Abstract
Senseless (Sens) is a conserved transcription factor required for normal development of the Drosophila peripheral nervous system. In the Drosophila retina, sens is necessary and sufficient for differentiation of R8 photoreceptors and interommatidial bristles (IOBs). When Sens is expressed in undifferentiated cells posterior to the morphogenetic furrow, ectopic IOBs are formed. This phenotype was used to identify new members of the sens pathway in a dominant modifier screen. Seven suppressor and three enhancer complementation groups were isolated. Three groups from the screen are the known genes Delta, lilliputian, and moleskin/DIM-7 (msk), while the remaining seven groups represent novel genes with previously undefined functions in neural development. The nuclear import gene msk was identified as a potent suppressor of the ectopic interommatidial bristle phenotype. In addition, msk mutant adult eyes are extremely disrupted with defects in multiple cell types. Reminiscent of the sens mutant phenotype, msk eyes demonstrate reductions in the number of R8 photoreceptors due to an R8 to R2,5 fate switch, providing genetic evidence that Msk is a component of the sens pathway. Interestingly, in msk tissue, the loss of R8 fate occurs earlier than with sens and suggests a previously unidentified stage of R8 development between atonal and sens.
SENS is a zinc-finger transcription factor that is necessary and sufficient for the development of the founding cell type of the Drosophila retina, the R8 photoreceptor (Frankfort et al. 2001; Frankfort and Mardon 2002). It is also necessary for development of the entire Drosophila peripheral nervous system (PNS). In the absence of Sens, sensory organ precursor (SOP) cells are specified but do not develop, and the corresponding sensory organs fail to form (Nolo et al. 2000). In addition, ectopic expression of Sens in the thorax generates a massive overproduction of mechanosensory bristles or macrochaete. These data indicate that sens acts near the top of the developmental cascade that specifies sensory organ fate. Furthermore, Sens is a member of a family of zinc-finger transcription factors termed the Gfi/Pag-3/Sens (GPS) proteins (Jafar-Nejad and Bellen 2004). These conserved proteins are required for normal neural development in many organisms including humans, mice (Bell et al. 1995), chicken (Fuchs et al. 1997), zebrafish (Dufourcq et al. 2004), Caenorhabditis elegans (Jia et al. 1996), and the house fly (Kasai and Scott 2001). In mammals, the Sens homolog Gfi-1 is involved in neurodevelopment of the inner ear hair cells (Wallis et al. 2003) and cerebellar Purkinje cells (Tsuda et al. 2005). Mice lacking Gfi-1 are deaf and exhibit ataxia typical of inner ear balance defects (Wallis et al. 2003).
Gfi-1 and its paralog Gfi-1b are also involved in hematopoiesis (Zeng et al. 2004; Duan and Horwitz 2005) and immune system function (Schmidt et al. 1998b; Karsunky et al. 2002; Zhu et al. 2002; Hock et al. 2003; Yucel et al. 2003; Zeng et al. 2004; Rathinam et al. 2005) and have been implicated in disease processes including neutropenia (Karsunky et al. 2002; Person et al. 2003), carcinogenesis (Gilks et al. 1993; Schmidt et al. 1996, 1998a; Hock et al. 2004; Kazanjian et al. 2004; Shin et al. 2004; Dwivedi et al. 2005), and neurodegenerative disease (Moroy 2005; Tsuda et al. 2005). The expression of both sens and Gfi-1 is positively regulated by the bHLH proneural gene atonal/Math-5. In the absence of atonal in Drosophila, no adult eye is formed due to the failure of R8 photoreceptor specification (Jarman et al. 1994). In mice, loss of Math-5 results in a severe reduction of Gfi-1 expression in the retina and loss of most retinal ganglion cells (Brown et al. 2001; Wang et al. 2001; Yang et al. 2003). The remarkable similarity in function and regulation within the GPS family of proteins suggests that they are part of an evolutionarily conserved neurodevelopment pathway. Surprisingly, in neural development, only the proneural genes have been identified as transcriptional targets of Sens. Regulation of proneural transcription by Sens provides amplification of the signal required for selection of the SOP cell (Nolo et al. 2001; Jafar-Nejad and Bellen 2004). Due to the importance of Sens in normal development, it is unlikely that proneural genes are its only targets in normal development. Previously, we described a phenotype generated by the expression of UAS-sens in undifferentiated cells by the lozenge-GAL4 driver (referred to as ls) that leads to the massive overproduction of interommatidial bristles (IOBs) (Frankfort et al. 2004). In addition, ectopic Sens is still capable of generating IOBs in the absence of the ac and sc proneural genes normally required for bristle development. This result supports the hypothesis that the proneural genes are not the only targets of Sens and provides us with a powerful tool for identifying new genes required in this developmental pathway. Identification of other genes acting with Sens in Drosophila will lead to a better understanding of normal neural development in the fly and provide insight for potential homologous pathway members in mice and humans.
A genomewide mutagenesis screen is a rapid and powerful method for generating and identifying new genes. The Drosophila eye, with its crystalline array of thousands of photoreceptor cells and bristle neurons (Cagan and Ready 1989), is an ideal system for screens that identify genes required for normal neuronal development. In the wild-type eye, each unit eye or ommatidium is a repeating unit with an invariant arrangement of eight photoreceptor cells, termed R1–R8, four lens-secreting cone cells, two primary pigment cells, six secondary pigment cells, and three tertiary pigment cells and three bristle complexes (each composed of four cells) situated at alternating vertices (see Figure 1 for a labeled example). The bristles and the secondary and tertiary pigment cells form a lattice, resulting in the hexagonal shape of each ommatidium and giving the eye a regular, tiled appearance. Perturbations of the normal structure of the eye are readily visible under the light microscope and make the eye particularly useful in genetic screens.
Figure 1.—
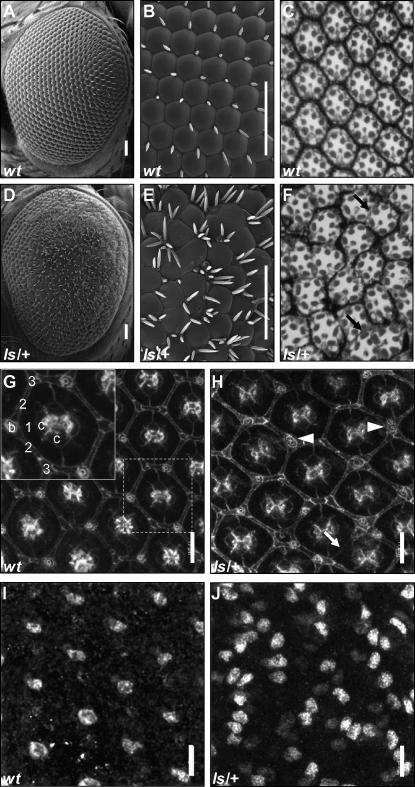
Ectopic Sens recruits cells to the interommatidial bristle fate. The wild-type adult Drosophila eye is composed of ∼800 ommatidia arranged in a crystalline-like lattice (A–C, G, and I). Each wild-type ommatidium has an interommatidial bristle (IOB) located at every other vertex. Scanning electron micrographs of adult Drosophila eyes 200× (A and D) or 1000× magnification (B and E) are shown. Bars in A–E, 50 μm. When UAS-sens is ectopically expressed using the lozenge-GAL4 driver (ls), extra IOBs are formed and the eye is roughened (D and E); note multiple bristles at a single vertex as well as ectopic bristles outside of the vertices. Thin plastic sections of adult wild-type eyes show the trapezoidal arrangement of normal photoreceptors (C). The normal number of photoreceptors is found in the ls eye (F); in contrast, normal morphology is disrupted by misrotation and ommatidial fusions (arrows in F and H) due to pigment cell loss. Staining of wild-type and ls 48-hr APF disc cell membranes with Armadillo (Arm) antibody shows the structure of the developing accessory cells (G and H). In the magnified image in G, each ommatidium has four cone cells (c) surrounded by two crescent-shaped primary pigment cells (1), four secondary pigment cells (2), three tertiary pigment cells (3), and three bristles (b). Cells on half the ommatidium are not labeled to aid visualization. Each ommatidium in G has a Sens-positive R8 cell shown in I. In the ls eye (H), some ommatidia show multiple bristles (arrowheads) and ommatidial fusions (arrow), as well as multiple Sens-positive cells (J). Bars in G–J, 11 μm.
The structure of the adult eye begins to emerge during the early third larval instar. At the posterior of the field of undifferentiated epithelial cells of the eye imaginal disc, a visible indentation termed the morphogenetic furrow is formed. Within and immediately ahead of the furrow, changes in gene expression required for the initiation of photoreceptor development occur (Pappu and Mardon 2004). The morphogenetic furrow progresses toward the anterior of the disc, leaving clusters of newly differentiating photoreceptor cells in its wake. At the molecular level, the proneural gene atonal (ato) is expressed in a stripe immediately anterior to the furrow and gradually resolves into regularly spaced groups of cells, the proneural clusters (PNCs). This expression is eventually resolved into single cells, corresponding to the nascent R8s (Jarman et al. 1994). At this stage, Sens expression is dependent on ato (Nolo et al. 2000; Frankfort et al. 2001) and strongly coincides with Ato at the three-cell stage (the R8 equivalence group) and persists beyond Ato expression in single R8 cells posterior to the furrow. sens in turn is necessary and sufficient for differentiation of the R8 cell (Frankfort et al. 2001). Later in eye development, sens is required for interommatidial bristle development (Frankfort et al. 2004). Expression of sens in this process is dependent on a different set of proneural genes, the genes of the achaete-scute complex. The interommatidial bristle is composed of four cells (the socket, shaft, glia, and neuron) that are clonally derived from a single cell that begins to express Sens 3–6 hr after puparium formation (APF) (Cagan and Ready 1989; Frankfort et al. 2004). By 24 hr APF all four cells of the bristle complex have been formed and express Sens, while at 48 hr APF Sens is no longer expressed in the bristle complex, but is still expressed in the developing R8 photoreceptor.
Here we describe a forward genetic screen using an ectopic Sens genotype (lz-GAL4 UAS-sens, termed “ls”) in which the ommatidia of the adult eye have a rough, irregular appearance and numerous extra IOBs. We crossed ls flies to ethyl methanesulfonate (EMS)-mutagenized w− flies and screened 1.1 × 105 progeny for enhancement or suppression of the rough eye and extra bristle phenotype. We isolated and mapped seven suppressor and three enhancer complementation groups. Three complementation groups were identified as the known genes lilliputian (lilli), Delta (Dl), and moleskin/dim-7 (msk). We discuss the relationship of these genes to the ectopic Sens phenotype, as well as the relationship between these genes and sens in normal development. Finally, we show that Msk, a homolog of the nuclear import factor Importin 7, is necessary for normal eye development and acts in R8 development at a previously unidentified stage upstream of Sens.
MATERIALS AND METHODS
Mutagenesis:
Isogenized w1118 males were collected and aged for 3 days. On day 3, males were starved for 8 hr and then added to bottles containing filter paper soaked with 25 mm EMS (Sigma, St. Louis) and 1% sucrose in water. Males were exposed to the mutagen for 12–15 hr and then allowed to recover on fresh media for 24 hr before mating to lz-GAL4,UAS-sens/FM7 females. Non-FM7 F1 progeny were scored for suppression or enhancement. Modified F1 progeny were crossed to lz-GAL4,UAS-sens/FM7 females or lz-GAL4,UAS-sens/Y males, and F2 male progeny that showed the same modification were then backcrossed to the lz-GAL4,UAS-sens/FM7 females for at least three generations to separate second-hit lethal mutations and weak modifiers from strong single-hit modifiers. Modification of ls caused by X chromosome mutations was scored in F1 females. In these flies, the mutation was located in trans to the ls chromosome. Due to the high rate of recombination that separates lz-GAL4 from UAS-sens, modified F1 females were always backcrossed to ls males to ensure that modification could be scored in the F2. In the F2 only modified males were selected. Then, in the F3, only ls females balanced by FM7 were scored and collected. Thus, a modifying mutation located on the X could be recovered only when a recombination event had occurred in the F1 female between the ls chromosome and the mutant chromosome. This recombination event happened either very rarely or not at all, because we did not recover any X chromosome mutants. After multiple backcrosses through females, F5 modified females were crossed to w; Bc,Elp/CyO and w; TM3/TM6B males. In the F6 single ls males that carried a balancer (CyO or TM3) and showed modification were crossed first to w− virgins and then to virgins of the corresponding balancer stock (w; Bc,Elp/CyO or w; TM3/TM6B). The female progeny of the w− cross were scored to determine segregation. Once the chromosome was determined, the progeny from the proper balancer stock cross were collected to generate the final balanced lines. Balanced mutations were then tested for failure to complement a set of Bloomington deficiencies that were able to modify ls. Complementation analysis was performed among all mutants on each chromosome.
Fly stocks and genetics:
Fly stocks used included w1118, Dl9P, DlB2, lilli00632, msk5, msk4, lz-GAL4,UAS-sens/FM7 (ls), w;ey-GAL4,UAS-flp/CyO,hs-hid; cl,GMR-hid,FRT79D/TM6B (EGUF79D), lz-GAL4/FM7; UAS-ro/CyO, sc10-1,lz-GAL4 (Frankfort et al. 2004), UAS-msk (gift from L. Perkins), UAS-Sens (on the second chromosome), sca-lacZ, RM104, BBO2, UAS-GFP, UAS-Dl, w; C96-GAL4,UAS-hrs/TM6B (gift from H. Bellen), and, from the Bloomington stock center, the first, second, and third chromosome deficiency kits. The molecularly mapped P elements used to fine map S(ls)1, S(ls)3, S(ls)5, S(ls)7, and E(ls)3 were obtained from the P-screen database (Bellen et al. 2004). Molecular information is available at http://flypush.imgen.bcm.tmc.edu/pscreen/. P elements flanking each group are as follows: S(ls)1, KG03264 and KG04159; S(ls)3, KG00038 and KG05698; E(ls)3 and S(ls)7, EY07182 and KG03062; and S(ls)5, KG05323 and KG00569.
Generation of mosaic retina and whole mutant eyes:
The msk5 allele was recombined onto a third chromosome containing an FRT element at either 79D or 80B. To generate pupal retinas mosaic for msk5, males of the genotype ls; msk5,FRT80B/TM6B were crossed with hsflp; ubi-GFP,FRT80B virgins. Progeny of this cross were heat-shocked for 1 hr at 37° 36–38 hr after egg laying. White prepupae were collected (0 hr APF) and aged at 18° for 96 hr (equivalent to 48 hr at 25°). To generate whole eyes mutant for msk, the EGUF system was utilized (Stowers and Schwarz 1999). Males of the genotype w; ey-GAL4,UAS-flp/CyO,hs-hid; cl,GMR-hid,FRT79D/TM6B were crossed to w; msk5, FRT79D/TM6B or w; FRT79D for control animals. w; msk5, RM104, FRT79D/TM6B or w; RM104, FRT79D/TM6B females were used for the experiments in Figure 10, A–H. To generate msk whole mutant eyes carrying the BBO2 or sca-lacZ reporters, females of the genotype w; reporter/CyO,hs-hid; msk5(or +) FRT79D/TM6B were used. Progeny were heat-shocked 72 hr after egg laying (AEL) for 1 hr at 37°. At 120–144 hr AEL non-tubby wandering third instar larvae were dissected for larval immunohistochemistry. Adults were collected for sectioning and scanning electron microscopy (SEM) analysis as described below.
Figure 10.—
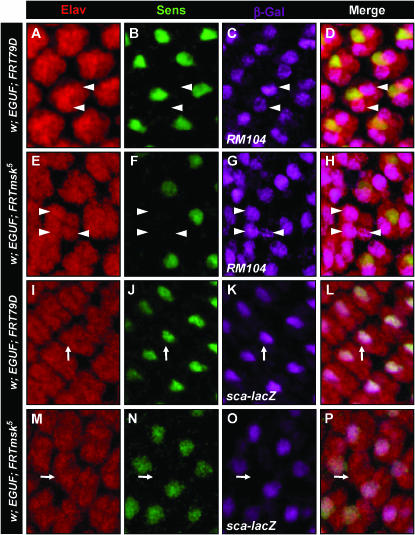
The mechanism of R8 cell loss is through a cell fate change to R2,5 that occurs earlier than in sens mutants. In a control eye (A–D), Sens (green) is expressed in one cell per cluster and the enhancer trap RM104 (purple) is expressed in the R2 and R5 cells (arrowheads in A–D). In the EGUF; msk5 disc (E–H), occasional clusters lacking Sens express RM104 in more than two cells (arrowheads in G and H). Elav staining is shown in red to illustrate ommatidial clusters. The enhancer trap sca-lacZ is normally expressed in R8 cells and coincides with Sens expression (I–L). In EGUF;msk5 eyes, clusters lacking Sens expression also lack sca-lacZ expression. See materials and methods for genotypes.
Antibody staining:
The following primary antibodies were used: rabbit anti-GFP (1:1000; Molecular Probes, Eugene, OR), guinea-pig anti-Sens (1:800, a gift from Hugo Bellen), rabbit anti-Atonal (1:5000, gift of Andrew Jarman), rabbit anti-β galactosidase (1:800, MP Biomedicals Cappel), rat anti-Elav (1:500, Developmental Studies Hybridoma Bank), mouse anti-Cut (1:100, Developmental Studies Hybridoma Bank), and mouse anti-Arm (1:500, Developmental Studies Hybridoma Bank). Goat anti-rat Cy3 and goat anti-rabbit Cy3 secondary antibodies were obtained from Jackson Laboratories (West Grove, PA). Goat anti-mouse Alexa, goat anti-guinea pig Alexa, and goat anti-rabbit Alexa secondary antibodies were obtained from Molecular Probes. All secondary antibodies were used at a 1:500 dilution.
For larval time points, eye imaginal discs were dissected from wandering third instar larvae into 1× PBS (0.1 m phosphate, pH 7.2, 150 mm NaCl) on ice. Discs were then fixed in 4% formaldehyde in 1× PBS for 25 min on ice. For Boss staining, discs were fixed in 2% paraformaldehyde, 10 mm NaIO4, 75 mm lysine, 3.5 mm NaPO4, pH 7.2 (PLP) for 40 min on ice. Discs were then washed for 10 min each in 1× PBS; in PBS with 1% BSA, 0.3% Triton X-100, and 0.3% sodium deoxycholate (PAXD); and in PAXD with 5% heat-inactivated and filtered normal goat serum (PAXDG) and then incubated with primary antibody diluted in PAXDG overnight at 4°. Discs were washed three times in PAXDG followed by incubation with secondary antibody for 2 hr at room temperature. Discs were then washed for 10 min each in PAXDG, PAXD, and PBS followed by fixation in 4% formaldehyde in PBS for 15 min at room temperature. Discs were washed one final time for 10 min in PBS and then equilibrated in Vectashield overnight at 4°. Fluorescent images were captured with a Zeiss LSM 510 confocal microscope. Images were processed using Image J and Adobe Photoshop software.
SEM:
Adult animals were collected and aged on fresh food for 2–3 days and then taken through an EtOH dehydration series. The flies were washed in 1 ml of 30, 50, 70, 90, 100, and 100% EtOH in water for 10–12 hr per step. Hexamethydisilizane (HMDS; Electron Microscopy Sciences, Fort Washington, PA) was then used to chemically dry the samples. The flies were washed for 30 min in 500 μl of the following solutions: 75% EtOH + 25% HMDS, 50% EtOH + 50% HMDS, 25% EtOH + 75% HMDS, 100% HMDS, 100% HMDS, and 100% HMDS. The final 100% HMDS wash was allowed to evaporate under vacuum in the presence of anhydrous calcium sulfate (Drierite; W. A. Hammond Drierite, Xenia, OH). The samples were then mounted on carbon-conductive tabs (Ted Pella, Redding, CA) on top of aluminum stubs (Electron Microscopy Sciences) and coated to 40 nm thickness with sputtered platinum. Images were captured with a JSM-5900 scanning electron microscope (Jeol, Tokyo) and processed with Adobe Photoshop software. The specific alleles pictured in Figure 5 are as follows: (Figure 5, B and F) EMF13, (Figure 5, C and G) EMC7, (Figure 5, D and H) SMB23, (Figure 5, I and M) SFB22, (Figure 5, J and N) SFF117, (Figure 5, K and O) SMD20, and (Figure 5, L and P) SFJ10.
Figure 5.—
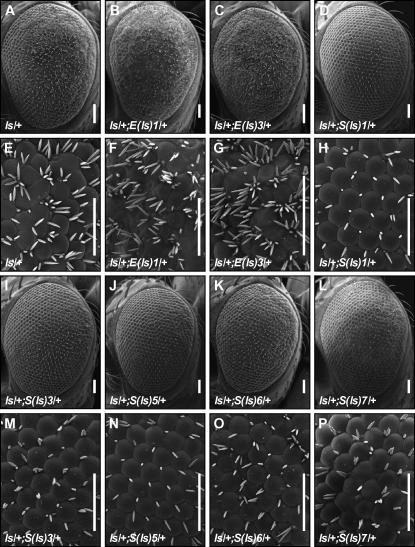
Dominant modification of ls by EMS alleles. Thirty-six independent mutations modify the ls phenotype (A and E) and form 10 complementation groups. The complementation group name is indicated. Complementation groups representing known genes are not shown in this figure (see Figures 2 and 7 and supplemental Figure 1 at http://www.genetics.org/supplemental/). B, C, F, and G show dominant enhancement. More bristles are formed and the external architecture of the eye is more disrupted. D and H–P show dominant suppression of ls. The number of bristles formed is decreased and the ommatidial architecture is less disrupted than in ls. Bars, 50 μm.
Light microscopy:
Adult wings were mounted on a 25-μl drop of xylene and then covered with Cytoseal XYL (Richard-Allan Scientific, Kalamazoo, MI) and a glass coverslip. Adult eye sections were prepared and sectioned as described in Tomlinson and Ready (1987). Images were captured using a Zeiss Axioplan2 microscope and a Zeiss Axiocam digital camera. Images were processed with Adobe Photoshop software.
Cell counting and intensity measurement:
Projections of confocal microscope stacks were made with ImageJ software (Abramoff et al. 2004). The area, in square micrometers, of twinspot (msk +/+) and heterozygous (msk −/+) tissue was measured with ImageJ and the number of Sens-positive cells in each area was counted using the ImageJ point selection tool. Data were imported into Microsoft Excel, which was used for all subsequent analysis. The number of cells counted was divided by the area of tissue to give the number of cells per square micrometer. Student's t-test was used to test for a significant difference in cell density between the twinspot and heterozygous tissue. To illustrate differences in pixel intensity graphically, the heat-mapping function of Image J was utilized. Images were then processed with Adobe Photoshop software as described above.
RESULTS
Ectopic expression of Sens in undifferentiated cells generates extra IOBs:
Misexpression of UAS-sens in the eye under the control of lozenge-GAL4 (ls) generates a roughened eye with many ectopic IOBs (see Figure 1, D and E). Expression of lozenge-GAL4 begins in undifferentiated cells posterior to the morphogenetic furrow (Crew et al. 1997). Later expression is found in the R1, -6, and -7 photoreceptor cells and in cone and pigment cells. Since Sens is sufficient for the R8 and IOB cell fate, we predicted that the ls phenotype would likely result from changes in normal ommatidial cell fate caused by ectopic Sens. To determine which cells are affected, we sectioned adult eyes and performed antibody staining on larval and pupal eye tissue with cell type-specific markers. Interestingly, no photoreceptor cell types are lost, and no ectopic R8 photoreceptor cells are formed despite the presence of ectopic Sens in the third larval instar disc (data not shown and compare Figure 1C to 1F). Therefore, the ls genotype is not affecting early cell fate decisions. The ectopic bristles caused by ls could be due to recruitment of undifferentiated cells to the bristle fate or a switch in cell fate of an accessory cell type to a bristle cell. Armadillo staining of ls pupal discs at 48 hr APF (Figure 1H) demonstrates that in most ommatidia all accessory cells are present, although secondary and tertiary pigment cell are slightly misshapen compared to wild type (Figure 1G; see inset in Figure 1G for cell-type identification). Occasional secondary pigment cells are absent, leading to ommatidial fusions (arrows in Figure 1, F and H). These missing secondary pigment cells may be recruited to the bristle fate, but their numbers cannot account for the massive number of extra IOBs. Therefore, most of the extra IOBs in the ls eye are likely recruited from undifferentiated cells that are usually removed by apoptosis during pupal development (Brachmann and Cagan 2003). To provide a comparison of Sens expression in the wild-type and ls eye, pupal discs were stained at 48 hr APF. In the wild-type eye, there is one Sens-positive cell in each ommatidium, corresponding to the R8 (Figure 1I). In contrast, the ls eye has many Sens-positive cells per ommatidium (Figure 1J).
Delta enhances the ls phenotype:
Ectopic expression of Sens by lz-GAL4 recruits many undifferentiated cells to the bristle fate. The increased number of neurons is similar to the neurogenic phenotype seen when Notch signaling is decreased. The Notch signaling pathway is responsible for limiting the number of cells in the PNC that achieve the neuronal fate (Bray 1998; Artavanis-Tsakonas et al. 1999; Baker 2000; Schweisguth 2004). When Notch signaling is lost, proneural gene expression fails to resolve into a single cell, thus allowing all cells of the PNC to become a neuron (Hartenstein and Posakony 1990; Skeath and Carroll 1992; Ruiz-Gomez and Ghysen 1993). Resolution of the PNC into a single SOP results from transcriptional repression of the proneural genes and their targets by the Enhancer of Split [E(spl)] proteins (Paroush et al. 1994; Jimenez and Ish-Horowicz 1997; Culi and Modolell 1998; Giagtzoglou et al. 2003). In addition, the ability of Sens to promote the neural fate is decreased when bound by E(spl)m8 (Jafar-Nejad et al. 2003). These known connections between lateral inhibition and sens led us to hypothesize that members of the Notch pathway should modify ls. Identification of modifying candidate genes in the Notch pathway would support the idea that genes interacting with sens could be identified in an EMS screen for modification of ls.
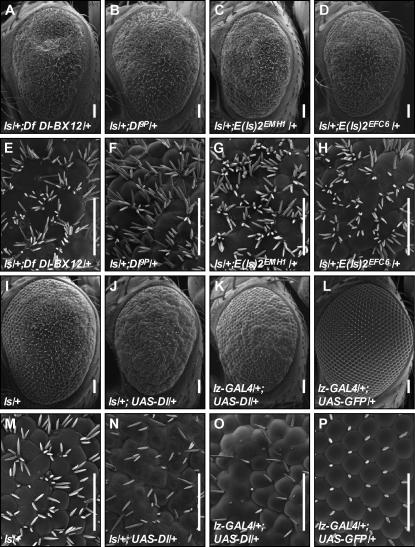
Multiple available alleles of Notch pathway members were crossed with ls virgins and F1 progeny were scored for enhancement or suppression of the ls phenotype. Loss of one copy of Dl causes strong enhancement of the number of ectopic bristles formed in ls and a marked increase in the disorganization of ommatidial architecture (see Figure 2). The enhancement is consistent for two amorphic alleles tested (Dl9P and DlB2) (see Figure 2, B and F, and data not shown). Consistent with the candidate gene screen that identified Dl as a modifier, in a test screen with the Bloomington deficiency kit, to be discussed later, Df(3R)Dl-BX12 was also identified as an enhancer of ls (see Figure 2, A and E). This deficiency uncovers the Delta locus. We were initially surprised that we did not see enhancement with other components of the Notch signaling pathway. However, since only dominant interactions were tested, this indicates only that the ls phenotype is more sensitive to loss of one copy of Delta than any other pathway member.
Figure 2.—
Delta modifies ls in a predicted manner. Df(3R) Dl-BX12 uncovers Delta and was identified as an enhancer of the ls phenotype (A and E). A previously identified mutation in Dl (DlP9) fails to complement the deficiency (data not shown) and also enhances the ls phenotype (B and F). Two EMS mutants were identified as alleles of Delta; these also enhance the ls phenotype (compare C and G and D and H to I and M). Correspondingly, overexpression of Delta (J and N) decreases the number of ectopic bristles produced by ls (compare J and N with I and M). Control animals are lz-GAL4/+; UAS-Dl/+ (K and O) and lz-GAL4/+;UAS-GFP/+ (L and P).
Decreasing Dl in the presence of ectopic Sens leads to an increased number of IOBs. Therefore, we hypothesized that increasing Dl in the ls background should decrease the number of IOBs formed. This was tested by expressing UAS-Dl with the lz-GAL4 driver along with UAS-sens. As predicted, in the ls/+; UAS-Dl/+ eye (Figure 2, J and N), a decreased number of bristles is formed compared to the ls eye (Figure 2, I and M). Together, these data demonstrate that the ls phenotype is modified in a manner predicted by known interactions between lateral inhibition and Sens-dependent neural development.
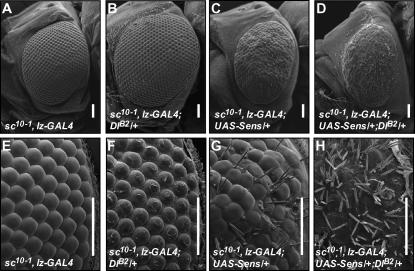
One possibility that these data do not address is that the effect of Dl on the ls phenotype could be mediated through the proneural genes and not by a direct effect on Sens. sens expression is downstream of proneural gene expression, and during SOP formation proneural gene expression is refined by lateral inhibition. With decreased expression of Delta, the resolution of the proneural field is impaired, and more than one SOP can be selected (Parks and Muskavitch 1993). This is due to the derepression of the proneural genes with decreased lateral inhibition (Parks et al. 1997). It is possible that the extra SOP formation that results from derepressed proneural expression explains the increased number of bristles seen in the Dl-enhanced ls eye. To determine if the enhancement of ls is dependent on the effect of Dl on proneural gene expression, we tested the ability of Dl to enhance ls in the absence of the relevant proneural genes. The sc10-1 deficiency removes both ac and sc, the proneural genes required for bristle development. Hemizygous males of this genotype can be collected as pharate pupae and completely lack IOBs (see Figure 3, A and E). With the addition of the ls chromosome, bristles can once again be formed even in the absence of ac and sc (Figure 3, C and G). As we have reported previously, the ability of ectopic Sens to form bristles in the absence of the ac/sc locus has regional dependence. The posterior region of the eye shows much stronger induction of bristles than the anterior of the eye (Frankfort et al. 2004). This is likely due to the progressive nature of the morphogenetic furrow that leads to a longer exposure of cells in the posterior region of the eye to ectopic Sens. When a single copy of Delta is removed in an ls fly that lacks ac and sc, the number of bristles formed is increased (Figure 3, D and H). Thus, we see an enhancement of the ls phenotype by Delta even in the absence of the bristle proneural genes. Anterior, middle, and posterior regions of the eye show enhancement in the presence of Dl consistent with the graded response to Sens. These data show that the enhancement of Dl on the ls phenotype is not dependent on the refinement of proneural gene expression and support previous evidence that lateral inhibition downregulates Sens function. This is consistent with a body of work that describes the effect of lateral inhibition not only at the level of proneural gene expression, but also at the level of regulation of proneural target gene expression and function (Culi and Modolell 1998; Giagtzoglou et al. 2003; Jafar-Nejad and Bellen 2004).
Figure 3.—
Delta modulates sens function in the absence of proneural genes. The sc10-1 deficiency removes the ac/sc locus, and hemizygous males lack IOBs (A and E). Loss of one copy of Dl does not affect this phenotype (B and F). In the sc10-1 background, ectopic Sens leads to the formation of IOBs and a roughening of the eye surface (C and G). Removal of one copy of Dl increases the number of bristles that are formed and leads to a further loss of normal ommatidial surface architecture (D and H). Images in A–D are shown at 200× magnification, and images in E–H are shown at 1000×. Bars, 50 μm.
Taken together, these data show that we have created an ectopic Sens phenotype that is dominantly modifiable in a predictable way by Dl, a key gene in a pathway known to interact with Sens in neural development. This suggested that a genomewide mutagenesis screen would identify new genes interacting with senseless by their ability to modify ls. The utility of the screen was confirmed as two new alleles of Delta were identified during the screen as enhancers of ls (Figure 2, C, D, G, and H).
An F1 dominant modifier screen identifies 10 complementation groups interacting with Sens:
An F1 dominant modifier EMS mutagenesis screen was performed in which 110,000 animals were screened. w1118 males were mutagenized and crossed to ls/FM7 virgins (Figure 4). F1 progeny were scored for suppression or enhancement of the ls phenotype. One thousand one hundred thirty-five suppressors and 501 enhancers were identified; these F1 animals were backcrossed to ls flies. After at least three generations of backcrosses of mutant females to ls males, 117 suppressors and 11 enhancers were isolated and mapped to the second or third chromosome (see Table 1). Once mutations were mapped to a chromosome, balanced stocks were generated. Thirty-two mutations mapped to the second chromosome while 96 mapped to the third. We did not identify any mutations on the X chromosome, in large part due to the location of the lz-GAL4 and UAS-sens P-element insertions on the X chromosome and our crossing scheme for recovering mutations (see materials and methods). This outcome was not considered a problem for the success of the screen since no X chromosome deficiency was identified as a modifier of ls, suggesting that few loci on the X would be dominant modifiers. Seventy-nine mutants were either lethal mutations or segregated with a second lethal hit as judged by their inability to homozygose, while 49 mutant chromosomes were homozygous viable. Thirty-six lethal mutations were assigned to 10 complementation groups on either the second or the third chromosome, while 43 lethal mutants were single hits. Only 1 of the 49 viable mutants was assigned to a complementation group; the other 48 are either single hits or multiple hits in genes with no readily detectable homozygous phenotype. Examples of modification of the ls phenotype by a representative member of each complementation group are shown in Figures 2, 5, and 7 and supplemental Figure 1 at http://www.genetics.org/supplemental/.
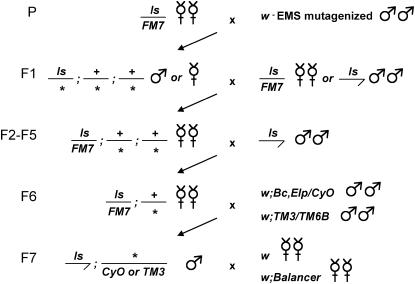
Figure 4.—
Crossing scheme for the F1 dominant modifier screen. Isogenized w1118 males were mutagenized with 25 mm EMS and crossed to females with the lz-GAL4,UAS-sens chromosome (ls). F1 progeny that showed enhancement or suppression of the ls phenotype were backcrossed to ls males or females. From F1 male backcrosses, F2 female progeny balanced by FM7 and carrying a modifying mutation were backcrossed to ls males for at least three generations. From F1 female backcrosses, F2 ls males carrying a modifying mutation were collected and crossed to ls/FM7 virgins. The F3 balanced female progeny carrying a modifying mutation were backcrossed to ls males for at least three generations. F6 ls/FM7 females still carrying a modifier were crossed in two separate batches to second and third chromosome balancer males. Single F7 ls males with a modifier and a balancer chromosome were crossed in series to w virgins and then to virgins from the corresponding balancer stock to map the mutation to a chromosome and produce a balanced stock.
TABLE 1.
Screen summary
| No. of flies | % recovery | |
|---|---|---|
| F1 flies screened | 110,000 | |
| F1 modified flies selected | 1,636 | 1.5 |
| Final balanced stocks | 128 | 0.1 |
| Complementation group name | Location | No. of alleles | Gene name |
|---|---|---|---|
| S(ls)1 | Centromeric 3 | 4 | |
| S(ls)2 | 66B6 | 15 | moleskin/dim-7 |
| S(ls)3 | 88E–88F | 3 | |
| S(ls)4 | 23C1–3 | 2 | lilliputian |
| S(ls)5 | 21A | 2 | |
| S(ls)6 | 35E5–6 | 1 | |
| S(ls)7 | 89B3–89B4 | 2 | |
| E(ls)1 | 42A1–42E6 | 1 | |
| E(ls)2 | 92A1–2 | 2 | Delta |
| E(ls)3 | 89B3–89B4 | 4 | |
| Single alleles | 92 |
Figure 7.—
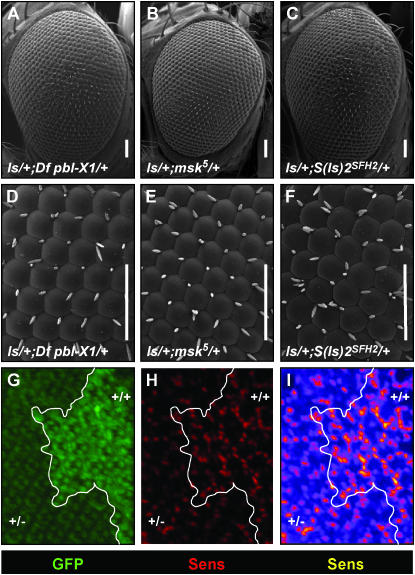
moleskin/dim-7 dominantly suppresses ectopic bristle formation and reduces ectopic Sens expression. Df pbl-X1 uncovers moleskin/dim-7 and was identified as a dominant suppressor of the ls phenotype (compare A and D with Figure 1, D and E). A previously identified mutation in msk (msk5) fails to complement the deficiency (data not shown) and also suppresses the ls phenotype (B and E). Fifteen EMS mutants were identified as alleles of msk; these also suppress the ls phenotype (C and F and data not shown). Consistent with this observation, ectopic expression of Sens in 48-hr APF discs heterozygous for msk5 (+/−) is reduced compared to that in the wild-type twinspot (+/+) (G–I). G shows GFP staining in green. The twinspot boundary is outlined in white. Sens staining is shown in red (H). To illustrate the differences in Sens staining between the twinspot and heterozygous tissue, pixel intensity from H was mapped to a heat scale (I); dim pixels are dark blue and bright pixels are yellow. Genotype of (+/+) is ls/+; ubi-GFP, FRT80B/ubi-GFP, FRT80B. Genotype of (+/−) is ls/+; ubi-GFP, FRT80B/msk5, FRT80B.
Mapping of the complementation groups utilized multiple strategies. Before the EMS screen was initiated, we performed a genomewide deficiency screen using the Bloomington deficiency kit to determine how many and which regions of the genome could modify the ls phenotype. At the time this screen was performed, the Bloomington kit contained 210 stocks that uncovered ∼70–80% of total euchromatin (Bloomington Stock Center 10/04/02). In this screen, ls/FM7 virgins were crossed to balanced deficiencies on the second, third, and fourth chromosomes while X chromosome deficiency virgins were crossed to ls males. This screen identified 12 deficiencies that modified the ls phenotype (see Table 2). For each complementation group from the EMS screen, a rough mapping position was determined either by failure to complement 1 of the 12 modifying deficiencies or by low-resolution meiotic mapping with a kit of molecularly mapped P{w+} elements (Zhai et al. 2003). Groups that failed to complement a deficiency were then tested against lethal genes located within the breakpoints of that deficiency. In this way, members of S(ls)2 were identified as alleles of moleskin/dim-7, S(ls)4 as alleles of lilliputian, and E(ls)2 as alleles of Delta. For groups that failed to complement a deficiency, but not a single gene within that deficiency, a round of rough meiotic mapping using five P{w+} elements spaced over the region of the deficiency was performed. Alternatively, for groups that did not map to a previously defined modifying deficiency, a rough map position was determined using the P{w+} elements in the kit outlined in Zhai et al. (2003), which are spaced every 1–3 Mb. Each complementation group was mapped to all the P{w+} elements for the appropriate chromosome and then subjected to a round of intermediate mapping using P{w+} elements spaced at ∼200-kb intervals between the flanking P{w+} elements. This round of mapping was followed by a final round of high-resolution P{w+} mapping as described in Zhai et al. (2003). This provided a predicted location within 50 kb. A summary of complementation group location by cytological position in the genome is presented in Figure 6.
TABLE 2.
Deficiencies that modify ls
| Bloomington deficiency no. | Symbol | Effect | Break points | Single genes identified |
|---|---|---|---|---|
| 90 | Df(2L)C144 | Su | 22F3–4; 23C3–5 | lilliputian/S(ls)4 |
| 1491 | Df(2L)r10 | Su | 35D1; 36A6–7 | S(ls)6 |
| 749 | In(2R)bwVDe2LCyR | Su | 42–43; 42A2–3 | |
| 3368 | Df(2R)cn9 | Su | 42E; 44C | |
| 3909 | Df(2R)59AD | Su | 59A1–3; 59D1–4 | |
| 1420 | Df(3L)pbl-X1 | Su | 65F3; 66B10 | moleskin/S(ls)2 |
| 3640 | Df(3L)brm11 | Su | 71F1–4; 72D1–10 | |
| 2998 | Df(3L)81k19 | Su | 73A3; 74F | |
| 823 | Df(3R)D605 | Su | 97E3; 98A5 | |
| 1007 | Df(2R)nap9 | En | 42A1–2; 42E6–F1 | E(ls)1 |
| 1931 | Df(3R)by416 | En | 85D8–12; 85E7–F1 | |
| 3012 | Df(3R)Dl-BX12 | En | 91F1–2; 92D3–6 | Delta/E(ls)2 |
Su, suppression; En, enhancement.
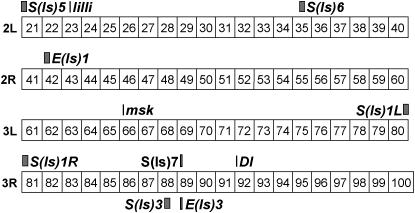
Figure 6.—
Ten complementation groups are distributed across both arms of chromosomes 2 and 3. Boxes indicate the interval containing a complementation group. Complementation groups identified as known genes or groups mapped to narrow intervals are indicated with vertical bars. Note that S(ls)1 could not be localized to one arm of chromosome 3 due to its extreme proximity to the centromere. Potential locations on either arm are denoted with an L or an R.
lilliputian is a suppressor of ls, but does not interact specifically with sens:
From the Bloomington deficiency kit screen, Df(2L)C144 was identified as a suppressor of ls. lilliputian is a gene within Df(2L)C144 that has been identified as a dominant suppressor of ectopic phenotypes in at least 10 screens (Dickson et al. 1996; Perrimon et al. 1996; Neufeld et al. 1998; Greaves et al. 1999; Li et al. 2000; Rebay et al. 2000; Su et al. 2001; Li and Li 2003; Luschnig et al. 2004; Anderson et al. 2005). Therefore, we tested lilli for its ability to modify the ls phenotype. lilli00632 showed strong dominant suppression and failed to complement Df(2L)C144. In addition, two independent suppressors of ls, corresponding to complementation group S(ls)4 from the EMS screen, were identified as alleles of lilli by failure to complement Df(2L)C144 and lilli00632. lilli is an AF4/FMR2 family transcription factor that is required for normal embryogenesis (Wittwer et al. 2001). Normal eye cell fate specification is not disrupted in lilli mutant clones, but the photoreceptor rhabdomeres are smaller in size than wild-type rhabdomeres (Tang et al. 2001; Wittwer et al. 2001).
To eliminate genes that modify ls due to nonspecific effects on the GAL4/UAS system or expression from the lozenge locus, all complementation groups were tested in two secondary screens. The first secondary screen tested the ability of mutants to modify a rough-eye phenotype generated by the ectopic expression of UAS-rough with the lz-GAL4 driver (lz-ro). Mutations that modify this phenotype as well as the ls phenotype could represent genes that are not specific to sens interaction, but rather could be affecting transcription from the lozenge (lz) locus. lz-Gal4 was generated by a P{GawB} insertion in the lz locus, so transcription of GAL4 in these flies is regulated by the factors that normally govern lz expression. All mutants and modifying deficiencies were tested against lz-ro. The lilliputian complementation group S(ls)4 comprised the only alleles that suppressed the lz-ro phenotype. Aside from effects at the lz locus, lilli could also be affecting some general aspect of transcription in the GAL4 system. To test for this possibility, another test was performed by crossing lilli to flies carrying the C96-GAL4 and UAS-hrs transgenes. This genotype leads to the formation of multiple notches along the wing margin, but has no known relationship to lz function or expression (see supplemental Figure 1I at http://www.genetics.org/supplemental/). All alleles of lilli tested, including Df(2l)C144 and the screen-generated mutants, strongly suppress the wing notching normally seen with C96-GAL4, UAS-hrs (supplemental Figure 1J at http://www.genetics.org/supplemental/). This strongly suggests that lilli suppresses all three gain-of-function phenotypes by suppressing the GAL4/UAS system. This conclusion is consistent with previous studies showing that lilli affects transcription from constructs utilizing hsp70 promoters for ectopic gene expression (Tang et al. 2001; Wittwer et al. 2001). The P{GAWB} element used to generate lz-GAL4 contains the GAL4 coding sequence under the control of the hsp70 promoter. Together with the data suggesting that lilli does not appear have a role in normal R8 or IOB development, it is likely that lilli does not play a specific role in the sens pathway, but rather affects transcription from the lz-GAL4 hsp70 promoter. Since no other complementation group failed the secondary screens, they are likely to have a functionally significant interaction with sens and do not modify ls due to nonspecific interactions.
Mutations in moleskin suppress the bristle and ommatidial phenotypes generated by ls and result in reduced expression of ectopic Sens:
Fifteen suppressor mutations form the largest complementation group identified in the screen. This group, S(ls)2, was mapped to the modifying deficiency Df(3L)pbl-X1. Df(3L)pbl-X1 is a strong dominant suppressor of ls (see Figure 7, A and D). This deficiency uncovers cytological position 65F3–66B10. To identify a single gene within this deletion that conferred suppression, lethal alleles within this region were tested for modification of ls. Two previously identified embryonic lethal alleles of the gene moleskin/dim-7, msk4 and msk5 (Baker et al. 2002), located at 66B6, suppress the ls phenotype and fail to complement Df(3L)pbl-X1 (see Figure 7, B and E, and data not shown). All 15 EMS alleles also failed to complement the alleles msk5 and msk4 identifying this complementation group as moleskin/DIM-7. The msk5 mutation is a slightly stronger suppressor of ls than two S(ls)2 alleles we examined by SEM (Figure 7, B, C, E, and F, and data not shown). This is probably due to the fact that the msk5 allele is a null due to a G > T point mutation that changes the second amino acid to a stop codon (Baker et al. 2002). Since msk5 is a molecularly characterized allele, and the molecular nature of the alleles isolated from the screen are not known, we decided to pursue further analysis of the interaction between sens and msk with this allele. Adult EGUF SEM and sectioning phenotypes generated with msk5 were also generated with two EMS-isolated alleles and the results were consistent (data not shown). Importantly, no allele of msk suppressed the phenotypes generated by the C96-GAL4,UAS-hrs or lz-GAL4,UAS-ro secondary screens (data not shown). This suggests that the suppression of ls by msk is due to a specific interaction of msk with the ectopic sens pathway.
Msk is a nuclear transport protein, or karyopherin, and is homologous to Importin 7. Importins are a family of proteins that act to transport macromolecules to and from the nucleus (Gorlich et al. 1997; Strom and Weis 2001). In Drosophila, moleskin has been implicated in the nuclear transport of proteins such as pMAPK (Lorenzen et al. 2001; Vrailas et al. 2006) and Caudal (Han et al. 2004). Sens is a 61-kDa protein (Nolo et al. 2001); this is just above the predicted size exclusion limit for passive entry into the nucleus (Bednenko et al. 2003; Weis 2003). This raised the possibility that msk dominantly suppresses the ls phenotype by limiting the entry of Sens into the nucleus. This model leads to two predictions. First, in the ls background we should be able to detect less nuclear Sens in tissue heterozygous for msk than in tissue with two wild-type copies of msk. Second, Sens and Msk should physically interact.
To test the first prediction, we generated mosaic eyes that contain neighboring wild-type (msk +/+) or heterozygous (msk +/−) tissue in an otherwise ls background (Figure 7, G–I). We predicted that these two types of tissue would behave differently with respect to nuclear localization of Sens and that in the heterozygous (+/−) tissue there would be a decrease in the number of Sens-expressing cells and a decrease in the levels of nuclear Sens detected. When we looked at 48-hr APF eye discs, this is indeed what we found. In the ls background, there were fewer Sens-positive cells in msk heterozygous (+/−) tissue than in wild-type tissue (+/+) (P < 0.014; see materials and methods). This represented an average 1.5-fold difference between the number of Sens-positive cells per square micrometer in wild-type vs. heterozygous tissue (standard deviation = 0.22). In addition, in all images examined Sens expression in heterozygous cells was less intense than in wild-type cells (Figure 7, G–I). An intensity map of Sens staining in the ls background in either twinspot (+/+) or heterozygous (+/−) msk tissue is shown in Figure 7I. Low levels of Sens staining are shown in blue, midrange levels in red, and high levels in yellow. Figure 7I illustrates that more intense Sens staining in individual cells occurs in the unmodified twinspots than in the heterozygous tissue. The simplest interpretation of these data is that in heterozygous msk tissue, nuclear entry of Sens or some factor necessary for ectopic Sens expression is decreased. With decreased nuclear transport, Sens function is reduced, and its ability to recruit cells to the bristle fate is decreased. This is consistent with the screen results that found strong functional suppression of the ls phenotype in heterozygous msk tissue. In contrast, control experiments with tissue lacking ectopic Sens (i.e., non-ls background) did not reveal a difference in Sens nuclear expression in (+/+) vs. (+/−) tissue (data not shown). In addition, bristles are formed normally in msk heterozygous tissue (data not shown). These data suggest that the amount of Msk function required to maintain wild-type levels of Sens function is lower than the amount of import required to produce the ls phenotype.
To test the second prediction, a direct interaction between Msk and Sens, GST-tagged Msk was generated and tested for its ability to pull down in vitro translated and transcribed Sens. Despite testing a range of wash conditions, a specific interaction between Msk and Sens could not be identified (data not shown). It is possible that the interaction is not direct or may require a cofactor that was not provided in the binding buffer. Thus, although a reduction in Sens nuclear import may contribute to the effect on staining and the suppression of the ls phenotype identified in the screen, it is more likely that Msk imports some other factor required for normal bristle development or Senseless pathway function.
moleskin is required for normal eye development:
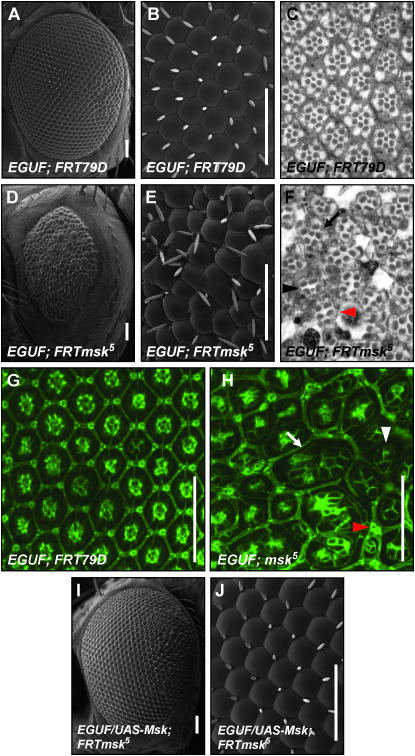
Interactions detected in modifier screens do not always accurately represent the biological processes present during normal development. Therefore, we next set out to assess the effect of loss of Msk on normal neural development and Sens function in the eye. To study the effect of a complete loss of Msk on normal development, we attempted to generate homozygous mutant eye tissue using mitotic recombination. Unfortunately, we found that msk clones are rare and small in larval eye tissue and are not detectable in pupal or adult tissue even when clones are made with a Minute chromosome (data not shown). This is consistent with previous observations in the wing and eye (Baker et al. 2002; Vrailas et al. 2006). The small clone size could be due to the inability of msk null tissue to compete with heterozygous tissue. To circumvent this problem, we made whole msk mutant eyes using the ey-GAL4, UAS-flp (EGUF) system, which removes competing heterozygous and wild-type cells due to the presence of the GMR-hid transgene (Stowers and Schwarz 1999). Using this system, small, disorganized adult eyes that are mutant for msk5 are recovered (compare Figure 8, A and B to D and E). Examination of the adult eye morphology by SEM reveals ommatidial fusions, irregular ommatidial shape and size, and occasional bristle loss or duplication (compare Figure 8B to 8E). IOB loss and duplication as well as ommatidial fusions are also apparent in 48-hr APF discs stained with anti-Armadillo antibody (compare Figure 8G to 8H). In addition, Armadillo staining reveals that all cell types are affected by the loss of msk. Some ommatidia have clusters containing more than four cone cells (white arrow in Figure 8H), but others with only two or three cone cells are observed, and many of these cone cells are an abnormal size. Primary pigment cells are often enlarged and abnormally shaped. Secondary and tertiary pigment cell number is often drastically reduced, as shown by multiple ommatidial fusions (arrow in Figure 8H). We examined Armadillo staining at earlier stages (third larval instar, 6 and 24 hr APF; data not shown), to determine at what stage loss of msk led to abnormal development. By third instar, abnormalities can already be seen in the spacing of ommatidial clusters as they emerge from the furrow (data not shown). To verify that the EGUF; msk5 phenotype is due specifically to the loss of msk, UAS-msk was expressed in EGUF; msk5 eyes (compare Figure 8, D and E to I and J). Eye development was rescued with the exception of minor disorganization at the posterior margin.
Figure 8.—
moleskin/dim-7 is required for normal eye development. An eye made entirely mutant for msk5 using the EGUF system is small and rough when compared with the control (D vs. A). At higher magnification ommatidial fusions and disruptions of the normal arrangement of bristles can be seen (compare E with B). Thin plastic sections of the EGUF; msk5 eye reveal a general disruption of normal ommatidial organization and numerous abnormal rhabdomeres (compare F with C). Some ommatidia have a normal number and arrangement of rhabdomeres (black arrow in F) while others have a decreased number of rhabdomeres per ommatidium (black arrowhead in F). Some ommatidia lack any small rhabdomeres (red arrowhead in F). This loss was verified by serial section (data not shown). At 48 hr APF the control eye has already formed the crystalline lattice seen in the adult eye (G). In the EGUF; msk5 eye, the cellular organization is clearly disrupted at 48 hr APF (H). The number of cone cells per ommatidium is often decreased to three (white arrowhead in H). Ommatidial fusions are seen where both primary and secondary pigment cells are absent (white arrow in H). Evidence of occasional bristle duplications seen by SEM in E are also apparent in the pupal disc (red arrowhead in H). The EGUF; msk5 mutant phenotype is rescued with the addition of UAS-msk (I and J). Bars, 50 μm. Genotype of EGUF; msk5 is w; ey-GAL4,UAS-flp/+; msk5, FRT79D/GMR-hid,Cl FRT79D. Genotype of EGUF control is w; ey-GAL4,UAS-flp/+; FRT79D/GMR-hid,Cl FRT79D. Genotype of EGUF/UAS-msk;FRTmsk5 is w; ey-GAL4,UAS-flp/UAS-msk; msk5,FRT79D/GMR-hid,Cl,FRT79D.
Loss of moleskin leads to the sporadic loss of R8 photoreceptors:
Thin plastic sections of the EGUF; msk5 eyes also demonstrate severe disorganization (Figure 8, C and F). There are occasional ommatidia that form with the normal number of rhabdomeres in the stereotypical trapezoidal pattern (arrow in Figure 8F), but most have fewer than normal numbers of rhabdomeres (arrowhead in Figure 8F). Normal rhabdomere morphology is also disrupted: some are abnormal in shape, appearing rectangular or oblong rather than the usual, more round aspect seen in wild-type eyes. In addition, multiple ommatidia lacking small rhabdomeres are seen. The small rhabdomeres of R7 and R8 are normally located in the center of each ommatidium, but at different depths. To test for the presence of both small rhabdomeres, serial sectioning of EGUF; msk5 eyes was performed through the entire thickness of the retina. We found that 22% of ommatidia in the EGUF; msk5 eye were completely missing small rhabdomeres (representative shown by red arrowhead in Figure 8F). This is interesting because this effect on small rhabdomeres is reminiscent of the sens mutant phenotype, which shows a complete loss of small rhabdomeres.
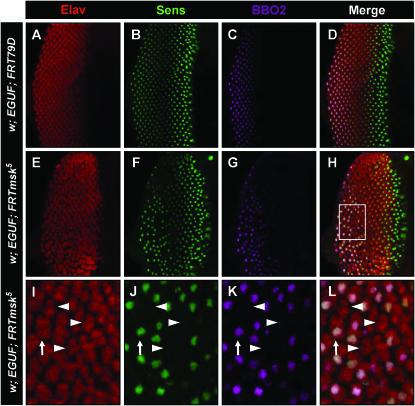
To further explore this phenotypic link between msk and sens, we investigated the effect of loss of msk on the expression of Sens in the developing R8 photoreceptor. Sens is required in the R8 cell to maintain the R8 fate by repressing the expression of the homeodomain transcription factor Rough (Frankfort et al. 2001). In the absence of Sens, the R8 adopts the R2,5 cell fate. Consistent with the sporadic loss of adult R8's, we found that Sens staining was lost sporadically throughout the EGUF; msk5 larval disc (Figure 9, F and J; compare to Figure 9B). This incomplete loss of Sens expression in developing clusters of the homozygous msk disc was surprising considering the strong effect of heterozygous msk loss on the screen phenotype. This indicates that msk is not the sole transport factor utilized by the Sens pathway and suggests that msk may affect the sens pathway differently in R8 and IOB development. A previous study exploring the role of msk in Drosophila eye development did not detect an effect of msk loss on Sens expression in R8 development (Vrailas et al. 2006). This is not surprising due to the sporadic effect of Msk loss on Sens expression. Consistent with our attempts to make mosaic msk eyes, Vrailas et al. were able to generate small clones of only a few cell widths. Detecting Sens loss with this method would require observation of potentially hundreds of such small clones. We are able to detect this interaction due to the large number of mutant ommatidia generated with the EGUF system. To determine if the loss of Sens staining in msk mutants also leads to the loss of the R8 fate, we tested for the expression of BBO2, an enhancer trap known to be expressed later in R8 development. BBO2 is normally expressed in the R8 photoreceptor by column 11–12 (Figure 9C) (Hart et al. 1990). In sens null tissue, BBO2 expression is lost in the putative R8 (Frankfort et al. 2001). We found that this marker is also lost in EGUF; msk5 cells that also lack Sens expression (Figure 9, G and K). Therefore, loss of msk leads to sporadic loss of R8 photoreceptors during larval development.
Figure 9.—
Loss of moleskin/dim-7 function leads to the sporadic loss of R8 photoreceptors. Expression of Elav (A, E, and I), Sens (B, F, and J), and BBO2 (C, G, and K) in control (A–D) vs. EGUF; msk5 (E–L) third instar larval eye discs is shown. I–L show a magnified view of the boxed area in H. Elav staining in wild-type third instar larval discs reveals the emerging crystalline order seen in the adult eye (A). Each wild-type Elav-positive cluster contains a Sens-positive R8 (B). The P{lacW} enhancer trap BBO2 is a later marker of the R8 photoreceptor. More mature R8 photoreceptors express both Sens and BBO2 (D and arrow in I–K). In contrast, the EGUF; msk5 eye (E–H) shows disruption of the wild-type organization with uneven spacing between Elav-positive clusters. Furthermore, some Elav-positive clusters lack Sens and BBO2 (arrowheads in I–L). Genotype of w; EGUF; msk5 is w; ey-GAL4,UAS-flp/BBO2; msk5, FRT79D/GMR-hid,Cl FRT79D. Genotype of w; EGUF; FRT79D is w; ey-GAL4,UAS-flp/BBO2; FRT79D/GMR-hid,Cl FRT79D.
There are three potential mechanisms for loss of the R8 photoreceptor: the R8 is never specified, the R8 is specified but then dies, or the R8 is specified but then changes fate. Our lab previously explored each of these possibilities while defining the sens mutant phenotype and determined that the presumptive R8 changed fate to an outer R2,5-type photoreceptor. We tested these possibilities in msk mutant discs to determine the mechanism for R8 loss in this genotype. In the case where an R8 is never specified, such as in an atonal mutant, no photoreceptors develop due to the lack of the EGFR ligand spitz, which is normally produced by R8 (Tio et al. 1994; Tio and Moses 1997). Therefore, if R8 is never specified, no other photoreceptors will develop or be detectable by staining for the neuronal antigen Elav (embryonic lethal, abnormal vision). As seen in Figure 9L, there are multiple Elav-positive, Sens-negative clusters in msk tissue, suggesting that the R8 cell was specified in the msk mutant and was able to initiate outer photoreceptor development before it either died or changed fate.
Due to the similarity of the msk and sens mutant phenotypes and the genetic interaction detected between msk and sens, we hypothesized that the msk R8 phenotype may be due to an effect on sens expression during normal development. This hypothesis predicts that the missing R8's in EGUF; msk5 eyes also switch fate to an R2,5-like photoreceptor. This hallmark of the sens phenotype is identified in the third instar disc by an Elav-positive cluster that lacks a Sens- or a BBO2-positive cell, but gains a third RM104-positive cell (Frankfort et al. 2001). RM104 is an enhancer trap specifically expressed in R2/5 in wild-type tissue (arrowheads in Figure 10, C and D). As seen in the sens mutant, EGUF; msk5 ommatidia lacking Sens expression also have three cells expressing RM104 (arrowheads in Figure 10, G and H). Therefore, loss of msk can lead to an early cell fate switch of R8 to an R2,5 photoreceptor and sporadically phenocopies the sens loss-of-function photoreceptor phenotype.
These data support a model where, in some developing clusters in msk mutant tissue, the level of nuclear transport stochastically fails to reach a critical threshold and leads to loss of both Sens expression and the R8 cell fate. However, the causal relationship and order of events is not clear: Does the loss of Sens lead to the loss of R8 fate, or is there an upstream effect on R8 fate that is the cause of the loss of Sens expression? To determine which event occurs first, we examined R8-less clusters for expression of the early R8 marker sca-lacZ. Previously, we demonstrated that in sens clones, sca-lacZ remains on and marks the cell that should have developed as an R8 (Frankfort et al. 2001). To determine when msk is required for R8 specification, we tested the Sens-negative clusters in EGUF; msk5 eyes for the expression of sca-lacZ (Figure 10, M–P). We found that clusters lacking Sens also lack sca-lacZ expression, suggesting that the R8 fate is compromised before the onset of Sens expression. The presence of three RM104-positive cells in R8-less Elav-positive clusters suggests that loss of msk causes a sporadic failure of R8 development that occurs after the initial specification by Atonal, but before the onset of sca-lacZ and Sens expression.
Superficially, loss of msk resembles the sens mutant phenotype with incomplete penetrance. However, these data also suggest that Msk has an independent role upstream of sens in normal eye development where it may be required to import other factors important in R8 fate maintenance. Interestingly, this is the first phenotype to identify a genetically separable developmental stage between Senseless and Atonal.
DISCUSSION
Sens, along with its homologs Gfi-1 and Pag-3, comprises a conserved family of proteins required for normal neural development. In Drosophila, sens is both necessary and sufficient for development of the PNS. In mice, loss of Gfi-1 leads to neurodegeneration of cerebellar Purkinje cells and sensoneural deafness due to loss of inner ear hair cells (Wallis et al. 2003; Tsuda et al. 2005). Despite the obvious importance of the GPS proteins in normal neural development and their place near the top of the neuronal development cascade, few targets of these proteins in the process of neurogenesis are known. To identify members of this pathway required in neurogenesis, we performed an F1 dominant modifier screen using an ectopic Sens phenotype in Drosophila. We took advantage of a dominant, modifiable phenotype generated by ectopic expression of Sens in undifferentiated cells posterior to the morphogenetic furrow. This ectopic Sens led to the recruitment of undifferentiated cells to the bristle fate.
We report here on both known and novel genes that have been identified as potential members of the sens pathway by their ability to modify an ectopic Sens phenotype. The Notch signaling pathway is known to regulate Sens function during the resolution of the proneural cluster (Jafar-Nejad et al. 2003). This interaction was identified in our screen by the ability of heterozygous loss of Dl to enhance the ectopic Sens phenotype. The nuclear import gene moleskin (msk) was able to strongly suppress the effect of ectopic Sens. We also show that msk plays a role in normal eye development and R8 photoreceptor differentiation. Identification of the genes that are represented in the remaining complementation groups will lead to a better understanding of the GPS pathway and normal neural development. It is likely that the remaining complementation groups represent components of the Sens pathway due to their specific effect on ls and not the secondary screens as well as their requirement for normal bristle development in adult thoracic clones (to be reported elsewhere). Further characterization of these genes will offer new insight into the highly conserved Sens pathway.
Alleles of msk were found to be suppressors of ls with the highest frequency of any complementation group in the EMS screen. Usually such high representation of alleles indicates that the gene has an important role in the phenotype being tested and/or is readily mutagenized. The results presented here suggest a model in which Msk plays a role in the sens pathway. Our initial observations of the effect of Msk on the ls phenotype suggested that Msk was needed to maintain high levels of Sens expression. It is possible that in this ectopic situation, Msk contributes to Sens import, but more likely Msk contributes to Sens expression indirectly by importing another component of the pathway that regulates Sens expression. Characterization of the EGUF; msk phenotype strongly suggests that Msk is not the only import factor involved in the Sens pathway during normal development. Clearly, there is functional redundancy with another importin since complete loss of Msk function during early eye development does not remove Sens expression in all R8 cells. In third instar discs, Msk appears to play a role in the maintenance of the R8 cell fate very early in development. Little is known about the early stages of R8 differentiation after specification by Atonal. Previous work on R8 specification and development outlined a hierarchy of events in which Atonal is expressed first and appears to simultaneously activate expression of the downstream targets sens and sca-lacZ (Mlodzik et al. 1990; Frankfort et al. 2001). Work on the sens phenotype determined that sca-lacZ expression is still present in sens clones, thereby establishing an epistatic relationship between sca-lacZ expression and sens (Frankfort et al. 2001). Our data indicate that there is yet another step in the relationship between Atonal and these two downstream factors. Our data suggest that in the msk eye, after specification of the R8 by Atonal but before the onset of sca-lacZ expression, R8 development is disrupted in some clusters, leading to an R2,5 fate switch. This is the first genetic evidence for factors positioned between ato and sca-LacZ/sens.
Nuclear transport is required for the viability of all cells. Interestingly, the loss or decrease in function of some importins can cause specific defects during development. For example, the nuclear exportin Dcas is required for the export of Importin α3 in Drosophila (Tekotte and Davis 2002). While null mutants in dcas are not viable, hypomorphs lead to specific cell fate changes in mechanosensory bristles. This phenotype is likely due to extreme sensitivity of Notch signaling to disruption of nuclear transport of one of its pathway members by Importin α3. It is possible that the Msk/Sens interaction was detectable for a similar reason. In the Sens gain-of-function situation, the high level of Sens required to generate ectopic bristles is very sensitive to decreased Msk levels, while during wild-type SOP differentiation, Sens is far less sensitive to Msk levels and exhibits only sporadic effects.
One question still remains: How does the EGUF; msk eye survive at all given the important cargo that Msk is known to transport? The functional redundancy in the Importin family likely provides the cell with enough transport for survival and development in the absence of Msk. However, this idea raises a new question: Why did we identify only Msk in our screen and no other importins? We propose a model in which Msk is the key importin utilized by the cell for high levels of signaling. The ls phenotype requires high levels of signaling to generate ectopic bristles, and this model would explain why we detected an effect with Msk and no other importin. The model does not preclude the ability of other importins to provide transport redundancy for Msk cargos, and in fact we see evidence for this redundancy in the ability of the EGUF; msk eye to survive and produce some normal ommatidia. Another importin must have the ability to import some level of Sens, pMAPK, and other unidentified factors into the nucleus. The recent article by Vrailas et al. (2006) provides data that indirectly support such a model for the role of Msk. In Vrailas et al. (2006) the authors show that in the Atonal intermediate groups within the morphogenetic furrow, Msk must be sequestered away from the nucleus to prevent the very high levels of cytoplasmic pMAPK from entering the nucleus. Although they do not test if other nuclear importins are also sequestered to block pMAPK nuclear entry, they do show that overexpression of Msk in the intermediate groups allows pMAPK to enter the nucleus and affect nuclear signaling. The fact that the cell needs to sequester Msk to prevent high levels of EGFR pathway signaling supports a model in which Msk is important for high levels of signaling.
It has been suggested in other developmental systems that importins are part of a mechanism that regulates the nuclear protein composition of transcription factors and chromatin remodeling factors (Hogarth et al. 2005). In Drosophila, Msk has been shown to import two other developmentally significant cargos, pMAPK and Caudal (Lorenzen et al. 2001; Han et al. 2004; Vrailas et al. 2006). In addition to these previously defined roles, here we show additional data that Msk and nucleocytoplasmic transport play an important role in Sens expression and R8 development. Perhaps more importantly, the fact that abnormalities seen in msk mutant eye discs arise between Atonal and Senseless expression suggests roles for as-yet undiscovered factors and new modes of regulation in this critical pathway.
Acknowledgments
We thank Kenneth Dunner, Jr., at the University of Texas M. D. Anderson Cancer Center High-Resolution Electron Microscopy Facility for exceptional assistance with SEM. We thank Hugo Bellen, Andrew Jarman, Lizbeth A. Perkins, the Developmental Studies Hybridoma Bank, and the Bloomington Stock Center (Indiana University, Bloomington, IN) for flies and antibodies. We especially thank Kartik Pappu, Mardelle Atkins, and Umesh Karandikar for comments on this manuscript, as well as past and present members of the Mardon lab for support and advice. This work was supported by the National Eye Institute, the Retina Research Foundation, and the Moran Foundation.
References
- Abramoff, M. D., P. J. Magelhaes and S. J. Ram, 2004. Image processing with ImageJ. Biophotonics Int. 11: 36–42. [Google Scholar]
- Anderson, J., R. Bhandari and J. P. Kumar, 2005. A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye. Genetics 171: 1655–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M. D. Rand and R. J. Lake, 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., 2000. Notch signaling in the nervous system. Pieces still missing from the puzzle. BioEssays 22: 264–273. [DOI] [PubMed] [Google Scholar]
- Baker, S. E., J. A. Lorenzen, S. W. Miller, T. A. Bunch, A. L. Jannuzi et al., 2002. Genetic interaction between integrins and moleskin, a gene encoding a Drosophila homolog of importin-7. Genetics 162: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednenko, J., G. Cingolani and L. Gerace, 2003. Nucleocytoplasmic transport: navigating the channel. Traffic 4: 127–135. [DOI] [PubMed] [Google Scholar]
- Bell, D. W., T. Taguchi, N. A. Jenkins, D. J. Gilbert, N. G. Copeland et al., 1995. Chromosomal localization of a gene, GF1, encoding a novel zinc finger protein reveals a new syntenic region between man and rodents. Cytogenet. Cell Genet. 70: 263–267. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., and R. L. Cagan, 2003. Patterning the fly eye: the role of apoptosis. Trends Genet. 19: 91–96. [DOI] [PubMed] [Google Scholar]
- Bray, S., 1998. Notch signalling in Drosophila: three ways to use a pathway. Semin. Cell Dev. Biol. 9: 591–597. [DOI] [PubMed] [Google Scholar]
- Brown, N. T. L., S. Patel, J. Brzezinski and T. Glaser, 2001. Math5 is required for retinal ganglion cell and optic nerve formation. Development 128: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan, R. L., and D. F. Ready, 1989. The emergence of order in the Drosophila pupal retina. Dev. Biol. 136: 346–362. [DOI] [PubMed] [Google Scholar]
- Crew, J. R., P. Batterham and J. A. Pollock, 1997. Developing compound eye in lozenge mutants of Drosophila: lozenge expression in the R7 equivalence group. Dev. Genes Evol. 206: 481–493. [DOI] [PubMed] [Google Scholar]
- Culi, J., and J. Modolell, 1998. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12: 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, B. J., A. van der Straten, M. Dominguez and E. Hafen, 1996. Mutations modulating Raf signaling in Drosophila eye development. Genetics 142: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Z., and M. Horwitz, 2005. Gfi-1 takes center stage in hematopoietic stem cells. Trends Mol. Med. 11: 49–52. [DOI] [PubMed] [Google Scholar]
- Dufourcq, P., S. Rastegar, U. Strahle and P. Blader, 2004. Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr. Patt. 4: 53–57. [DOI] [PubMed] [Google Scholar]
- Dwivedi, P. P., P. H. Anderson, J. L. Omdahl, H. L. Grimes, H. A. Morris et al., 2005. Identification of growth factor independent-1 (GFI1) as a repressor of 25-hydroxyvitamin D 1-alpha hydroxylase (CYP27B1) gene expression in human prostate cancer cells. Endocr. Relat. Cancer 12: 351–365. [DOI] [PubMed] [Google Scholar]
- Frankfort, B. J., and G. Mardon, 2002. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129: 1295–1306. [DOI] [PubMed] [Google Scholar]
- Frankfort, B. J., R. Nolo, Z. Zhang, H. Bellen and G. Mardon, 2001. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 32: 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfort, B. J., K. L. Pepple, M. Mamlouk, M. F. Rose and G. Mardon, 2004. Senseless is required for pupal retinal development in Drosophila. Genesis 38: 182–194. [DOI] [PubMed] [Google Scholar]
- Fuchs, B., T. Wagner, N. Rossel, M. Antoine, H. Beug et al., 1997. Structure and erythroid cell-restricted expression of a chicken cDNA encoding a novel zinc finger protein of the Cys + His class. Gene 195: 277–284. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou, N., P. Alifragis, K. A. Koumbanakis and C. Delidakis, 2003. Two modes of recruitment of E(spl) repressors onto target genes. Development 130: 259–270. [DOI] [PubMed] [Google Scholar]
- Gilks, C. B., S. E. Bear, H. L. Grimes and P. N. Tsichlis, 1993. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 13: 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., M. Dabrowski, F. Bischoff, U. Kutay, P. Bork et al., 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, S., B. Sanson, P. White and J. Vincent, 1999. A screen for identifying genes interacting with armadillo, the Drosophila homolog of β-catenin. Genetics 153: 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S., J. Ryu, C. Oh, K. Nam, H. Nam et al., 2004. The moleskin gene product is essential for Caudal-mediated constitutive antifungal Drosomycin gene expression in Drosophila epithelia. Insect. Mol. Biol. 13: 323–327. [DOI] [PubMed] [Google Scholar]
- Hart, A. C., H. Kramer, D. L. Van Vactor, Jr., M. Paidhungat and S. L. Zipursky, 1990. Induction of cell fate in the Drosophila retina: the bride of sevenless protein is predicted to contain a large extracellular domain and seven transmembrane segments. Genes Dev. 4: 1835–1847. [DOI] [PubMed] [Google Scholar]
- Hartenstein, V., and J. W. Posakony, 1990. A dual function of the Notch gene in Drosophila sensillum development. Dev. Biol. 142: 13–30. [DOI] [PubMed] [Google Scholar]
- Hock, H., M. J. Hamblen, H. M. Rooke, D. Traver, R. T. Bronson et al., 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18: 109–120. [DOI] [PubMed] [Google Scholar]
- Hock, H., M. J. Hamblen, H. M. Rooke, J. W. Schindler, S. Saleque et al., 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431: 1002–1007. [DOI] [PubMed] [Google Scholar]
- Hogarth, C., C. Itman, D. A. Jans and K. L. Loveland, 2005. Regulated nucleocytoplasmic transport in spermatogenesis: A driver of cellular differentiation? BioEssays 27: 1011–1025. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad, H., and H. J. Bellen, 2004. Gfi/Pag-3/senseless zinc finger proteins: A unifying theme? Mol. Cell. Biol. 24: 8803–8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafar-Nejad, H., M. Acar, R. Nolo, H. Lacin, S. M. Parkhurst et al., 2003. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 17: 2966–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman, A. P., E. H. Grell, L. Ackerman, L. Y. Jan and Y. N. Jan, 1994. Atonal is the proneural gene for Drosophila photoreceptors. Nature 369: 398–400. [DOI] [PubMed] [Google Scholar]
- Jia, Y., G. Xie and E. Aamodt, 1996. pag-3, a Caenorhabditis elegans gene involved in touch neuron gene expression and coordinated movement. Genetics 142: 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, G., and D. Ish-Horowicz, 1997. A chimeric enhancer-of-split transcriptional activator drives neural development and achaete-scute expression. Mol. Cell. Biol. 17: 4355–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge et al., 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30: 295–300. [DOI] [PubMed] [Google Scholar]
- Kasai, S., and J. G. Scott, 2001. A house fly gene homologous to the zinc finger proto-oncogene Gfi-1. Biochem. Biophys. Res. Commun. 283: 644–647. [DOI] [PubMed] [Google Scholar]
- Kazanjian, A., D. Wallis, N. Au, R. Nigam, K. J. Venken et al., 2004. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res. 64: 6874–6882. [DOI] [PubMed] [Google Scholar]
- Li, J., and W. X. Li, 2003. Drosophila gain-of-function mutant RTK torso triggers ectopic Dpp and STAT signaling. Genetics 164: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., E. Noll and N. Perrimon, 2000. Identification of autosomal regions involved in Drosophila Raf function. Genetics 156: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen, J. A., S. E. Baker, F. Denhez, M. B. Melnick, D. L. Brower et al., 2001. Nuclear import of activated D-ERK by DIM-7, an importin family member encoded by the gene moleskin. Development 128: 1403–1414. [DOI] [PubMed] [Google Scholar]
- Luschnig, S., B. Moussian, J. Krauss, I. Desjeux, J. Perkovic et al., 2004. An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics 167: 325–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik, M., N. E. Baker and G. M. Rubin, 1990. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 4: 1848–1861. [DOI] [PubMed] [Google Scholar]
- Moroy, T., 2005. The zinc finger transcription factor Growth factor independence 1 (Gfi1). Int. J. Biochem. Cell Biol. 37: 541–546. [DOI] [PubMed] [Google Scholar]
- Neufeld, T. P., A. H. Tang and G. M. Rubin, 1998. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148: 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo, R., L. A. Abbot and H. J. Bellen, 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362. [DOI] [PubMed] [Google Scholar]
- Nolo, R., L. A. Abbott and H. J. Bellen, 2001. Drosophila Lyra mutations are gain-of-function mutations of senseless. Genetics 157: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu, K. S., and G. Mardon, 2004. Genetic control of retinal specification and determination in Drosophila. Int. J. Dev. Biol. 48: 913–924. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., and M. A. Muskavitch, 1993. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev. Biol. 157: 484–496. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., S. S. Huppert and M. A. Muskavitch, 1997. The dynamics of neurogenic signalling underlying bristle development in Drosophila melanogaster. Mech. Dev. 63: 61–74. [DOI] [PubMed] [Google Scholar]
- Paroush, Z., R. L. Finley, Jr., T. Kidd, S. M. Wainwright, P. W. Ingham et al., 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805–815. [DOI] [PubMed] [Google Scholar]
- Perrimon, N., A. Lanjuin, C. Arnold and E. Noll, 1996. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics 144: 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person, R. E., F. Q. Li, Z. Duan, K. F. Benson, J. Wechsler et al., 2003. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 34: 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam, C., R. Geffers, R. Yucel, J. Buer, K. Welte et al., 2005. The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity 22: 717–728. [DOI] [PubMed] [Google Scholar]
- Rebay, I., F. Chen, F. Hsiao, P. A. Kolodziej, B. H. Kuang et al., 2000. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154: 695–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., and A. Ghysen, 1993. The expression and role of a proneural gene, achaete, in the development of the larval nervous system of Drosophila. EMBO J. 12: 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T., M. Zornig, R. Beneke and T. Moroy, 1996. MoMuLV proviral integrations identified by Sup-F selection in tumors from infected myc/pim bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 24: 2528–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T., H. Karsunky, E. Gau, B. Zevnik, H. P. Elsasser et al., 1998. a Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene 17: 2661–2667. [DOI] [PubMed] [Google Scholar]
- Schmidt, T., H. Karsunky, B. Rodel, B. Zevnik, H. Elasser et al., 1998. b Evidence implicating Gfi-1 and Pim-1 in pre-T-cell differentiation steps associated with beta-selection. EMBO J. 17: 5349–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth, F., 2004. Notch signaling activity. Curr. Biol. 14: 129–138. [PubMed] [Google Scholar]
- Shin, M. S., T. N. Fredrickson, J. W. Hartley, T. Suzuki, K. Agaki et al., 2004. High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 64: 4419–4427. [DOI] [PubMed] [Google Scholar]
- Skeath, J. B., and S. B. Carroll, 1992. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development 114: 939–946. [DOI] [PubMed] [Google Scholar]
- Stowers, R. S., and T. L. Schwarz, 1999. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, A. C., and K. Weis, 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2: REVIEWS3008. [DOI] [PMC free article] [PubMed]
- Su, M. A., R. G. Wisotzkey and S. J. Newfeld, 2001. A screen for modifiers of decapentaplegic mutant phenotypes identifies lilliputian, the only member of the fragile-X/Burkitt's lymphoma family of transcription factors in Drosophila melanogaster. Genetics 157: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, A. H., T. P. Neufeld, G. M. Rubin and H. A. Muller, 2001. Transcriptional regulation of cytoskeletal functions and segmentation by a novel maternal pair-rule gene, lilliputian. Development 128: 801–813. [DOI] [PubMed] [Google Scholar]
- Tekotte, H., and I. Davis, 2002. Intracellular mRNA localization: motors move messages. Trends Genet. 18: 636–642. [DOI] [PubMed] [Google Scholar]
- Tio, M., and K. Moses, 1997. The Drosophila TGF alpha homolog Spitz acts in photoreceptor recruitment in the developing retina. Development 124: 343–351. [DOI] [PubMed] [Google Scholar]
- Tio, M., C. Ma and K. Moses, 1994. spitz, a Drosophila homolog of transforming growth factor-alpha, is required in the founding photoreceptor cells of the compound eye facets. Mech. Dev. 48: 13–23. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., and D. F. Ready, 1987. Cell fate in Drosophila ommatidia. Dev. Biol. 123: 264–275. [DOI] [PubMed] [Google Scholar]
- Tsuda, H., H. Jafar-Nejad, A. J. Patel, Y. Sun, H. K. Chen et al., 2005. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 122: 633–644. [DOI] [PubMed] [Google Scholar]
- Vrailas, A. D., D. R. Marenda, S. E. Cook, M. A. Powers, J. A. Lorenzen et al., 2006. smoothened and thickveins regulate Moleskin/Importin 7-mediated MAP kinase signaling in the developing Drosophila eye. Development 133: 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, D., M. J. Hamblen, Y. Zhou, K. J. Venken, A. Schumacher et al., 2003. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130: 221–232. [DOI] [PubMed] [Google Scholar]
- Wang, S. W., B. S. Kim, K. Ding, H. Wang, D. Sun et al., 2001. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15: 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K., 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441–451. [DOI] [PubMed] [Google Scholar]
- Wittwer, F., A. van der Straten, K. Keleman, B. J. Dickson and E. Hafen, 2001. Lilliputian: an AF4/FMR2-related protein that controls cell identity and cell growth. Development 128: 791–800. [DOI] [PubMed] [Google Scholar]
- Yang, Z., K. Ding, L. Pan, M. Deng and L. Gan, 2003. Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol. 264: 240–254. [DOI] [PubMed] [Google Scholar]
- Yucel, R., H. Karsunky, L. Klein-Hitpass and T. Moroy, 2003. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 197: 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H., R. Yucel, C. Kosan, L. Klein-Hitpass and T. Moroy, 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23: 4116–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, R. G., P. R. Hiesinger, T. W. Koh, P. Verstreken, K. L. Schulze et al., 2003. Mapping Drosophila mutations with molecularly defined P element insertions. Proc. Natl. Acad. Sci. USA 100: 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., L. Guo, B. Min, C. J. Watson, J. Hu-Li et al., 2002. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity 16: 733–744. [DOI] [PubMed] [Google Scholar]