. Furthermore, temperature-sensitive mutations of five of these genes also cause meiotic metaphase I arrest when L4 hermaphrodites are shifted to restrictive conditions. Taken together, these results were the first demonstration of a requirement for the APC at the metaphase-to-anaphase transition in meiosis I.
To identify regulators and targets of the APC in C. elegans, we performed a suppressor screen on a mat-3 temperature-sensitive mutant. MAT-3 is a regulatory, TPR-repeat-containing subunit of the APC, orthologous to Cdc23 in Saccharomyces cerevisiae. The mat-3(or180) mutation was selected because the defect in this mutant is confined to meiosis. While other temperature-sensitive mat-3 alleles display pleiotropic somatic and germline defects, mat-3(or180) two-cell embryos shifted to the restrictive temperature exhibit no developmental effects. Thus, in this screen, suppressor mutations only had to be sufficient to increase the APC activity at the meiotic divisions to restore viability. For this reason, we anticipated that we would recover global regulators of the APC as well as meiosis-specific factors.
In this study, we describe the recovery of 27 mat-3(or180) suppressors and give them the gene name som (suppressor of metaphase arrest). All strains carrying both the mat-3(or180) and som mutations are viable and fertile at restrictive temperatures, although most double mutants display phenotypes suggestive of meiotic chromosome segregation defects. Three of the suppressors are recessive, while the remainder are dominant or semidominant. Each suppressor has been mapped to a small genetic interval and altogether they define a minimum of nine genes. We have identified the molecular lesion in 18 of these suppressors, which correspond to five genes. These include three spindle checkpoint genes, mdf-1, mdf-2, and mdf-3/san-1, as well as the Cdc20 ortholog fzy-1. We have genetically and phenotypically characterized these suppressors and show that fzy-1(av15) and the mdf-2 and mdf-3/san-1 alleles display embryonic lethality and/or brood size defects, while animals carrying the mdf-1 or fzy-1(av4) allele are wild type. We also found that four of the suppressors are able to suppress the synthetic lethality observed when MDF-1 is depleted by RNAi in a mat-3(or180) animal. The remaining uncloned suppressors do not map near known spindle checkpoint genes and thus are likely to provide novel insight into the regulation of the APC.
MATERIALS AND METHODS
Nematode strains and culture techniques:
Except for the mapping strain AG153, all strains were derivatives of N2 (Bristol). AG153 was derived from CB4856 (Hawaii). We constructed AG153 by crossing CB4856 to a mat-3(or180) dpy-1(e1) N2 derivative. Dumpy (Dpy) F2 progeny of this cross were tested for the presence of the mat-3(or180) phenotype and were then backcrossed to CB4856 six times, allowing for the loss of dpy-1(e1). In the last backcross, non-Dpy progeny displaying the Mat phenotype were selected. These progeny were tested for the presence of CB4856 single nucleotide polymorphisms (SNPs) by the snip-SNP method (Wicks et al. 2001). SNP markers for the left arm, right arm, and center of each linkage group were CB4856 specific except for the SNP marker pkP3045, which is on the left arm of LG III in the vicinity of mat-3 (http://www.wormbase.org). Nematodes were cultured by standard methods.
Mutagenesis:
Mixed-stage mat-3(or180) hermaphrodites were suspended in M9 containing 50 mm ethyl methanesulfonate at 15° for 4 hr. The animals were washed in M9 three times and allowed to recover on a modified Youngren's, only Bacto-peptone (MYOB) plate at 15° for 1 hr. Twenty-five L4-stage worms were transferred to each of 50 10-cm-diameter MYOB plates. Plates were incubated at 15° until the F1 generation had begun producing F2 embryos. Animals and F2 embryos were washed off of the plates and treated with hypochlorite solution to isolate embryos. The F2 embryos were plated on MYOB plates without food and allowed to hatch at 15°. The synchronized L1 larvae were washed off of the plates and transferred to high growth plates (MYOB plates containing 2% peptone and the Escherichia coli strain NA22). These F2 animals were incubated at 15° until the L4 larval stage. Plates were then transferred to 25.5° for 2 weeks and screened for visible larvae. Larvae from a single plate were treated as one independent suppressor line and were transferred to fresh plates. Candidate suppressors were retested for suppression at 25.5° at the L4 larval stage. Each suppressor was then outcrossed by mating with the original mat-3(or180) strain at least three times. Further investigation revealed that the Mat phenotype of mat-3(or180) animals was completely penetrant at 24°, and for that reason all experiments that follow the completion of the screen were done at 24° and not at 25.5°.
Genetic mapping:
To facilitate mapping, mat-3(or180) was linked to dpy-1(e1) and then a triple mutant was made with each suppressor. The suppressor was then mapped to a chromosome by mating a mat-3(or180) dpy-1(e1); suppressor hermaphrodite to the CB4856-mat-3(or180) derivative strain AG153 at the permissive temperature of 20°. Non-Dpy F1 worms were picked and grown at 20°. Non-Dpy and Dpy F2 worms were picked as L4-stage larvae at a ratio of 3:1 and arrayed into individual wells of 24-well dishes at 24° for 2 days to score for suppression. For mapping of av4, av14, and av20, 100 suppressed (“mutant”) and 100 unsuppressed (“wild-type”) worms were picked into individual tubes and lysed as described (Wicks et al. 2001). We found that the chromosomal assignments for some of the suppressors could be defined with as few as 20 each of mutant and wild-type F2 segregants. An equal portion of each single-worm lysate was pooled into one tube to create a bulk lysate. Chromosomal assignments were made using the snip-SNP/bulked segregant analysis method as described (Wicks et al. 2001). We further refined the genetic map position of the recessive av20 suppressor by analyzing SNP markers in 500 individual mutant (suppressed) F2 segregants.
Once each mutant was assigned to a specific linkage group, we used classical genetic mapping with morphological markers to further refine the map position of each suppressor. The following strains were used for genetic mapping: AG154 mat-3(or180); dpy-5(e61) unc-13(e450), AG155 mat-3(or180); dpy-10(e128) unc-4(e120), AG162 mat-3(or180) dpy-17(e164) unc-32(e189), AG163 mat-3(or180) unc-93(e1500) dpy-17(e164), AG164 mat-3(or180) unc-32(e189) dpy-18(e364), AG50 daf-7(e1372ts) dpy-1(e1), AG156 mat-3(or180); lin-1(e1777) unc-17(e245), AG157 mat-3(or180); unc-17(e113) dpy-13(e184), AG158 mat-3(or180); dpy-13(e184) unc-24(e138), AG159 mat-3(or180); unc-24(e138) dpy-20(e1282), and AG161 mat-3(or180); dpy-11(e224) unc-42(e270). All strains used for the construction of these mapping strains were obtained from the Caenorhabditis Genetics Center (Minneapolis). In general, mat-3(or180); suppressor males were mated into mapping strain mat-3(or180); m1m2. Non-M1M2 F1 cross progeny hermaphrodites were selected and allowed to self. M1–non-M2 or M2–non-M1 homozygotes were generated from F2 recombinants and shifted to 24° to determine if the suppressor was present. Map distance was calculated on the basis of the percentage of recombinants in which the suppressor had recombined onto the same chromosome as the marker. Suppressors on LGIII were initially mapped in the region near mat-3 to determine if these suppressors might be alleles of mat-3. These mapping experiments required a slightly different strategy due to the proximity of the original mutation. mat-3(or180) suppressor males were mated to a daf-7(e1372ts) dpy-1(e1) line at 20°. Non-Dpy non-Dauer cross progeny were selected and allowed to reproduce at 24° where daf-7(e1372ts) animals are constitutively dauer. Dpy non-Dauer recombinants were picked at 24°. By definition, since mat-3 maps to the left of the daf-7 dpy-1 interval, these animals pick up mat-3(or180) upon recombination. Homozygotes for the marker dpy-1 [and thus mat-3(or180)] were generated from these recombinants and tested for the presence of the suppressor. Map distance was then calculated.
A complementation test was performed with av6 and dpy-10(e128). Male mat-3(or180); av6 animals were mated into dpy-10(e128) hermaphrodites. The success of this mating was confirmed by the presence of males in the F1 progeny. The F1 were then scored for the production of non-Dpy progeny, indicative of complementation.
To test for genetic interactions between fzy-1 and mat-3, mat-3(or180); him-8(e1489) males were mated with fzy-1(h1983) dpy-10(e128) hermaphrodites. Non-Dpy F1 cross progeny were picked and allowed to self. Non-Dpy F2 were picked to single plates and individuals homozygous for mat-3(or180) were determined. Dpy F3 mat-3(or180); fzy-1(h1983) dpy-10(e128) hermaphrodites were assessed at permissive temperature (15°) for fertility and production of viable progeny.
The mdf-3/san-1 deletion strain mdf-3/san-1(ok1580) was outcrossed 10 times to dpy-5(e61) unc-13(e450). In the final mating, non-Dpy non-uncoordinated (Unc) F1 hermaphrodites were picked to individual plates. Hermaphrodites that did not segregate DpyUncs were genotyped with PCR to confirm the presence of mdf-3/san-1(ok1580). mdf-3/san-1(ok1580) was marked by picking Dpy non-Unc and Unc non-Dpy progeny from the 10th outcross, screening for marked homozygotes in the next generation. PCR genotyping was used to confirm the presence of the deletion. We determined the endpoints of the deletion by PCR and sequencing. The breakpoint was determined to be between nucleotides 414 and 1406, which deletes all of exon 3 through the middle of intron 7.
To genetically test if our alleles of mdf-1 and mdf-3/san-1 behaved as gain- or loss-of-function mutations, the mdf-1(gk2) and mdf-3/san-1(ok1580) deletion alleles were utilized. A mat-3(or180); dpy-11(e224) mdf-1(gk2) strain was generated from the parent strain KR3876 fzy-1(h1983) dpy-10(e128); unc-46(e177) mdf-1(gk2). To balance the lethal unc-46(e177) mdf-1(gk2) chromosome, KR3876 hermaphrodites were mated with dpy-11(e224) unc-42(e270)/+ males. F1 cross progeny were selfed and balanced F2 progeny that did not segregate Dpy-10 animals were selected to obtain animals wild type at the fzy-1 locus [unc-46(e177) mdf-1(gk2)/dpy-11(e224) unc-42(e270)]. These hermaphrodites were selfed and dpy-11 non-unc-42 recombinants were selected and mated with mat-3(or180); dpy-11(e224) unc-42(e270)/+ males. F1 cross progeny of this mating were selfed and Dpy F2 progeny that segregate Dpy Uncs were genotyped for the presence of mat-3(or180) (see materials and methods). The final strain was also PCR genotyped to confirm the presence of mdf-1(gk2). The strain was maintained as mat-3(or180); dpy-11(e224) mdf-1(gk2)/dpy-11(e224) unc-42(e270) until it was determined that the mat-3(or180); dpy-11(e224) mdf-1(gk2) strain was nonlethal at 20°. mat-3(or180); him-8(e1489) males or mat-3(or180); mdf-1(av19) males were mated with the marked deletion strain mat-3(or180); dpy-11(e224) mdf-1(gk2). Non-Dpy F1 were shifted to 24° and tested for suppression. Dpy F2 were also tested by this assay at 24°. Similarly for mdf-3/san-1, mat-3(or180); him-8(e1489), mat-3(or180); mdf-3/san-1(av20), or mat-3(or180); mdf-3/san-1(av31) males were mated to mat-3(or180); mdf-3/san-1(ok1580) unc-13(e450) hermaphrodites. Non-Unc F1 L4 progeny were shifted to 24° and tested for embryonic viability of their progeny.
Sequencing:
For sequencing, candidate genes were PCR amplified from genomic DNA isolated from mutant strains. PCR products were then purified and submitted for sequencing by SeqWright (Houston).
Suppression:
To assess embryonic hatching, a minimum of 20 L4 mat-3(or180); suppressor hermaphrodites were shifted to the restrictive temperature (24°) for 24 hr. Hermaphrodites were then removed and embryos were allowed to develop for an additional 24 hr. At that time, unhatched embryos and larvae were counted and the percentage that hatched was calculated as the number of larvae/brood (larvae plus embryos). Males were also counted and incidence of males was calculated as the number of males/total hatched larvae.
Dominance:
Each suppressor was determined to be dominant or recessive by mating mat-3(or180); suppressor males into a mat-3(or180) dpy-1(e1) line. Five non-Dpy L4 hermaphrodites were shifted to restrictive temperature (24°) and the percentage of embryos hatching was determined as above. If no viable progeny was observed, the suppressor was designated as recessive. Suppressors producing any hatching larvae when heterozygous were considered dominant. Suppressors were considered weakly dominant if heterozygous suppression was <10% of the homozygous suppression, semidominant if between 10 and 70% of the homozygous suppression, and fully dominant if >70% of the homozygous suppression.
Suppression of mat-3 RNAi:
To test if any of the suppressors could suppress a strong loss of function in mat-3, we combined the mat-3(or180) allele with mat-3 RNAi. Five L4 hermaphrodites with the genotype mat-3(or180); suppressor were grown on mat-3 RNAi at 24° for 48 hr. The percentage of embryonic hatching was determined for the 24- to 48-hr period, when RNAi is most effective. For further examination of som-3(av29), animals were treated as above, using singly plated animals to assess worm-to-worm variation. Control experiments indicate that mat-3(or180) som-3(av29) animals are slightly resistant to RNAi.
RNAi:
Bacteria containing RNAi constructs either were taken from the C. elegans bacterial feeding library (Fraser et al. 2000; Kamath et al. 2003) (Geneservices, Cambridge, UK) (mat-3, mdf-1, mdf-3/san-1, fzy-1) or were generated in the laboratory (mdf-2). The constructs from Geneservices were sequenced to confirm that they included the correct gene insert. The mdf-2 construct consisted of a genomic fragment (410 bp) spanning from exon 1 to exon 3. Briefly, a PCR product from the mdf-2 gene was ligated into the L4440 (Timmons et al. 2001) vector and then transformed into HT115 (DE3) bacteria. To induce RNAi, bacteria were seeded on plates containing carbenicillin (25 μg/ml; Sigma, St. Louis) and IPTG (2 mm; Roche) and allowed to grow over 2–3 days at 22°. For all experiments, L4 hermaphrodites were grown on RNAi plates and the percentage of embryos hatching was determined for embryos laid during the 24- to 48-hr period.
Attempts to suppress the mat-3(or180) one-cell arrest by mdf-3/san-1 RNAi feeding were not successful, so RNAi injections were performed. Briefly, the RNAi construct was purified from bacteria, and double-stranded RNAi was synthesized using the Ambion Megascript T7 kit (Ambion, Austin, TX). RNA was microinjected into the body cavity of young adult hermaphrodites. Hermaphrodites recovered at 16° for 6–20 hr and then were shifted to restrictive temperature. Plates were monitored for the production of larvae or multicellular embryos.
Phenotypic analysis of spindle checkpoint mutants:
Spindle checkpoint supressors were linked to morphological markers and crossed into a wild-type background to separate the suppressor from the mat-3(or180) mutation. A single HinfI restriction site is eliminated by the point mutation in the mat-3(or180) strain and this snip-SNP marker was used to genotype the resulting F2 progeny for loss of the mat-3(or180) mutation. To determine brood size, 10 L4 hermaphrodites were shifted to the restrictive temperature (24°) and allowed to lay their entire brood; embryonic hatching was calculated. We accounted for the death of P0 hermaphrodites during brood experiments by noting the number of hermaphrodites on the plate at the beginning of the day and calculating progeny per animal for each day. At the conclusion of the experiment, these values were added together for the total brood size. Reported brood size is the average of three experiments. To observe phenotypes of individual animals, 100 larvae (born at 24°) were picked at the L1 and L2 stages onto single plates at 24°. Three days after the shift of the F1 L1 larvae, these individuals were assessed for any phenotypic abnormalities such as larval arrest or lethality, somatic defects, production of F1 males, or sterility. Control strains contained only the morphological marker.
Suppression of other APC mutants:
To test for suppression of other APC subunit mutations, several alleles of five APC subunits and one allele of emb-1 were combined with a single allele of each spindle checkpoint mutant linked to a visible marker. Double mutants of marked APC or emb-1 and suppressors were generated with these strains in all combinations (m1 mat-X; m2 sup-X). Control strains contained both markers and the APC mutant allele but were wild type at the suppressor locus (m1 mat-X; m2 +). The lowest restrictive temperature (at which all embryos were arrested at the one-cell stage) for each mat/emb allele was determined by shifting control strains to 23°, 24°, and 25°. Double-mutant L4 hermaphrodites were shifted to the lowest restrictive temperature and allowed to lay embryos for 2 days, and embryonic hatching was calculated. If multicellular embryos or larvae were present on the plate, the double mutant was scored as suppressed. A minimum of 200 embryos and larvae were counted for each double mutant. All control strains produced one-cell embryos at the restrictive temperature with the exception of mat-1(ax212) and emb-30(or420), which produce multicellular embryos even at 25.5°.
RESULTS
Recovery and characterization of suppressors:
To identify potential regulators and targets of the APC, we performed a suppressor screen with the temperature-sensitive mat-3(or180) allele, which has a meiosis-specific phenotype. mat-3(or180) L4 hermaphrodites shifted to 24° produce one-cell embryos arrested at metaphase of meiosis I (Golden et al. 2000). Chemical mutagenesis of 3.5 × 105 haploid genomes identified 29 suppressors (av4-av32) that were able to rescue the one-cell arrest phenotype of mat-3(or180) mutants and produce fertile adult progeny. Two of these mutants, av12 and av24, were extremely weak suppressors and were not characterized further. All mat-3(or180); suppressor mutants were morphologically wild type except for av6, which was Dpy. The suppressors exhibited a range of effectiveness in suppressing the embryonic lethality of mat-3(or180) at 24° (Table 1). The strongest suppressor had an embryonic viability of 98%, while in the weakest suppressor, only 18% of the embryos hatched. The percentage of embryos hatching was also assessed at 25° and in all cases suppression was less robust at this temperature (data not shown). Although many of the embryos produced by weaker suppressor lines did not hatch, the majority of these embryos complete meiosis and arrest as multicellular embryos. This indicates that the one-cell meiotic metaphase arrest phenotype was strongly suppressed in all cases. Multicellular embryonic arrest or lethality might be attributed to the weakness of the suppressor or, alternatively, the suppressor mutation itself may be moderately lethal at this stage.
TABLE 1.
Embryonic viability and incidence of males in mat-3(or180); suppressor strains
| Strain | Hatching (%) | Males (%) | n |
|---|---|---|---|
| N2 | 100 | 0.3 | 1831 |
| mat-3(or180) | 0 | 0 | 2901 |
| mat-3(or180av26) | 98 | 0.1 | 1936 |
| mat-3(or180); mdf-1(av19) | 68 | 1.7 | 2034 |
| mat-3(or180); mdf-2(av7) | 53 | 3.4 | 1132 |
| mat-3(or180); mdf-2(av8) | 57 | 2.9 | 2294 |
| mat-3(or180); mdf-2(av13) | 48 | 2.3 | 2316 |
| mat-3(or180); mdf-2(av14) | 53 | 1.6 | 1932 |
| mat-3(or180); mdf-2(av16) | 33 | 2.8 | 1643 |
| mat-3(or180); mdf-2(av17) | 47 | 2.9 | 1476 |
| mat-3(or180); mdf-2(av18) | 77 | 3.6 | 2435 |
| mat-3(or180); mdf-2(av22) | 55 | 2.9 | 822 |
| mat-3(or180); mdf-2(av25) | 64 | 1.4 | 1475 |
| mat-3(or180); mdf-2(av27) | 62 | 3.5 | 1261 |
| mat-3(or180); mdf-2(av30) | 50 | 2.0 | 1119 |
| mat-3(or180); mdf-3/san-1(av20) | 85 | 2.6 | 2489 |
| mat-3(or180); mdf-3/san-1(av31) | 58 | 4.2 | 1565 |
| mat-3(or180); fzy-1(av4) | 53 | 3.4 | 1326 |
| mat-3(or180); fzy-1(av15) | 74 | 1.3 | 1227 |
| mat-3(or180); fzy-1(av23) | 52 | 1.2 | 1535 |
| mat-3(or180); som-1(av5) | 59 | 3.1 | 2060 |
| mat-3(or180); som-2(av9) | 64 | 0.7 | 1480 |
| mat-3(or180); som-2(av21) | 18 | 8.1 | 1360 |
| mat-3(or180); som-3(av10) | 79 | 1.6 | 1553 |
| mat-3(or180); som-3(av11) | 91 | 0.4 | 2038 |
| mat-3(or180); som-3(av29) | 95 | 1.2 | 1055 |
| mat-3(or180); som-3(av32) | 71 | 2.3 | 1719 |
| mat-3(or180); som-4(av28) | 42 | 3.2 | 1911 |
| mat-3(or180); dpy-10(av6) | 72 | 2.7 | 347 |
A minimum of 20 L4 hermaphrodites were shifted to restrictive temperature (24°) for 24 hr and the percentage of progeny hatching was assessed. The incidence of males was calculated as a percentage of hatched progeny. N2 is the wild-type strain. n is the total number of embryos and larvae assayed to determine embryonic viability and incidence of males.
In C. elegans, nondisjunction in meiosis can result in the loss of an X chromosome, producing XO progeny, which are male. Thus, the generation of males in a population is an indicator that errors are occurring in chromosome segregation. We observed an incidence of males in mat-3(or180); suppressor lines up to 25 times that observed in wild-type animals (Table 1). This phenotype was enhanced at 25° (data not shown). Suppression in our screen required only that the APC function enough to allow chromosome segregation to occur. The fidelity of this segregation is likely not error free and defects in chromosome segregation during the meiotic and subsequent mitotic divisions likely account for the Him phenotype and embryonic lethality that we observe with most of the mat-3(or180); suppressor lines. It is unlikely that the Him phenotype is attributable exclusively to the suppressor mutations; mat-3(or180) animals are Him at semipermissive temperatures and the mat-3(or180); suppressor double mutants probably mimic these conditions.
Of the 27 suppressors characterized, only 3 were recessive: av6, av20, and av31 (Figure 1). All other suppressors were able to suppress mat-3(or180) when heterozygous, although a range of dominance was observed. Two suppressors (av10 and av32) were very weakly dominant; embryonic hatching of heterozygotes (av10/+, av32/+) was <10% of that observed when these suppressors are homozygous. Nine suppressors were semidominant and 13 were fully dominant.
Figure 1.—
Most suppressors are dominant suppressors of mat-3(or180). mat-3(or180); suppressor males were mated into the mat-3(or180) dpy-1 strain. Hermaphrodite non-Dpy F1 cross progeny were shifted to 24° at the L4 stage and embryonic viability of the F2 generation was monitored and compared to viability of the homozygous mat-3(or180); suppressor strain. ▪, homozygous; □, heterozygous.
Because the majority of the suppressors were dominant, we tested whether any of these suppressors were able to suppress a severe reduction of MAT-3. Because null alleles of mat-3 result in hermaphrodite sterility (Garbe et al. 2004), we sought an alternative method to reduce the function of MAT-3. mat-3 RNAi is very effective at eliminating MAT-3 function in wild-type animals and results in 96% embryonic lethality. To produce a strong loss of function, we combined mat-3 RNAi with the mat-3(or180) allele at 24°. Under these conditions, all but one of the suppressors produced broods that exhibited <4% embryonic viability. In contrast, som-3(av29) produced larvae at significant levels (30% hatching). However, this strain was also observed to be slightly resistant to RNAi of control constructs. Therefore, the suppressors isolated in our screen are unable to suppress a strong reduction in MAT-3 and thus do not act as bypass mutations.
Suppressor mapping:
Each suppressor was assigned to a chromosome by utilizing snip-SNP mapping (Wicks et al. 2001). Classical genetic mapping was employed to further refine the location of each suppressor on the chromosome (see materials and methods). In all, we identified a minimum of nine genes that map to five chromosomes (Figure 2). Seven dominant suppressors map to chromosome III. Mapping experiments with the daf-7 dpy-1 interval indicated that only one of these suppressors, av26, mapped to the left of daf-7, very close to mat-3. The mat-3 locus was sequenced from av26 animals and was found to contain a single point mutation corresponding to a leucine-to-phenylalanine substitution at amino acid 436. Thus, we have isolated one intragenic suppressor of mat-3. Of the remaining six suppressors on chromosome III, four (av10, av11, av29, and av32) mapped between dpy-17 and unc-32, while av9 and av21 mapped to the right of dpy-18 (Figure 2). We have tentatively designated the four suppressors between dpy-17 and unc-32 as alleles of som-3 due to predicted map position, although phenotypic differences (see below) indicate that these suppressors may define more than one gene. Similarly, we have made the conservative assignment of av9 and av21 as two alleles of a single gene, but these mutations may also represent two separate genes. Due to the dominance of these alleles, complementation tests cannot be performed to further clarify this issue.
Figure 2.—
Suppressors of mat-3(or180) map to a minimum of nine genes, five of which have been molecularly identified. Thick horizontal lines represent linkage groups I–V (LG X is not shown). Thick vertical lines indicate molecularly identified suppressors. The number of alleles recovered for a given gene is indicated in brackets following the gene name. Markers shown below the line represent morphological markers used for mapping. Thin vertical lines indicate the location of uncloned suppressors based on genetic mapping and phenotypic characterization. Bar, 5 cM.
Our genetic data suggested that the suppressor av6 is an allele of dpy-10. av6 mapped to chromosome II, was recessive, and was morphologically Dpy. In addition, av6 failed to complement dpy-10(e128). dpy-10(e128) was able to suppress mat-3(or180) and an additional allele of mat-3 (data not shown). Alleles of two other dpy genes, dpy-11 and dpy-20, also weakly suppressed mat-3(or180) (data not shown). Other reports indicate that some dpy mutations are able to suppress a variety of unrelated mutations, possibly by inducing a physiological stress response in the animal that induces chaperones, leading to a higher level of functional protein (Maine and Kimble 1989; Kramer and Johnson 1993).
The remaining suppressors were mapped to small genetic regions. som-1(av5) was mapped to a defined interval of LG IV (Figure 2). We are currently working toward its identification. Mapping indicated that we had also isolated two recessive alleles of a gene on LG I, three alleles of a gene on LG II, 11 (possibly 12; see discussion of som-4 below) alleles of a gene on LG IV, distinct from som-1(av5), and a single suppressor on LG V. Once each of these suppressors was mapped to a genetic interval of ≤3 cM, we began to look at the genomic sequence of C. elegans (http://www.wormbase.org) for candidate genes in these regions that might be anticipated to have a role in the regulation of the APC. Sequencing of candidate genes from the appropriate mutants was carried out to confirm the presence or absence of mutations.
Molecular identification of mutations in spindle checkpoint genes:
Using the approach detailed above, we identified a molecular lesion in 18 of the 27 suppressors, which define five genes (Table 2). In addition to an intragenic mat-3 mutation (above), the remaining 17 suppressors are mutations in the spindle checkpoint genes mdf-1, mdf-2, mdf-3/san-1, and the APC activator fzy-1. The mutations in mdf-1, mdf-2, and mdf-3/san-1 are the first to be recovered in a C. elegans forward genetic screen. Thus, mdf-1, mdf-2, mdf-3/san-1, and fzy-1 are likely to act as regulators of the APC during meiosis, consistent with the well-defined role for these genes in meiosis and mitosis of other organisms (Chan et al. 2005).
TABLE 2.
Nucleotide and amino acid changes in suppressors of mat-3(or180)
| Gene | Allele | Mutation |
|---|---|---|
| mat-3 | or180 | S505F |
| av26 | L436F | |
| mdf-1 | av19 | A508V |
| mdf-2 | av7 | E121K |
| av8 | S187L | |
| av13 | G112E | |
| av14 | Intron 5 splice donor G → A | |
| av16 | E97K | |
| av17 | R98K | |
| av18 | T136I | |
| av22 | A172T | |
| av25 | L142F | |
| av27 | G14R | |
| av30 | E127K | |
| mdf-3/san-1 | av20 | Intron 5 splice donor G → A |
| av31 | Intron 5 splice acceptor G → A | |
| fzy-1 | av4 | T144A |
| av15 | A138V | |
| av23 | S143N |
Eighteen suppressors were sequenced and found to have mutations in five genes. The single amino acid change that results from the nucleotide point mutation is listed. When no amino acid changes result from the mutation, the site of the splice mutation is indicated. Also included is the amino acid change found in the parent strain, mat-3(or180).
mdf-2:
We recovered 11 alleles of mdf-2 in our screen, all of which acted as dominant suppressors of mat-3(or180). The majority of the alleles (10/11) are single nucleotide substitutions that resulted in an amino acid change (Table 2). Most of these were nonconservative mutations and several resulted in a charge alteration. One mutant allele, mdf-2(av14), contained a mutation at the splice donor site for intron 5 (GT → AT). Overall, the mutations are enriched in the C-terminal half of the protein (10/11), indicating that this region of the protein may be important in the regulation of the APC. An additional allele, som-4(av28), mapped to the mdf-2 region of chromosome IV, but sequencing of the mdf-2 exons, splice junctions, 102 bp 3′ of the stop codon, and 2 kb upstream of the initiator methionine revealed no nucleotide changes. We thus assign som-4(av28) a separate gene name at this time (Figure 2).
In other organisms, the mdf-2 ortholog, such as Mad2 in S. cerevisiae, acts as a direct negative regulator of Cdc20/FZY-1 (Hwang et al. 1998; Kim et al. 1998). Thus, we predicted that the mdf-2 alleles result in reduced inhibition of FZY-1, thereby increasing APC activity in the mat-3(or180) mutants. In C. elegans, RNAi can often recapitulate loss-of-function mutations. We found that mdf-2 RNAi was effective in suppressing mat-3(or180) at 24°: ∼60% of mat-3(or180) embryos survived to hatching (data not shown), comparable to the embryonic viability observed with the mdf-2 alleles. Therefore, the mutations recovered in our suppressor screen are likely loss-of-function mutations. Our genetics experiments suggest that reducing the dose of MDF-2 by ≤50% is enough to allow a compromised APC mutant to function; this attenuation of negative regulation is sufficient to restore the metaphase-to-anaphase transition in embryos. Thus, the role of mdf-2 in meiosis is consistent with the current understanding of the role of MDF-2 as an inhibitor of APC activity.
mdf-3/san-1:
Two recessive alleles of mdf-3/san-1 were recovered. Sequencing revealed point mutations at the splice donor (av20) and acceptor (av31) sites of intron 5 (Table 2). It is likely that mdf-3/san-1(av20) and mdf-3/san-1(av31) generate mdf-3/san-1 transcripts with a variety of 3′-ends, as work on other splice-site mutants has shown that alternative donor or acceptor sites can be utilized (Aroian et al. 1993). The translation of these aberrantly spliced transcripts may result in MDF-3 proteins with altered or truncated C termini. Alternatively, the aberrant transcripts may be detected and removed by the smg system (Mango 2001), yielding a reduction in mdf-3/san-1 message and protein. In parallel to the analysis of the mdf-3/san-1 alleles generated in this screen, the mdf-3/san-1(ok1580) deletion was also studied. The endpoints of the deletion were determined by sequencing and it was found that the deletion removes 992 nucleotides between exon 3 and intron 7. Thus, mdf-3/san-1(ok1580) is likely to be a null mutation, although severely truncated forms of the protein might be produced.
The identification of mdf-3/san-1(av20) and mdf-3/san-1(av31) as recessive mutations suggests that they result in a loss of function. We attempted to corroborate this hypothesis by testing whether the mdf-3/san-1 deletion was able to suppress mat-3(or180) at 24°. The mdf-3/san-1 deletion suppressed the one-cell arrest caused by mat-3(or180), although not as effectively as either point mutant (Figure 3). We also examined the phenotype of mdf-3/san-1(av20)/mdf-3/san-1(ok1580) and mdf-3/san-1(av31)/mdf-3/san-1(ok1580) and found that the embryonic viability of these lines at 24° was intermediate between that of the deletion and the two other alleles. The discrepancies in the degree of suppression provided by the three mutations might be ascribed to differences in strength or in the genetic background of these strains. We also tested whether RNAi of mdf-3/san-1 could suppress mat-3(or180). Modest suppression was observed upon double-strand RNA injection into hermaphrodites (data not shown). Together, these data indicate that the mdf-3/san-1 mutations are reduction-of-function mutations.
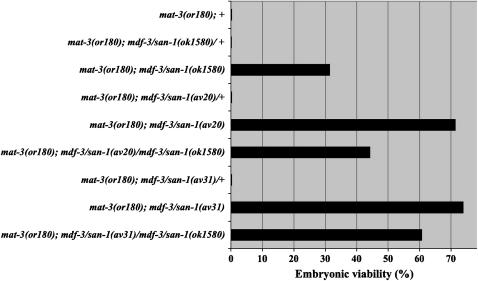
Figure 3.—
mdf-3/san-1(av20) and mdf-3/san-1(av31) are loss-of-function mutations. Genetic experiments with three mdf-3/san-1 alleles, mdf-3/san-1(ok1580), mdf-3/san-1(av20), and mdf-3/san-1(av31) indicate that mat-3(or180); mdf-3/san-1 suppression at 24° is recessive. Embryonic viability for all three mdf-3/san-1 strains was determined. For these experiments, an mdf-3/san-1(ok1580) deletion marked with unc-13 was used.
mdf-1:
One semidominant allele of mdf-1 was recovered. Sequencing revealed a point mutation resulting in an alanine-to-valine change in amino acid 508 (Table 2). This mutation occurs in close proximity to the region that mediates the Mad1/Mad2 interaction in the human protein (Sironi et al. 2002) (Figure 4A). To characterize the mdf-1(av19) mutation, we employed a genetic approach. We first tested whether an mdf-1 deletion, mdf-1(gk2), could suppress mat-3(or180). The mdf-1(gk2) deletion has been previously characterized (Kitagawa and Rose 1999) and animals that are homozygous for this mutation exhibit a reduced brood size, extensive embryonic and larval lethality, sterility, and a high incidence of males. The cumulative effect of these phenotypic defects is death of the homozygous strain after two or three generations. mdf-1(gk2); mat-3(or180) homozygotes produced no more live progeny than the control strain at 24° (Figure 4B). However, close examination of the embryos revealed that they were arrested at a multicellular stage of development. Thus, mdf-1(gk2) suppressed the one-cell arrest phenotype of mat-3(or180) animals at 24° but resulted in embryonic lethality in this background. mdf-1 RNAi suppressed the mat-3(or180) mutation to the same extent. While these data indicate that loss of MDF-1 function is sufficient to suppress the meiosis I arrest, they also suggest that MDF-1 function is absolutely required for embryogenesis in mat-3(or180) animals. This result is distinct from the effects of mdf-1 RNAi or the mdf-1 deletion in a wild-type background, in which the majority of embryos develop into larvae at 24° (Figure 4C; data not shown). These data suggest that the role of MDF-1 in embryonic development is distinct from its role in meiosis and is essential when the APC is compromised.
Figure 4.—
mdf-1(av19) is a separation-of-function allele. (A) mdf-1(av19) contains a point mutation in a conserved alanine at residue 508, near the defined Mad2-binding region (underlined). The red residues indicate conserved amino acids. The green residue indicates the mutation in the mdf-1(av19) allele. The alignment is adapted from Sironi et al. (2002). C.e., C. elegans; D.m., Drosophila melanogaster; S.c., S. cerevisiae. (B) Genetic experiments with mdf-1 strains including the deletion strain mdf-1(gk2) and the mutant allele mdf-1(av19) to assess embryonic viability at 24°. All strains are homozygous for mat-3(or180). (C) RNAi has weak effects on the wild-type N2 strain at 24°, but greatly reduces hatching of mat-3(or180); mdf-1(av19). Diamonds indicate conditions where the majority of embryos were arrested at a multicellular stage. OP50 is the normal food source for C. elegans.
To address the nature of the mdf-1(av19) allele, we compared it with the mdf-1 deletion. mdf-1(av19) is not likely to be a loss-of-function mutation because mdf-1(gk2)/+ does not suppress mat-3(or180) embryos to viability above background; these hermaphrodites produce mostly one-cell embryos (Figure 4B). We assessed the embryonic viability of mdf-1(av19)/+ and mdf-1(av19)/mdf-1(gk2) trans-heterozygotes in a mat-3(or180) background. mdf-1(av19)/+ heterozygotes are able to suppress mat-3(or180) to viability at ∼50% of the homozygous suppression levels. In mat-3(or180); mdf-1(av19)/mdf-1(gk2) hermaphrodites, suppression was restored to mat-3(or180); mdf-1(av19) levels. This finding indicates that the mutant MDF-1 product may compete with wild-type MDF-1, suggesting an antimorphic function. Thus, while both mdf-1(av19) and mdf-1(gk2) suppress mat-3(or180) meiotic metaphase arrest, the mechanism of this suppression may be different as these mutations result in distinctive fates for the embryo. Together, these data indicate that mdf-1(av19) is a separation-of-function mutant. This may reflect two different functions of MDF-1 or a similar function utilized in two distinctive ways (see discussion).
Interestingly, while the mdf-1(gk2) deletion fails to suppress mat-3(or180) to viability at 24°, mat-3(or180) is able to suppress the genetic instability phenotype of mdf-1(gk2) homozygotes at 20°. The mdf-1(gk2) homozygous strain becomes inviable after a few generations, but mat-3(or180); mdf-1(gk2) homozygotes are viable and can be maintained at 20° indefinitely (data not shown). This observation is consistent with those of earlier studies indicating that reduction of APC activity ameliorates phenotypes caused by mdf-1(gk2) at 20° (Furuta et al. 2000; Kitagawa et al. 2002). Thus, these experiments indicate that mat-3(or180) is a suppressor of mdf-1(gk2) at 20°, while acting as an enhancer of mdf-1(gk2) at 24°.
fzy-1:
The three mutant alleles of fzy-1 recovered in the suppressor screen result in single amino acid substitutions (Table 2). The sites of these three mutations are separated by only five amino acids and the clustered nature of the mutations suggests that this small region may have a role in positive regulation of APC. However, a function for this region of Cdc20 has not been previously described. On the basis of the known role of Cdc20 in the spindle checkpoint as an activator of APC, as well as the observation that fzy-1 RNAi results in an embryonic one-cell arrest (Kitagawa et al. 2002), we reasoned that fzy-1(av4), fzy-1(av15), and fzy-1(av23) were gain-of-function (gf) mutations.
If gf mutations of fzy-1 suppress mat-3(or180), a loss-of-function mutation would be predicted to enhance mat-3(or180). Only one loss-of-function fzy-1 mutation has been isolated in C. elegans, fzy-1(h1983) (Kitagawa et al. 2002). We tested whether this fzy-1 allele was an enhancer of mat-3(or180). At the permissive temperature (16°), fzy-1(h1983); mat-3(or180) double mutants produce only dead multicellular embryos, despite the fact that neither mutant has an embryonic phenotype at this temperature (Golden et al. 2000; Kitagawa et al. 2002). These data further support the assignment of fzy-1(av4), fzy-1(av15), and fzy-1(av23) as gf suppressors because these alleles function in a manner opposite to fzy-1(h1983).
Suppressors do not exhibit APC subunit specificity:
Is the ability of the suppressors to restore APC activity specific to the mat-3(or180) allele, the MAT-3 subunit, or do the suppressors function via a more general mechanism? This question was addressed by constructing double mutants of one mutated APC subunit and one suppressor of mat-3(or180). For each of four genes, mdf-1, mdf-2, mdf-3/san-1, and fzy-1, the allele that exhibits the strongest suppression of mat-3(or180) was tested in this experiment (Table 3). In turn, the suppressors were combined with APC subunit mutations of different strengths. To date, temperature-sensitive mutants in five subunits of the APC have been isolated: mat-1, mat-2, mat-3, emb-27, and emb-30 (Furuta et al. 2000; Golden et al. 2000). Temperature-sensitive mutant hermaphrodites lay broods of one-cell embryos when shifted to the restrictive temperature during the L4 larval stage. emb-1 mutant animals were also tested because they also produce one-cell embryos at the restrictive temperature, suggesting that this gene may have a role in APC function (Golden et al. 2000). The emb-1 gene has not yet been molecularly identified.
TABLE 3.
Suppressors do not exhibit APC subunit specificity
| Gene | Temperature | mdf-1(av19) | mdf-2(av18) | mdf-3/san-1(av31) | fzy-1(av15) |
|---|---|---|---|---|---|
| mat-1(ax212) | 25° | Sup (90) | Sup (81) | ND | Sup (88) |
| mat-2(ax78) | 24° | Sup (MC) | Sup (1.5, MC) | Sup (0.5, MC) | ND |
| mat-2(or170) | 23° | Sup (MC) | Sup (2.5, MC) | Sup (44) | ND |
| mat-3(or180) | 24° | Sup (43) | Sup (72) | Sup (66) | Sup (83) |
| mat-3(or148) | 24° | Sup (14) | Sup (5.9) | Sup (5.6) | Not sup |
| mat-3(or192) | 24° | Not sup | Not sup | Not sup | Not sup |
| emb-27(ax80) | 23° | Not sup | Not sup | Not sup | ND |
| emb-27(ax81) | 23° | Not sup | ND | Not sup | ND |
| emb-27(ax348) | 25° | Sup (32) | ND | Sup (68) | ND |
| emb-30(ax69) | 23° | Sup (MC) | Sup (5) | Sup (21) | Not sup |
| emb-30(or420) | 25° | Sup (69) | Sup (78) | Sup (53) | Sup (97.3) |
| emb-1(hc57) | 24° | Sup (32) | ND | Sup (16) | Not sup |
Double mutants were generated and assayed for suppression. Percentage of embryonic viability is in parentheses. mat-1(ax212) dpy-5 exhibited 25–35% background hatching even at the most restrictive temperature, due to suppression by the dpy-5 marker. Sup, suppressed; MC, multicellular embryos that failed to hatch were observed; ND, not determined.
All four suppressor strains were able to suppress at least one mutant allele of each APC subunit mutant. Each mat/emb mutation was either uniformly suppressed by the four suppressors or uniformly not suppressed, with three exceptions: fzy-1(av15) did not suppress mat-3(or148), emb-30(ax69), or emb-1(hc57). Thus, we found that suppression of the APC by spindle checkpoint mutants does not appear to be specific in regard to either allele or subunit. Rather, the ability of the spindle checkpoint mutants to suppress each APC mutant probably reflects the strength of the APC mutation itself. This supports the hypothesis that these suppressors modify the overall activity of the APC (via FZY-1) rather than mediate direct interactions with specific APC subunits.
Phenotypes of spindle checkpoint mutants:
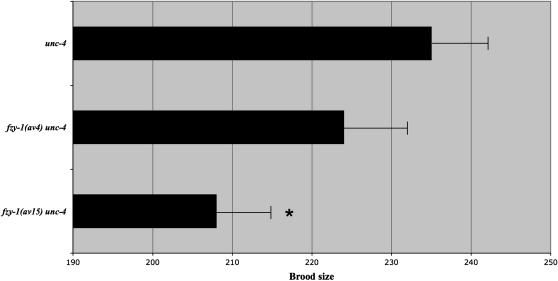
To evaluate the roles for mdf-1, mdf-2, mdf-3/san-1, and fzy-1 in C. elegans development and fertility, we asked if these mutants exhibited any phenotypes in an otherwise wild-type background. The average embryonic viability and brood size at 24° was evaluated for each mutant. In addition, 100 F1 larvae of temperature-shifted P0 hermaphrodites were individually evaluated for any observable defects, including larval arrest or lethality, somatic defects, production of F1 males, or sterility. With this assay, mdf-1(av19) and fzy-1(av4) did not display any apparent defects in development. Brood size and embryonic viability were indistinguishable from control animals and no defects were observed in F1 progeny (Figure 5; data not shown). fzy-1(av15) animals were normal in all respects except for a mild reduction in brood size (Figure 5).
Figure 5.—
fzy-1(av15) has a reduced brood at 24°. unc-4, fzy-1(av4) unc-4, and fzy-1(av15) unc-4 hermaphrodites were grown at 24° and assessed for brood size. Asterisk indicates a P-value of <0.05 by Student's t-test. Error bars indicate SEM.
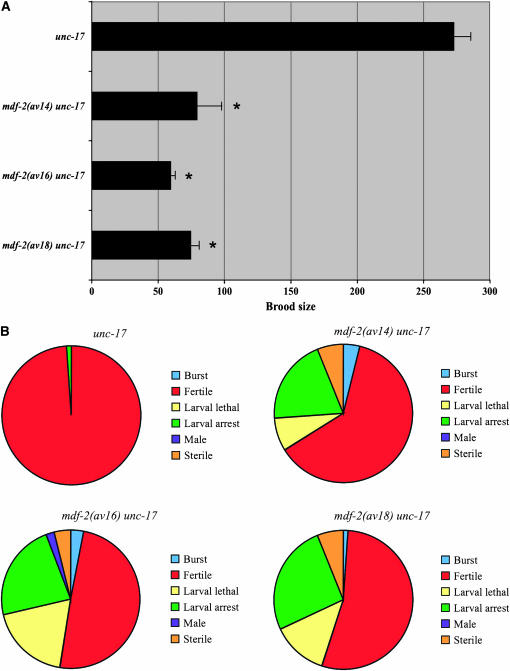
To assess whether the mdf-2 mutants have a phenotype, we chose three alleles, av14, av16, and av18, that, respectively, provided weak, moderate, and strong suppression of mat-3(or180), which we considered might correspond to increasing loss of MDF-2 function. Notably, all three alleles have a significantly reduced brood size, ∼25% of control levels (Figure 6A). In addition, ∼10% embryonic lethality was observed (data not shown). Thus, the strength of the phenotypes of mdf-2 mutants alone does not correlate with the strength of mat-3(or180) suppression. To examine the phenotypes in more detail, the fate of 100 hatched larvae was followed (Figure 6B). Only about half of these animals became fertile adults. The remainder of the F1 larvae were sterile, larval lethal, or larval arrested and exhibited protruding vulva (Pvl), a feature commonly associated with cell cycle defects. Fertile animals produced a brood of severely reduced size. The reduction in brood size may be due to effects on the germline, but may also result from the observation that Pvl animals often retain embryos (bag) or rupture at the vulva (burst). These defects result in the early death of the mother, which reduces reproductive potential.
Figure 6.—
Brood size and F1 phenotypes of mdf-2 mutants. (A) unc-17, mdf-2(av14) unc-17, mdf-2(av16) unc-17, and mdf-2(av18) unc-17 hermaphrodites were grown at 24° and assessed for brood size. All three strains have a significantly smaller brood size than unc-17 (P < 0.05). Error bars indicate SEM. (B) F1 progeny were phenotypically classified. Pie charts depict the distribution of phenotypes in each of the four indicated genotypes.
We observed another interesting phenotype uniformly present in the mdf-2 mutants. All three alleles exhibited slow growth. Development of the mutants lagged behind wild-type animals, due to an extended duration of each larval stage. As stated above, about half of these delayed animals were fertile, but it took some L1 larvae grown at 24° >7 days to mature and lay embryos, while wild-type animals accomplished this in 2 days.
Phenotypic analysis was also performed on the mdf-3/san-1 alleles. When compared to control animals, the mdf-3/san-1(av20), mdf-3/san-1(av31), and mdf-3/san-1(ok1580) deletion strains all have similarly mild phenotypes. The only observed phenotype in the shifted P0 is a reduction in brood size to ∼75% of wild type (Figure 7). Embryonic lethality was not observed. In the analysis of the F1 generation, phenotypes included larval arrest and lethality. Additionally, an increased occurrence of bagging and bursting in adult hermaphrodites was observed. This may be due to subtle abnormalities in vulval development, as mdf-1(gk2) deletion and mdf-2 mutants also exhibit egg-laying defects.
Figure 7.—
Brood size and F1 phenotypes of mdf-3/san-1 mutants. (A) unc-13, mdf-3/san-1(ok1580) unc-13, mdf-3/san-1(av20) unc-13, and mdf-3/san-1(av31) unc-13 hermaphrodites were grown at 24° and assessed for brood size. Asterisks denote a P-value of <0.05. Error bars reflect SEM. (B) F1 progeny from unmarked control and mutant strains were phenotypically classified. Pie charts depict the distribution of phenotypes in each of the four genotypes: wild type, mdf-3/san-1(ok1580), mdf-3/san-1(av20), and mdf-3/san-1(av31). Egg-laying defective (Egl) refers to animals that retain embryos in their uterus.
Some suppressors suppress the synthetic lethality observed in MDF-1-depleted mat-3(or180) animals:
In yeast, Mad1 functions in the spindle checkpoint pathway upstream of Mad2 and Cdc20 (Nasmyth 2005). As described above, depletion of MDF-1 by genetic deletion or RNAi is sufficient to suppress the meiotic phenotype of mat-3(or180), but at 24° the reduction of MDF-1 and APC activity is synthetically lethal at the multicellular stage of embryogenesis. To evaluate whether any of the suppressors suppress this embryonic lethality, we determined whether suppression to viability was observed when MDF-1 was depleted. L4 hermaphrodites of 17 mat-3(or180);suppressor lines representing all extragenic genes were fed mdf-1 RNAi-expressing bacteria for 2 days at 24° and embryonic viability was quantitated for the 24- to 48-hr period following the temperature shift. About half of the mat-3(or180); suppressor lines failed to suppress the mat-3(or180); mdf-1 RNAi lethality (Table 4). However, some of the mat-3(or180); suppressor strains remained viable upon mdf-1 RNAi. Thus, at least to some extent, these suppressors were able to bypass the requirement for wild-type MDF-1 in embryos with a defective APC. The two mat-3(or180); mdf-3/san-1 strains and two of the mat-3(or180); som-3 strains were observed to have moderate viability. Four additional strains had normal or near-normal viability. Of these, dpy-10(av6) is hypothesized to act as a suppressor of mat-3(or180) due to the induction of heat shock and the upregulation of chaperones (Kramer and Johnson 1993). It is possible that som-2(av9), som-3(av11), and som-3(av29) also suppress mat-3(or180) by mechanisms that modify transcription or stability of mat-3(or180). Alternatively, these genes may encode proteins that act downstream of the APC. In summary, dpy-10(av6), som-2(av9), som-3(av11), and som-3(av29) suppress the embryonic lethal phenotype that results from depletion of MDF-1 in mat-3(or180) mutants.
TABLE 4.
Suppression of the synthetic lethality of MDF-1-depleted mat-3(or180) animals
| Strong | Medium | Fail to suppress |
|---|---|---|
| som-2(av9) | mdf-3/san-1(av20) | mdf-1(av19) |
| som-3(av11) | mdf-3/san-1(av31) | mdf-2(av14) |
| som-3(av29) | som-3(av10) | mdf-2(av16) |
| dpy-10(av6) | som-3(av32) | mdf-2(av18) |
| fzy-1(av4) | ||
| fzy-1(av15) | ||
| fzy-1(av23) | ||
| som-1(av5) | ||
| som-2(av21) | ||
| som-4(av28) |
mat-3(or180); suppressor L4 hermaphrodites were fed mdf-1 RNAi and their F1 progeny were evaluated for embryonic viability. As a control, mat-3(or180) L4 hermaphrodites were also fed mdf-1 RNAi. This control results in suppression of the metaphase arrest in F1 embryos, but results in death at the multicellular stage. All feeding experiments were performed at the nonpermissive temperature of 24°. Strong suppressors had embryonic lethality rates similar to their counterparts not subjected to mdf-1 RNAi. Strains that were medium suppressors had significant embryonic hatching but less than their control counterparts not subjected to mdf-1 RNAi. Most strains failed to suppress the embryonic lethality when MDF-1 was depleted.
DISCUSSION
In this report we have described 27 suppressor mutations that restore viability to embryos deficient in APC function. Eighteen of these suppressors have been molecularly identified and here we show that most of these genes encode regulators of the APC, including components of the conserved spindle checkpoint. We have also shown that loss of the spindle checkpoint function provided by mdf-2 and mdf-3/san-1 is detrimental to the organism, whereas the mdf-1 and fzy-1 mutations recovered in this study do not have an effect on the health of the animal. Finally, we have discovered a unique aspect of mdf-1 function during embryogenesis.
Our data argue that the APC is regulated by spindle checkpoint components and their targets in the absence of any spindle damage during C. elegans meiosis. Mutants in the APC are not expected to trigger the spindle checkpoint because the defect is downstream of spindle formation. Microscopic observations of the meiotic spindle in mat mutants show wild-type spindle assembly (Golden et al. 2000; Shakes et al. 2003). Thus, our mutant suppressors result in the upregulation of APC activity independently of spindle defects. This is underscored by the observation that mdf-2 and mdf-3/san-1 mutants have phenotypes in a mat-3(+) background, which suggests a role for these genes during cell divisions throughout development. These data are consistent with the model that there is a certain threshold of APC activity required for progression through the cell cycle. This threshold appears to be highest at meiosis because very weak loss-of-function APC mutants exhibit metaphase arrest at meiosis but not at any mitotic division. The suppressors isolated in this study may work by increasing APC activity so that it is above the threshold limit by weakening inhibitory interactions or by strengthening positive interactions with the APC.
Our experiments generating animals mutant for both a mat/emb mutant and a spindle checkpoint component revealed that the suppressors restore viability to emb-1 mutants. These data implicate emb-1 in the spindle checkpoint/APC pathway. The molecular identity of emb-1 is not known, but it could act as a novel APC component or a positive regulator of the metaphase-to-anaphase transition. emb-1 does not map near any known APC subunit orthologs, and thus its molecular identification is likely to reveal a new player in the metaphase-to-anaphase transition.
Dominant mutations of FZY-1 may perturb interactions with MDF-2:
Several dominant gf mutants in Cdc20 have been identified in budding and fission yeast (Hwang et al. 1998; Kim et al. 1998). These mutants do not arrest the cell cycle in response to spindle damage by chemical agents such as nocodazole and they are unable to bind to Mad2 or Mad3 (Hwang et al. 1998; Kim et al. 1998). Sequence analysis demonstrates that Cdc20(gf) mutants contain point mutations in a region N-terminal of the first WD domain, a motif found in all Cdc20-related proteins. This region has been identified as the site of Mad2 interaction by structural studies (Luo et al. 2000; Sironi et al. 2002). While mutations in fzy-1(av4), fzy-1(av15), and fzy-1(av23) are in a clustered location, they do not fall within the 10 amino acids that directly mediate the Mad2/Cdc20 interaction. Yet, the fact that the three mutations lie within a few amino acids of this domain implicates this region as important for enhancement of Cdc20 function. In C. elegans, fzy-1 mutations in this region may interfere with MDF-2 binding while retaining the ability to activate the APC and so thus would be equivalent to the loss of MDF-2 or MDF-3/SAN-1. Alternatively, the mutant protein may provide a more productive interaction with the APC that results in an increased level of ubiquitin ligase activity. We currently favor the first possibility as most likely for two reasons. First, the ability to have a more productive interaction with APC would be predicted to result in a global ability to suppress APC temperature-sensitive mutants; however, this is not observed in our studies. Second, in yeast, a dominant Cdc20(gf) allele can be mimicked by overexpressing wild-type Cdc20, indicating that no neomorphic function is required to increase APC activity (Schott and Hoyt 1998).
MDF-2 and MDF-3 loss-of-function mutations suppress APC:
Based on studies in yeast and vertebrates, Mad2 is positioned at the nexus of APC regulation. Mad2 receives the checkpoint signal from upstream checkpoint proteins and directly inhibits Cdc20. Thus, loss-of-function mdf-2 mutations are predicted to relieve negative regulation of APC activity. Mad2 deletions in yeast bypass the spindle checkpoint and are able to suppress APC mutants (Kim et al. 1998). Most of the C. elegans mdf-2 mutations (10/11) are found in the C-terminal half of the protein but do not define a particular domain. The structure of the Mad2-binding pocket for Cdc20 and Mad1 has been solved, but only one of the mdf-2 alleles contains a mutation at a residue directly involved in this interaction. Thus, we think it is most likely the mdf-2 mutations result in a loss of structural integrity that leads to a reduction in protein levels or function. The abundance of mdf-2 alleles recovered in this screen probably can be explained by the fact that loss-of-function mutations are much easier to obtain than gf mutations. In the same way, mdf-3/san-1 loss-of-function alleles are also sufficient to suppress mat-3(or180). The ability of the mdf-3/san-1 deletion and the two recessive alleles to suppress mat-3(or180) supports the model that MDF-3 acts as a negative regulator of APC.
MDF-1 has two functions:
Our genetic studies with mdf-1 have revealed two discrete functions of MDF-1 during C. elegans development. We have shown that loss of function of MDF-1 (using the gk2 deletion or mdf-1 RNAi) results in highly penetrant embryonic lethality when the APC is compromised. However, the multicellular lethality indicates that the metaphase I arrest typical of mat-3(or180) embryos has been suppressed. Lethality in these double mutants may be a result of the accumulation of chromosome segregation errors. In contrast, the majority of mat-3(or180); mdf-1(av19) embryos develop normally and hatch, suggesting that chromosome segregation is largely error free during the meiotic and mitotic divisions in this strain. mat-3(or180); mdf-1(av19)/mdf-1(gk2) animals also have similarly high rates of viability. Thus, in the complete absence of functional MDF-1, the mitotic divisions are severely compromised in a mat-3(or180) background. The semidominant av19 allele, however, appears to be defective in its meiotic role, but wild type for its mitotic role, even when it is the only copy of MDF-1. These data suggest two contexts for mdf-1 function.
Several models can account for differential activity of MDF-1. First, MDF-1 protein may possess two different functions. In this scenario, one domain may provide spindle checkpoint function, whereas the other domain may regulate a novel function required for embryogenesis that is sensitized in a mat-3(or180) background. This novel function could include nuclear transport, as there is some evidence that MDF-1 may participate in this process (Iouk et al. 2002). In this model, av19 is altered for the spindle checkpoint but not the other function. Second, MDF-1 may have the same interaction with two populations of proteins. For example, MDF-1 may interact with meiosis-specific APC subunits or regulators of the APC. These interactions would be predicted to be disrupted in the mdf-1(av19) mutant, while mitotic interactions are maintained. Finally, MDF-1 may be required at two different levels: a high level of MDF-1 function may be required at meiosis, consistent with the threshold hypothesis, while a lower level would be needed for mitotic division. In this case, activity of the mutant MDF-1 is not sufficient for meiosis but satisfies the mitotic requirement.
The hypothesis that MDF-1 may function in more than one context is not inconsistent with observations in the literature. In yeast, it has been established that Mad1 has a seemingly conflicting biochemical role as both a competitive inhibitor and an obligate activator of Mad2 (Nasmyth 2005). This is because Mad1 is required to modify the structure of Mad2 so that Mad2 can efficiently bind to Cdc20. However, Mad1 also competes with Cdc20 for binding to Mad2. Due to its location, the subtle av19 mutation may result in an alteration in the interactions between MDF-1 and MDF-2 in C. elegans. Because of the dual role of Mad1, loss-of-function and gf mutations of mdf-1 may lead to the same net result: reduction in active MDF-2. A loss of function of mdf-1 would be anticipated to suppress mat-3(or180) by reducing the levels of activated MDF-2, leading to timely passage through meiosis. A gf in MDF-1 may have an enhanced binding interaction with MDF-2, which may titrate MDF-2 and relieve inhibition on FZY-1, activating the APC. Future biochemical studies are required to determine the nature of mdf-1(av19) suppression of APC. Thus, by taking a genetic approach, we have obtained a separation-of-function allele that would not have been uncovered by RNAi experiments.
Novel regulators of APC:
We are beginning to understand how the eight uncharacterized som suppressors might regulate the APC. We have ruled out the possibility that any of the som genes encode Bub1 or Bub3 orthologs because they do not map to these chromosomal locations. Thus, these suppressors may define novel APC regulators or substrates. We have begun to functionally categorize these suppressors on the basis of genetic mapping, dominance, and ability to suppress the synthetic lethality of mat-3(or180); mdf-1 RNAi. mat-3(or180); som-1(av5) animals do not suppress mat-3(or180) animals depleted of MDF-1, suggesting that SOM-1 may function in the spindle checkpoint pathway. In addition, som-1(av5) double mutants with other APC mutant subunits exhibit suppression of lethality (data not shown) in a pattern similar to our currently identified spindle checkpoint suppressor mutants (Table 3).
The RNAi experiments with mdf-1 in a mat-3(or180) background have subdivided the suppressors into three categories, which may reflect different relationships with the spindle checkpoint pathway. Suppression of mat-3(or180) by most suppressors is dependent on MDF-1 function. This genetic interaction with mdf-1 RNAi suggests participation in a similar pathway. While mapping experiments place four of the six uncloned LG III suppressors into one genetic cluster, functional assays reveal that the suppressors fall into two classes, which may indicate that they represent two genes. This is based on analogy to mdf-2, mdf-3/san-1, and fzy-1, where a link between function in the mdf-1 RNAi assay and gene assignment is observed (Table 4). All alleles of a given gene behave in the same way. Specifically, mdf-2 and fzy-1 are unable to suppress the embryonic lethality that results from MDF-1 depletion in the mat-3(or180) background, but mdf-3/san-1 alleles support some viability. By this logic, som-3(av10) and som-3(av32) are likely to be alleles of a single gene. These are the only two suppressors that are weakly dominant for mat-3(or180) suppression and both suppressors moderately suppress mat-3(or180); mdf-1 RNAi. A similar argument can be made for the assignment of som-2(av9) and som-2(av21) to two loci as som-2(av9) is a very effective suppressor of mdf-1 RNAi while som-2(av21) is the only LG III suppressor that does not suppress mat-3(or180) animals depleted of MDF-1.
It was not surprising that most suppressors were not functional when mdf-1 was depleted in mat-3(or180); suppressor strains, considering that embryonic divisions occur within cells bearing a compromised APC and a defective spindle checkpoint. However, it was surprising that the suppressor mutations dpy-10(av6), som-2(av9), som-3(av11), and som-3(av29) suppressed the lethality of MDF-1-depleted mat-3(or180) embryos (Table 4). These MDF-1-depleted mat-3(or180); suppressor strains are exceedingly healthy and resemble wild-type animals. Suppression may be achieved by restoring APC to near wild-type activity independent of the spindle checkpoint pathway. Alternatively, these genes may encode proteins that act downstream of the APC.
Recent studies have shown that the spindle checkpoint may have an important role in maintaining the nuclear integrity of mammalian oocytes (Wassmann et al. 2003; Homer et al. 2005). It has also been shown that mice containing a hypomorphic BubR1/Mad3 allele are sterile, due to chromosome instability in gametes (Baker et al. 2004). C. elegans is a useful system in which the role of these components during meiosis and throughout development can be further dissected.
Acknowledgments
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources, including deletion strains produced by the C. elegans Gene Knockout Consortium at the Oklahoma Medical Research Foundation and the University of British Columbia. The strains containing fzy-1(h1983) dpy-10(e128) and unc-46(e177) mdf-1(gk2) were gratefully obtained from Ann Rose. We thank Ben Ross for assistance with construction of double-mutant control strains for the APC subunit–suppressor matrix and Julia S. Lindenberg for sequencing the mat-3 locus of av26. We thank Paula Fearon and members of our lab for their critical reading of the manuscript, the Bethesda and Baltimore/DC worm clubs for helpful suggestions, and Kevin O'Connell for many invaluable discussions and suggestions. This work was supported by the Intramural Research Program at NIH's National Institute of Diabetes and Digestive and Kidney Diseases.
References
- Aroian, R. V., A. D. Levy, M. Koga, Y. Ohshima, J. M. Kramer et al., 1993. Splicing in Caenorhabditis elegans does not require an AG at the 3′ splice acceptor site. Mol. Cell. Biol. 13: 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. J., K. B. Jeganathan, J. D. Cameron, M. Thompson, S. Juneja et al., 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36: 744–749. [DOI] [PubMed] [Google Scholar]
- Baker, D. J., K. B. Jeganathan, L. Malureanu, C. Perez-Terzic, A. Terzic et al., 2006. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto, R., R. Gomes and R. E. Karess, 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol. 2: 939–943. [DOI] [PubMed] [Google Scholar]
- Bharadwaj, R., and H. Yu, 2004. The spindle checkpoint, aneuploidy, and cancer. Oncogene 23: 2016–2027. [DOI] [PubMed] [Google Scholar]
- Chan, G. K., S. T. Liu and T. J. Yen, 2005. Kinetochore structure and function. Trends Cell Biol. 15: 589–598. [DOI] [PubMed] [Google Scholar]
- Chu, T., G. Henrion, V. Haegeli and S. Strickland, 2001. Cortex, a Drosophila gene required to complete oocyte meiosis, is a member of the Cdc20/fizzy protein family. Genesis 29: 141–152. [DOI] [PubMed] [Google Scholar]
- Cooper, K. F., M. J. Mallory, D. B. Egeland, M. Jarnik and R. Strich, 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 97: 14548–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, E. S., L. Wille, B. A. Chestnut, P. L. Sadler, D. C. Shakes et al., 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160: 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, I. A., S. Roth and S. Artavanis-Tsakonas, 1995. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J. Cell Biol. 129: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles, M., V. Liberal, M. L. Scott, R. Benezra and P. K. Sorger, 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101: 635–645. [DOI] [PubMed] [Google Scholar]
- Encalada, S. E., J. Willis, R. Lyczak and B. Bowerman, 2005. A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 16: 1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., H. Yu and M. W. Kirschner, 1998. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2: 163–171. [DOI] [PubMed] [Google Scholar]
- Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann et al., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330. [DOI] [PubMed] [Google Scholar]
- Furuta, T., S. Tuck, J. Kirchner, B. Koch, R. Auty et al., 2000. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11: 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe, D., J. B. Doto and M. V. Sundaram, 2004. Caenorhabditis elegans lin-35/Rb, efl-1/E2F and other synthetic multivulva genes negatively regulate the anaphase-promoting complex gene mat-3/APC8. Genetics 167: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, A., P. L. Sadler, M. R. Wallenfang, J. M. Schumacher, D. R. Hamill et al., 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky, G. J., R. H. Chen and A. W. Murray, 1998. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 141: 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J. W., J. L. Burton and M. J. Solomon, 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16: 2179–2206. [DOI] [PubMed] [Google Scholar]
- Homer, H. A., A. McDougall, M. Levasseur, K. Yallop, A. P. Murdoch et al., 2005. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 19: 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M. A., L. Totis and B. T. Roberts, 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517. [DOI] [PubMed] [Google Scholar]
- Hwang, L. H., L. F. Lau, D. L. Smith, C. A. Mistrot, K. G. Hardwick et al., 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science 279: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai and R. W. Wozniak, 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger, S., 2006. Preventing fatal destruction: inhibitors of the anaphase-promoting complex in meiosis. Cell Cycle 5: 405–415. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kim, S. H., D. P. Lin, S. Matsumoto, A. Kitazono and T. Matsumoto, 1998. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science 279: 1045–1047. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., and A. M. Rose, 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1: 514–521. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., E. Law, L. Tang and A. M. Rose, 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12: 2118–2123. [DOI] [PubMed] [Google Scholar]
- Kramer, J. M., and J. J. Johnson, 1993. Analysis of mutations in the sqt-1 and rol-6 collagen genes of Caenorhabditis elegans. Genetics 135: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., and A. W. Murray, 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531. [DOI] [PubMed] [Google Scholar]
- Luo, X., G. Fang, M. Coldiron, Y. Lin, H. Yu et al., 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7: 224–229. [DOI] [PubMed] [Google Scholar]
- Luo, X., Z. Tang, J. Rizo and H. Yu, 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9: 59–71. [DOI] [PubMed] [Google Scholar]
- Maine, E. M., and J. Kimble, 1989. Identification of genes that interact with glp-1, a gene required for inductive cell interactions in Caenorhabditis elegans. Development 106: 133–143. [DOI] [PubMed] [Google Scholar]
- Mango, S. E., 2001. Stop making nonSense: the C. elegans smg genes. Trends Genet. 17: 646–653. [DOI] [PubMed] [Google Scholar]
- Millband, D. N., and K. G. Hardwick, 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22: 2728–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K., 2005. How do so few control so many? Cell 120: 739–746. [DOI] [PubMed] [Google Scholar]
- Nystul, T. G., J. P. Goldmark, P. A. Padilla and M. B. Roth, 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science 302: 1038–1041. [DOI] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham and A. A. Hyman, 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153: 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch, O., E. Schwob, F. de Fraipont, A. Camasses, R. Bordonne et al., 1994. RPK1, an essential yeast protein kinase involved in the regulation of the onset of mitosis, shows homology to mammalian dual-specificity kinases. Mol. Gen. Genet. 243: 641–653. [DOI] [PubMed] [Google Scholar]
- Rudner, A. D., and A. W. Murray, 2000. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 149: 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, E. J., and M. A. Hoyt, 1998. Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics 148: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes, D. C., P. L. Sadler, J. M. Schumacher, M. Abdolrasulnia and A. Golden, 2003. Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130: 1605–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn, M. A., R. McCarroll and A. W. Murray, 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289: 300–303. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., W. A. Michaud and P. Hieter, 1993. p62cdc23 of Saccharomyces cerevisiae: a nuclear tetratricopeptide repeat protein with two mutable domains. Mol. Cell. Biol. 13: 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi, L., M. Mapelli, S. Knapp, A. De Antoni, K. T. Jeang et al., 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. S., and F. McKeon, 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89: 727–735. [DOI] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Wassmann, K., T. Niault and B. Maro, 2003. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr. Biol. 13: 1596–1608. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Wirth, K. G., R. Ricci, J. F. Gimenez-Abian, S. Taghybeeglu, N. R. Kudo et al., 2004. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 18: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]