Abstract
The peroxisome, sole site of β-oxidation in Saccharomyces cerevisiae, is known to be required for optimal growth in the presence of fatty acid. Screening of the haploid yeast deletion collection identified ∼130 genes, 23 encoding peroxisomal proteins, necessary for normal growth on oleic acid. Oleate slightly enhances growth of wild-type yeast and inhibits growth of all strains identified by the screen. Nonperoxisomal processes, among them chromatin modification by H2AZ, Pol II mediator function, and cell-wall-associated activities, also prevent oleate toxicity. The most oleate-inhibited strains lack Sap190, a putative adaptor for the PP2A-type protein phosphatase Sit4 (which is also required for normal growth on oleate) and Ilm1, a protein of unknown function. Palmitoleate, the other main unsaturated fatty acid of Saccharomyces, fails to inhibit growth of the sap190Δ, sit4Δ, and ilm1Δ strains. Data that suggest that oleate inhibition of the growth of a peroxisomal mutant is due to an increase in plasma membrane porosity are presented. We propose that yeast deficient in peroxisomal and other functions are sensitive to oleate perhaps because of an inability to effectively control the fatty acid composition of membrane phospholipids.
bβ-OXIDATIVE catabolism of fatty acids occurs in peroxisomes of all eukaryotes, yet there is tremendous species variation in other functions carried out by this organelle, also called the microbody, the glycosome, and the glyoxisome (van den Bosch et al. 1992). While β-oxidation occurs in all peroxisomes, only in some species, such as the yeasts, is it a uniquely peroxisomal function (Hiltunen et al. 2003). Animals, by contrast, also perform β-oxidation in the mitochondrion using a separate set of enzymes (Bartlett and Eaton 2004). A variety of elegant methods using human cells, Saccharomyces cerevisiae, and other yeast species have identified 33 Pex proteins, or peroxins, that play a wide range of roles in peroxisome biogenesis and function (Purdue and Lazarow 2001; Eckert and Erdmann 2003; Veenhuis et al. 2003; Moyersoen et al. 2004). Most Pex proteins are highly conserved throughout eukaryotes; human disorders that result from mutations in any one of a number of pex genes have been studied extensively (Eckert and Erdmann 2003). Only 2 of the 27 known Saccharomyces peroxins, Pex3 and Pex19, are required to maintain the organelle (Hohfeld et al. 1991; Gotte et al. 1998; Hettema et al. 2000).
Despite the extensive knowledge of the roles of many of these proteins, the specific biological functions of the peroxisome, as well as the molecular basis of pathologies that result from peroxisomal dysfunction, remain poorly understood. Most pointedly, the cellular role of peroxisomal β-oxidation has not been established. Mitochondrial β-oxidation, found only in animals, is thought to be responsible for catabolism of dietary fatty acid (Bartlett and Eaton 2004). Peroxisomal β-oxidation, found in every eukaryote examined (Kunau et al. 1988), is likely to play a different cellular role. In S. cerevisiae, β-oxidation appears to be the only complete biochemical pathway carried out in the peroxisome (Hiltunen et al. 2003). Such relative simplicity, as well as the unsurpassed genetic utility of this yeast, makes it an attractive system for helping to elucidate the organelle's cellular function. Soon after it was established that Saccharomyces could be used to study the peroxisome (Veenhuis et al. 1987), a genetic screen for mutants that grow slowly in the presence of oleic acid (oleate), an 18-carbon, cis-monounsaturated fatty acid, led to the identification of PEX1, PEX3, and PEX4 (Erdmann et al. 1989). Slow growth was attributed to the inability of yeast with defective peroxisomes to carry out β-oxidation and to therefore be incapable of utilizing fatty acids as a source of carbon.
As part of an effort to identify proteins required for peroxisome inheritance, we sought to identify additional proteins that are required for its maintenance. A set of yeast deletion strains constructed by a consortium of laboratories (Winzeler et al. 1999) has been screened extensively for a broad range of phenotypes. In this work, haploid deletion strains that grow poorly in the presence of oleate were identified. Most Pex proteins previously shown to be required for optimal growth in the presence of oleate were identified by the screen, yet the screen also identified genes whose products have nonperoxisomal functions. Contrary to previous interpretations, strains identified in the screen grew poorly in the presence of oleate because of the inhibition of their growth by oleate rather than because they were incapable of using oleate as a carbon source. We present evidence suggesting that the toxicity of oleate to pex and other mutants may be the result of changes in the plasma membrane. These findings implicate the peroxisome, an organelle whose biological function has remained obscure, in membrane function.
MATERIALS AND METHODS
Yeast media, strains, and growth:
Synthetic media with tergitol and yeast extract (STY) contains 0.67% yeast nitrogen base (Difco 291940); 0.05% yeast extract (Difco 212750); 1% tergitol [Sigma (St. Louis) NP40S]; a mixture of amino acids, uracil, and adenine at one-tenth the concentration used previously (Sherman 1991); 0.01% ampicillin and 0.01% G418 (both ampicillin and G418 are omitted from liquid media). Oleate and palmitoleate from Nu-Chek Prep were dissolved in warm 10% tergitol before being added to the other components. Plates contained 2% bactoagar (Difco 214010). Nourseothricin (clonNAT) was provided by Werner BioAgents.
All yeast strains were derived from BY4741 and BY4742 (Brachmann et al. 1998). The oleate screen was carried out using the set of MATα haploid deletion strains (Open Biosystems YSC1049) maintained in the supplier's 96-well format. Frozen stocks were thawed and inoculated into 0.1 ml of YPD + G418 medium in 50, 96-well dishes using a pinning device mounted on the Beckman Biomek 2000, also used for all further deletion set manipulations. After growth for 2 days at 30° without shaking, cultures were diluted ∼100-fold in water and 1μl was pinned onto rectangular agar plates containing STY + oleate (0.1%) and STY + glycerol (3%) medium for growth at 25°. STY + glycerol plates were grown for 4 days and stored at 4° until sufficient growth of the STY + oleate plates (13 days) allowed growth data to be visually recorded. Strain data and growth properties were managed using Filemaker software. To combine a gene deletion with Pex11-GFP, the MATα deletion strain from the commercial set was mated to the Pex11-GFP fusion derivative of BY4741 constructed by the Yeast GFP Fusion Localization Database (Huh et al. 2003). The diploid was then sporulated, and His+ G418R spore colonies were identified from tetrads in which these two markers each segregated 2:2. Flow cytometric analysis of cell cycle distribution was performed as described (Foss 2001).
Microscopy, Western blotting, and Sytox green staining:
Fluorescence microscopy was carried out using a Zeiss Axiovert 200M with FITC filters to detect GFP. Western blots of yeast protein prepared (Kornitzer 2002) from midlog cultures were probed with mouse monoclonal antibody to GFP (Roche 1814460). The secondary anti-mouse monoclonal antibody, conjugated to horseradish peroxidase, was reacted with an acridinium ester [Amersham (Buckinghamshire, UK) RPN2132], and the product was detected on the Amersham Storm 860 phosphoimager. Blots were then probed with rabbit polyclonal antibody to yeast actin (kindly provided by Alex Merz), further probed with anti-rabbit monoclonal antibody conjugated to horseradish peroxidase, and reacted and visualized as above.
To assay the influence of oleate on cell permeability, yeast grown in STY liquid medium with or without 0.1% oleate to midlog were resuspended in 0.5 μm Sytox green (BioVision K201) as suggested by the supplier for 10 min at room temperature and visualized immediately on the Zeiss microscope with FITC filters.
RESULTS
A screen for proteins that permit normal growth in the presence of oleate:
Two previous studies led to the isolation of mutants of S. cerevisiae that grow slowly in the presence of oleate (Erdmann et al. 1989; Marzioch et al. 1994). Such growth impairment was attributed to the inability to utilize oleate as a carbon source. Six of these mutants fell into four complementation groups, which eventually came to be known as PEX1 (Erdmann et al. 1991), PEX3 (Hohfeld et al. 1991), PEX4 (Wiebel and Kunau 1992), and PEX7 (Marzioch et al. 1994). To carry out a screen of the yeast deletion strains using this strategy, a growth medium, STY, was devised (based on the variety of oleate media formulations used since 1989) that in our study maximized the growth difference between Pex+ and pexΔ strains on STY + oleate relative to growth on STY + glycerol. The rationale for using glycerol, a nonfermentable carbon source, as a control in the current and previous screens was that acetyl-CoA produced by β-oxidation can serve as an energy source only in respiratory-competent strains, i.e., those capable of growing on glycerol. Respiratory-deficient mutants, incapable of growth on both glycerol and oleate, could thereby be distinguished from those that grow slowly only on oleate.
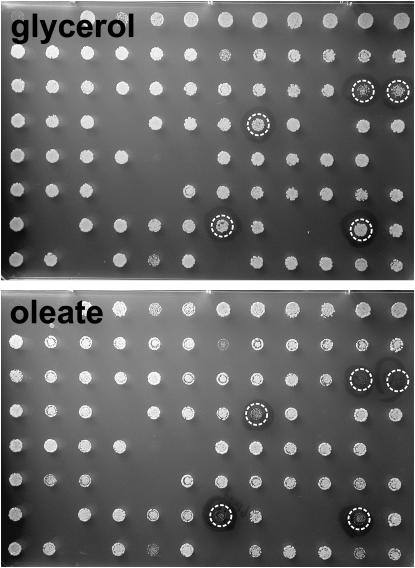
The haploid (MATα) deletion collection (4773 strains) was grown on STY + oleate and STY + glycerol to give 50 pairs of plates, one of which is shown in Figure 1. A total of 136 strains deleted in the genes listed in Table 1 were identified. Twenty-six of these compose 13 pairs of chromosomally adjacent genes. Since five of the six dubious ORFs (predicted to encode proteins that have no orthologs in other fungi) identified by the screen were adjacent in chromosomal location to bona fide genes listed in Table 1, it is likely that sequence in these dubious ORFs affects expression of adjacent oleate-sensitive deletions. These six genes—SPF1, PEX14, OPT1, KNS1, NCS1, and GAL11—were thus in effect identified twice. For the seven other pairs of chromosomally adjacent genes, the identities of the true hits are ambiguous. As has also been observed by others (J. Aitchison and P. Lazarow, personal communications), we observed variability in the oleate phenotype when retesting a number of the strains identified in the screen. This variability is undoubtedly due to changes in cellular physiology and possibly to subtle changes in growth conditions. We are currently attempting to determine the cause of this variability. Nevertheless, retesting the MATα deletion strains identified in the screen confirmed an oleate phenotype for each strain listed in Table 1. In addition, the MATa versions of most of the strains identified in the screen were also tested and found to correlate well with the phenotypes listed in Table 1.
Figure 1.—
A genomewide screen for oleate sensitivity. One of 50 pairs of plates that were compared to identify deletion strains that exhibit the oleate growth phenotype. The five circled strains are med2Δ, ptc1Δ, rpn4Δ, pex19Δ, and cbs1Δ. Deletion strains were applied to all but 2 of the 96 positions of this plate pair; the absence of growth at the other positions is due to the inability of these strains to grow on either oleate or glycerol. For example, the cox9Δ strain, known to be respiratory deficent, is located in the position second from the bottom, fourth from the right.
TABLE 1.
Deletion strains identified in the oleate screen
| ORF | Gene | Phenotype | Functional category | Adjacent hit |
|---|---|---|---|---|
| YJR105W | ADO1 | ++ | Adenosine kinase | |
| YNL197C | WHI3 | +++ | Cell cycle | |
| YCR077C | PAT1 | +++ | Cell wall | |
| YDR017C | KCS1 | +++ | Cell wall | |
| YDR162C | NBP2 | +++ | Cell wall | |
| YHL011C | PRS3 | +++ | Cell wall | |
| YKL212W | SAC1 | +++ | Cell wall | |
| YLR438W | CAR2 | ++ | Cell wall | |
| YMR238W | DFG5 | +++ | Cell wall | |
| YOL081W | IRA2 | +++ | Cell wall | |
| YOR360C | PDE2 | +++ | Cell wall | |
| YNL215W | IES2 | +++ | Chromatin remodeling | PEX17 |
| YLR357W | RSC2 | ++ | Chromatin remodeling | |
| YOR038C | HIR2 | ++ | Chromatin remodeling | |
| YOL004W | SIN3 | ++ | Chromatin remodeling (HDAC) | |
| YOL012C | HTZ1 | +++ | Chromatin remodeling (H2A variant) | |
| YDR334W | SWR1 | + | Chromatin remodeling (SWR-C) | |
| YLR085C | ARP6 | +++ | Chromatin remodeling (SWR-C) | |
| YML041C | VPS71 | +++ | Chromatin remodeling (SWR-C) | |
| YNL107W | YAF9 | +++ | Chromatin remodeling (SWR-C) | |
| YEL033W | ++ | Dubious ORF | SPF1 | |
| YGL152C | +++ | Dubious ORF | PEX14 | |
| YJL211C | +++ | Dubious ORF | OPT1 | |
| YLL020C | ++ | Dubious ORF | KNS1 | |
| YNL120C | ++ | Dubious ORF | NCS1 | |
| YOL050C | +++ | Dubious ORF | GAL11 | |
| YPR087W | ++ | Dubious ORF | ||
| YJL101C | GSH1 | +++ | Glutathione synthesis | |
| YOR080W | DIA2 | + | Invasive growth | |
| YCR034W | FEN1 | +++ | Lipid biosynthesis | |
| YKR067W | GPT2 | +++ | Lipid biosynthesis | |
| YLR362W | STE11 | ++ | Mating | |
| YOR297C | TIM18 | ++ | Mitochondrial protein import | |
| YAL010C | MDM10 | ++ | Mitochondrial inheritance | SPO7 |
| YLR368W | MDM30 | +++ | Mitochondrial inheritance | |
| YOL076W | MDM20 | +++ | Mitochondrial inheritance | |
| YMR060C | SAM37 | +++ | Mitochondrial (Mdm10 cofactor) | |
| YPR131C | NAT3 | ++ | Mitochondrial (Mdm20 cofactor) | |
| YML120C | NDI1 | +++ | Mitochondrial (oxidoreductase) | |
| YMR145C | NDE1 | +++ | Mitochondrial NADH dehydrase | |
| YDL198C | GGC1 | ++ | Mitochondrial transport | |
| YPL060W | LPE10 | +++ | Mitochondrial transport | |
| YPL270W | MDL2 | ++ | Mitochondrial transport | |
| YHR191C | CTF8 | ++ | Mitosis | |
| YOR073W | SGO1 | +++ | Mitosis | |
| YJR074W | MOG1 | +++ | Nuclear protein import | |
| YDR264C | AKR1 | +++ | Palmitoyltransferase | PEX10 |
| YAL051W | OAF1 | +++ | Peroxisome | |
| YAL055W | PEX22 | +++ | Peroxisome | |
| YDL065C | PEX19 | +++ | Peroxisome | |
| YDR142C | PEX7 | +++ | Peroxisome | |
| YDR244W | PEX5 | +++ | Peroxisome | |
| YDR265W | PEX10 | +++ | Peroxisome | AKR1 |
| YDR329C | PEX3 | +++ | Peroxisome | |
| YEL031W | SPF1 | +++ | Peroxisome | YEL033W |
| YGL153W | PEX14 | +++ | Peroxisome | YGL152C |
| YGL205W | POX1 | + | Peroxisome | |
| YGR077C | PEX8 | +++ | Peroxisome | PAC10 |
| YGR133W | PEX4 | +++ | Peroxisome | PHB1 |
| YIL160C | POT1 | +++ | Peroxisome | |
| YIR004W | DJP1 | + | Peroxisome | |
| YKL197C | PEX1 | +++ | Peroxisome | |
| YKR009C | FOX2 | + | Peroxisome | |
| YLR191W | PEX13 | +++ | Peroxisome | |
| YMR026C | PEX12 | +++ | Peroxisome | |
| YNL214W | PEX17 | +++ | Peroxisome | IES2 |
| YNL329C | PEX6 | +++ | Peroxisome | |
| YOL044W | PEX15 | +++ | Peroxisome | |
| YPL112C | PEX25 | + | Peroxisome | |
| YBR106W | PHO88 | +++ | Phosphate transport | |
| YNL248C | RPA49 | ++ | Pol I subunit | |
| YBL025W | RRN10 | +++ | Pol I transcription | |
| YGR104C | SRB5 | ++ | Pol II mediator (head) | |
| YHR041C | SRB2 | +++ | Pol II mediator (head) | |
| YDL005C | MED2 | +++ | Pol II mediator (tail) | PTC1 |
| YGL025C | PGD1 | +++ | Pol II mediator (tail) | |
| YOL051W | GAL11 | +++ | Pol II mediator (tail) | YOL050C |
| YBL058W | SHP1 | +++ | Protein degradation | |
| YDL020C | RPN4 | ++ | Protein degradation | |
| YGR135W | PRE9 | +++ | Protein degradation | |
| YHR111W | UBA4 | +++ | Protein degradation | |
| YIL008W | URM1 | +++ | Protein degradation | |
| YLR024C | UBR2 | +++ | Protein degradation | |
| YNL119W | NCS2 | ++ | Protein degradation | YNL120C |
| YDL090C | RAM1 | +++ | Protein farnesyltransferase | |
| YEL003W | GIM4 | ++ | Protein folding | |
| YGR078C | PAC10 | ++ | Protein folding | PEX8 |
| YHR064C | SSZ1 | +++ | Protein folding | |
| YOR265W | RBL2 | ++ | Protein folding | |
| YLL019C | KNS1 | ++ | Protein kinase | YLL020C |
| YNL298W | CLA4 | + | Protein kinase | |
| YDL006W | PTC1 | +++ | Protein phosphatase | MED2 |
| YER054C | GIP2 | +++ | Protein phosphatase | |
| YBL027W | RPL19B | +++ | Ribosomal protein | |
| YBL072C | RPS8A | +++ | Ribosomal protein | |
| YDR025W | RPS11A | +++ | Ribosomal protein | |
| YDR418W | RPL12B | +++ | Ribosomal protein | |
| YHL033C | RPL8A | ++ | Ribosomal protein | |
| YIL052C | RPL34B | +++ | Ribosomal protein | |
| YGR070W | ROM1 | + | Signal transduction | |
| YPL213W | LEA1 | +++ | Splicing | |
| YJR104C | SOD1 | ++ | Superoxide dismutase | |
| YBR084W | MIS1 | ++ | Tetrahydrofolate metabolism | SPT7 |
| YLR423C | ATG17 | ++ | Trafficking | |
| YPR032W | SRO7 | ++ | Trafficking | |
| YPR139C | VPS66 | +++ | Trafficking | |
| YPR173C | VPS4 | ++ | Trafficking | |
| YIL128W | MET18 | +++ | Transcription | |
| YDR216W | ADR1 | ++ | Transcription | |
| YMR179W | SPT21 | ++ | Transcription (histones) | |
| YBR081C | SPT7 | ++ | Transcription (SAGA) | MIS1 |
| YJL140W | RPB4 | +++ | Transcription (stress tolerance) | |
| YJL056C | ZAP1 | +++ | Transcription (phospholipid synthesis) | |
| YHR206W | SKN7 | + | Transcription/oxidative stress | |
| YDL069C | CBS1 | +++ | Translation | |
| YPR163C | TIF3 | ++ | Translation | |
| YJL212C | OPT1 | +++ | Transporter | YJL211C |
| YAL009W | SPO7 | +++ | Unclear | MDM10 |
| YAR014C | BUD14 | ++ | Unclear | |
| YCR047C | BUD23 | +++ | Unclear | |
| YCR063W | BUD31 | + | Unclear | |
| YFR035C | ++ | Unclear | ||
| YGL127C | SOH1 | +++ | Unclear | |
| YGL173C | KEM1 | +++ | Unclear | |
| YGR132C | PHB1 | +++ | Unclear | PEX4 |
| YHR100C | +++ | Unclear | ||
| YJL184W | GON7 | +++ | Unclear | |
| YJR118C | ILM1 | +++ | Unclear | |
| YKR028W | SAP190 | +++ | Unclear | |
| YLR021W | ++ | Unclear | ||
| YLR114C | AVL9 | + | Unclear | |
| YMR014W | BUD22 | + | Unclear | |
| YMR032W | HOF1 | + | Unclear | |
| YMR100W | MUB1 | +++ | Unclear | |
| YNR018W | + | Unclear | ||
| YOL072W | THP1 | +++ | Unclear |
A rating of the ratio of growth on STY + glycerol relative to growth on STY + oleate of strains deleted in the indicated ORFs is indicated by “+” (smallest oleate effect) to “+++” (largest oleate effect). “Adjacent hit” refers to the appearance in this table of a gene that is adjacent in chromosomal location to any other gene listed in the table. A line under the ORF indicates that the effect of its deletion on the localization of Pex11 was examined (see text).
The screen was judged to be successful because it identified 23 peroxisomal proteins, including 16 Pex proteins that have been reported to be required for optimal growth on oleate. A number of other functional categories are also overrepresented relative to their prominence in the yeast proteome. The Pol II mediator complex is one such category: five of its eight components (Bjorklund and Gustafsson 2005) were identified (one other of its components is required for respiration and another is essential). Another functional category involves H2AZ, the alternative histone whose substitution into chromatin locally influences transcription (Kobor et al. 2004). Four of nine components of SWR-C (four other SWR-C proteins are essential), the complex that substitutes histone H2AZ for H2A in chromatin, were identified, as was H2AZ itself.
Oleate inhibits the growth of peroxisomal mutants and other oleate-sensitive strains:
In isolating Saccharomyces pex mutants, Erdmann et al. (1989) attributed their poor growth in the presence of oleate to an inability to use fatty acid as a carbon source. On the contrary, we find that oleate inhibited the growth of pex mutants. Figure 2 compares the growth of pex6Δ and Pex+ strains on STY containing glycerol, no added carbon source, or oleate. In contrast to the results of Erdmann et al. (1989), which showed wild-type Saccharomyces to be unable to grow unless both 0.05% yeast extract and oleate are present on plates, we found growth to occur without the addition of any carbon source as long as 0.05% yeast extract was included. Tergitol, the detergent in STY medium added for fatty acid solubilization, had no effect on growth (data not shown). Note the slight enhancement in colony size of both strains by the addition of glycerol and of the Pex+ strain by the addition of oleate. Growth of the pex6Δ strain, however, is severely inhibited by oleate addition. While there is a slight augmentation in growth of the Pex+ strain by the addition of oleate, the poorer growth on oleate of the strains identified in our screen is mainly due to the inhibition of their growth by oleate rather than to the failure of their growth to be enhanced by oleate. By comparing the growth of all positives obtained in the screen on STY, STY + glycerol, and STY + oleate, oleate inhibition was found to account for the oleate phenotype of all but one of the strains (the exception, pck1Δ, was capable of growth on STY + glycerol but not on STY and therefore does not appear in Table 1). Because the mutants are oleate inhibited rather than unable to be fed by oleate, the screen in retrospect could have used STY rather than STY + glycerol as a control.
Figure 2.—
Oleate inhibits growth of the pex6Δ strain. Ten-fold dilutions of strain YD14044 (Pex+) and YD11115 (pex6Δ) were spotted onto STY medium supplemented with 3% glycerol, nothing, or 0.1% oleate and grown for 1 week at 23°.
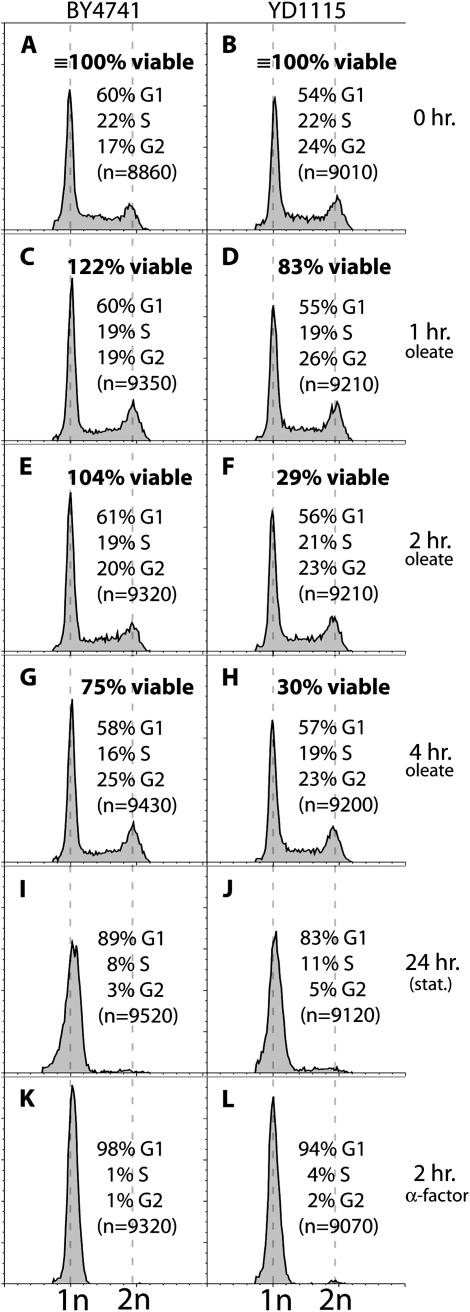
STY medium was used to conduct the screen since this minimal medium allows the detection of even weakly oleate-inhibited strains. The possibility therefore arose that the strains appearing to be less strongly inhibited by oleate were exhibiting this phenotype because of starvation. We addressed this possibility with a series of control experiments that examined the effect of oleate on the cell cycle distribution of cells from Pex+ and pex6Δ cultures incubated in STY + oleate. Starvation of yeast is well established to cause cells to accumulate in G1 (Johnston et al. 1977). However, at 1, 2, and 4 hr after the shift from glucose- to oleate-containing STY medium, no shift in cell cycle profile of either the Pex+ (Figure 3, A, C, E, and G) or the pex6Δ (Figure 3, B, D, F, and H) strains was detectable. While the viability of the Pex+ strain is largely unaffected by this 4-hr oleate treatment, the progressive decrease in viability of the pex6Δ cells during this interval demonstrates the expected toxic effect of oleate on this strain. An accumulation of G1 cells can indeed be observed in stationary cultures of both strains (Figure 3, I and J) or when they are arrested with α-factor (Figure 3, K and L). Since the pex6Δ strain was not starved while oleate was exerting a toxic effect, there is no indication that starvation contributes to the toxic effect of oleate on the pex6Δ strain. It seems reasonable to extend this conclusion to the other strains that exhibit oleate sensitivity in STY medium but not in rich medium.
Figure 3.—
The oleate sensitivity of the pex6Δ strain is not due to starvation on STY medium. (A–H) BY4741 and YD1115 (pex6Δ) were grown to early log phase in STY + 2% glucose and washed with water. Cells were resuspended in STY + 0.1% oleate to their previous density, incubated at 30° for the indicated times, and then analyzed by flow cytometry and plated to score for viable cells. (I–L) The two strains were grown in YPD + 1% tergitol to early log phase, supplemented with 30 μm α-factor and after 2 hr subject to flow cytometry (K and L) or allowed to grow unarrested for 24 hr to stationary phase and then analyzed by flow cytometry (I and J).
Peroxisome structure and function in oleate-sensitive strains:
The original goal of the screen was the identification of proteins that, like Pex3 and Pex19, are responsible for the maintenance of the peroxisome. Seventy of the deletions identified by the screen (underlined in Table 1) were therefore tested for their ability to cause the loss of peroxisomes. Versions of these 70 strains were constructed that express Pex11-GFP in place of Pex11 (this fusion does not influence oleate sensitivity; data not shown). Of these 70 strains, 68 showed punctate GFP fluorescence; the pex3Δ and pex19Δ strains were the only two strains that failed to show punctate GFP fluorescence (data not shown). Thus, the oleate screen identified no additional proteins that are required for maintaining the peroxisome.
While many genes identified by the screen encode peroxisomal components, others have no known connection to the organelle. The relationship of several of these gene products to the peroxisome was addressed by determining whether deletion influenced oleate-induced enhancement of Pex11-GFP expression. Numerous previous studies have shown oleate to cause a substantial induction in the size and/or number of Saccharomyces peroxisomes (Veenhuis et al. 2003). Oleate-induced expression of PEX11 and the other PEX genes depends on Oaf1 and Pip2, transcription factors that bind as a heterodimer to the oleate-responsive element (Luo et al. 1996; Rottensteiner et al. 1996; Karpichev and Small 1998). Adr1, the transcription factor first identified as the inducer of yeast alcohol dehydrogenase (Ciriacy 1975), is also important for PEX gene induction (Simon et al. 1991; Gurvitz et al. 2001).
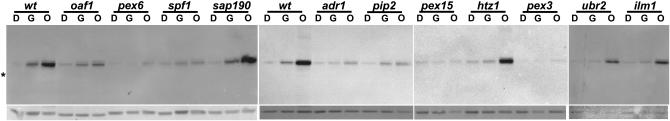
To determine the change in Pex11-GFP level in a strain grown under various conditions, total protein was analyzed on GFP-probed Western blots. Figure 4 shows the results from 12 such strains, grown first in glucose-containing liquid medium, after a subsequent shift to glycerol, and after shifting from glycerol to oleate. The wild-type strain shows a substantial induction in Pex11-GFP expression upon shift to glycerol; a subsequent shift to oleate shows an additional substantial induction. As expected, deletion of either OAF1 or PIP2 permits the initial glycerol induction but blocks the oleate induction, and deletion of ADR1 blocks the initial glycerol induction but permits the subsequent oleate induction. Deletion of PEX3, PEX6, or PEX15, as anticipated, allows little if any Pex11-GFP to accumulate even after oleate treatment. Spf1, identified in the oleate screen, is reported to be involved in ER function, calcium homeostasis (Cronin et al. 2002), and the orientation of protein insertion into membranes (Tipper and Harley 2002). It was also shown by mass spectrometry to be associated with the peroxisome (Marelli et al. 2004). Figure 4 shows that neither glycerol nor oleate cause Pex11-GFP levels to increase in a spf1Δ background. Spf1 is thus grouped into the “peroxisome” functional category of Table 1.
Figure 4.—
Induction of Pex11-GFP by glycerol and oleate in oleate-sensitive deletion strains. Protein extracts of strains containing Pex11-GFP and deleted in the indicated genes were made from portions of midlog cultures after growth in STY + 2% glucose (D). The remaining cells were washed and transferred to STY + 3% glycerol. After 24 hr a portion of these cells were harvested (G). The remaining cells were then transferred to STY + 0.1% oleate and harvested after 16 hr (O). Western blots were probed for GFP (top) and subsequently probed for actin (bottom). The asterisk indicates the position of the actin band.
In contrast, other deletions show glycerol and oleate induction of Pex11-GFP that is indistinguishable from wild type. Figure 4 shows this to be the case for sap190Δ and ilm1Δ, genes that encode proteins of unknown function. akr1Δ, dfg5Δ, pox1Δ, ptc1Δ, and sac1Δ strains are also unaffected in Pex11-GFP induction (data not shown). These results imply that, although these eight proteins participate in preventing oleate inhibition of growth, they are unlikely to have a role in the maintenance or induction of the peroxisome. Indeed, aside from Pox1, which is a peroxisomal protein but which has no known role in organelle maintenance, these proteins have no known connection to the peroxisome. An intermediate degree of induction is seen in Figure 4 for ubr2Δ, implicating Ubr2 perhaps tangentially in peroxisome function. In summary, these Pex11-GFP induction data confirm, as shown in Table 1, that both peroxisomal and nonperoxisomal cellular processes are required to protect S. cerevisiae from the toxic effects of oleate.
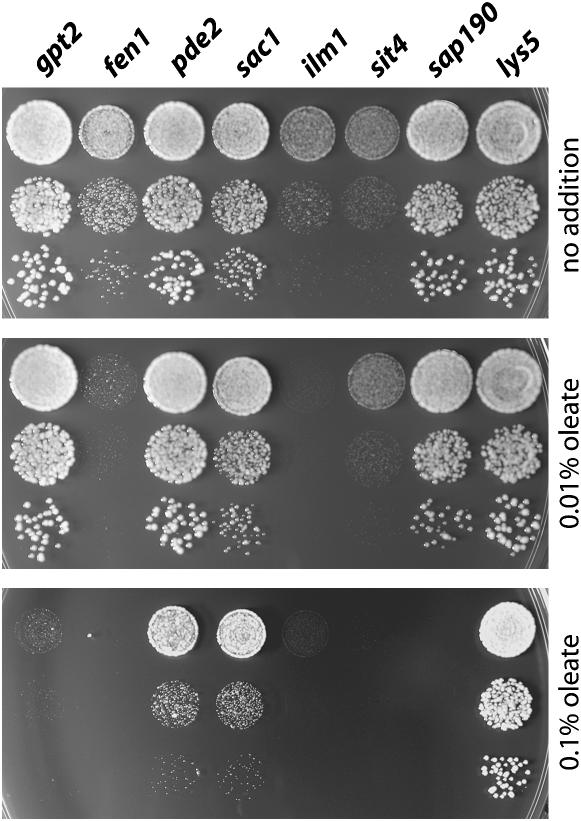
The strains identified as being most inhibited by oleate also showed inhibition when rich, glucose-containing medium (YPD) instead of STY medium was used (Figure 5). Gpt2 and Fen1 have roles in lipid metabolism but are unlikely to be related to the peroxisome. Gpt2 is one of two acyl transferases that produce lysophosphatidic acid, a phospholipid precursor (Zheng and Zou 2001; Zaremberg and McMaster 2002; Sorger and Daum 2003). Its deletion causes substantial growth inhibition by oleate. In contrast, a strain lacking Sct1, the other glycerol-3-phosphate acyl transferase, fails to show oleate sensitivity (data not shown). Fen1 is one of three yeast fatty acyl elongases that lengthen the C14, C16, and C18 fatty acyl CoA esters that are produced by the fatty acyl synthetase complex (Oh et al. 1997). Figure 5 shows the fen1Δ strain to be among the most strongly inhibited by oleate, yet strains deleted in genes that encode either of the other elongases, Elo1 (Toke and Martin 1996) and Sur4 (Oh et al. 1997), were rechecked and showed no oleate phenotype (data not shown).
Figure 5.—
Oleate inhibition of growth on YPD medium. The indicated MATα strains from the deletion collection were suspended in water at 10-fold dilutions, spotted onto YPD + 1% tergitol plates containing the indicated amounts of oleate, and grown at 30° for 40 hr. The lys5Δ strain served as a wild-type control.
Proteins that are implicated in cell-wall function are highly represented among the oleate-inhibited deletion strains. Data for two of these, pde2Δ and sac1Δ, are also presented in Figure 5. Deletion of PDE2, which encodes the yeast high-affinity cAMP phosphodiesterase, has major effects on cell-wall integrity (Jones et al. 2003). Sac1, one of several yeast phosphoinositide phosphatases, influences the cell wall by controlling chitin synthase trafficking (Schorr et al. 2001). As indicated by the following examples, there is a substantial concurrence among proteins found from previous studies to influence the cell wall and those reported here that prevent oleate inhibition: mutants in PEX6/PAS8 and FOX2 have altered sensitivity to calcofluor white, a gauge of cell-wall strength (Lussier et al. 1997); pex12Δ causes resistance to killer toxin, another measure of cell-wall integrity (Page et al. 2003); and FEN1, the elongase discussed above, was first described as GNS1, mutations in which cause severe defects in synthesis of 1,3-β-glucan, a cell-wall component (el-Sherbeini and Clemas 1995). The connection between oleate sensitivity and the cell wall is discussed in more detail in the discussion. As is also shown in Figure 5, the protein required for resistance to the lowest level of oleate (0.01% in YPD) is Ilm1, a 203-residue protein of unknown function predicted to contain four trans-membrane domains. The sensitivity of a number of deletion strains to oleate even in the presence of 2% glucose further supports our conclusion that the reason for slow growth of mutants in the presence of oleate is growth inhibition rather than the inability to consume oleate as a carbon and energy source.
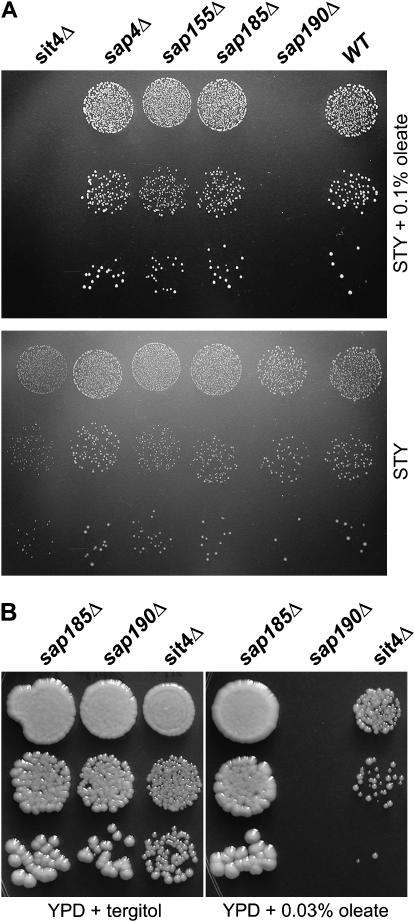
The sap190Δ and ilm1Δ strains are highly sensitive to oleate yet insensitive to palmitoleate:
The sap190Δ strain was also particularly sensitive to oleate; Figure 6B shows it to be inhibited by 0.03% oleate on YPD. Sap190 was first identified as one of two large proteins that co-immunoprecipitate in a cell-cycle-specific manner with Sit4 (Sutton et al. 1991), one of five yeast PP2A-like protein phosphatases (Duvel and Broach 2004). Sap190 and Sap155, the other Sit4-associated protein that was initially identified, were subsequently shown by a combination of homology, immunoprecipitation, and genetic criteria, to be two of three (possibly four) Sap proteins of Saccharomyces. Additional genetic evidence from that study suggested that Sap4, Sap155, Sap185, and Sap190 all have distinct but uncharacterized functions related in some way to Sit4 (Luke et al. 1996). Strains lacking SAP4, SAP155, or SAP185 were not identified in the oleate screen and their reexamination confirmed them all to be oleate insensitive (Figure 6A). As expected on the basis of the relationship of Sap190 and Sit4, however, deletion of SIT4 did indeed cause sensitivity to oleate (Figures 5 and 6). Our screen failed to identify the sit4Δ strain probably because of its slow growth even in the absence of oleate.
Figure 6.—
Oleate inhibition of sap190Δ and related strains. The indicated MATα strains from the deletion collection were suspended in water at 10-fold dilutions, spotted on the indicated plates, and grown for (A) 7 days at 30° and (B) 2 days at 30°.
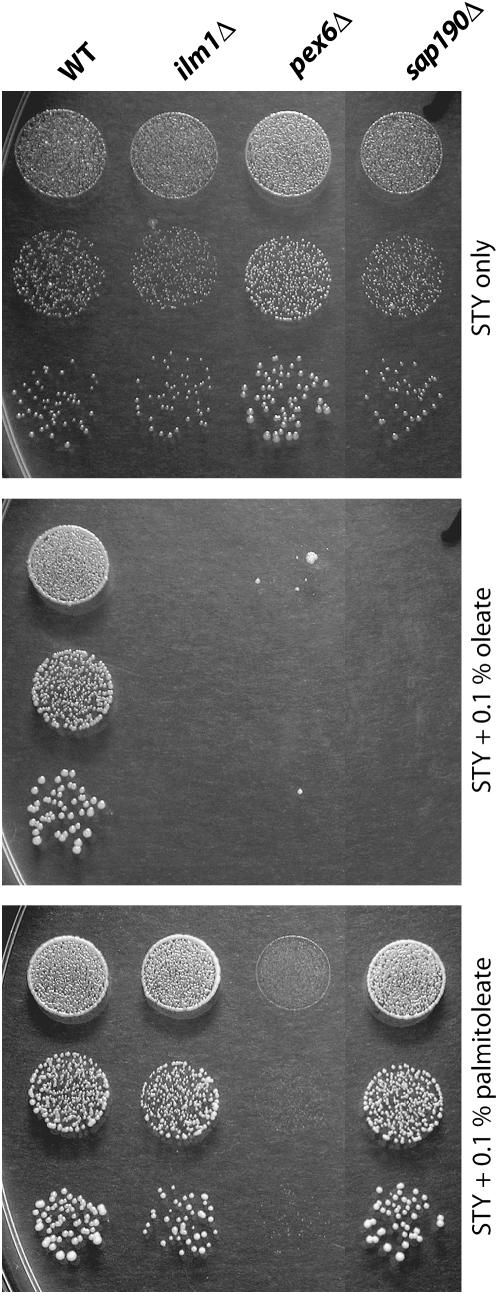
In addition to oleate, the other main unsaturated fatty acid found in S. cerevisiae is palmitoleate (Daum et al. 1999), which is two carbons shorter but otherwise identical to oleate. A comparison of the ability of the two fatty acids to inhibit growth of several strains is shown in Figure 7. Palmitoleate, instead of severely inhibiting growth, promotes the same slight stimulation of growth of the sap190Δ strain as seen when the wild-type strain is supplemented with oleate. The ilm1Δ strain is also selectively inhibited by oleate but not by palmitoleate. Growth of the pex6Δ strain, in contrast, is inhibited by palmitoleate almost as much as by oleate. The specificity with which fatty acids inhibit growth of the sap190Δ and ilm1Δ strains reinforces our assumption that oleate inhibition is a physiologically relevant phenomenon rather than a more generalized detergent-like effect.
Figure 7.—
Oleate and palmitoleate differ in ability to inhibit growth. The indicated MATα strains from the deletion collection were suspended in water at 10-fold dilutions, spotted on the indicated plates, and grown for 7 days at 30°.
Mutations augment the ability of oleate to permeabilize the plasma membrane:
What can account for the toxicity of oleate to strains in which peroxisomal and other functions are impaired? We reasoned that addition of oleate to these mutant strains might cause membrane changes that wild-type strains are capable of resisting. The double bond in oleate's acyl chain introduces a kink that disrupts acyl chain packing in the lipid bilayer. An increase in oleate incorporation into phospholipids therefore can increase membrane fluidity (Hazel 1995). Too great an increase in fluidity compromises membrane integrity which, at least for the plasma membrane, has lethal consequences. One possible effect of removal of a protein that permits yeast to grow normally in the presence of oleate could be the impairment of the cell's ability to properly control the fatty acid content of its membrane phospholipid. Addition of unsaturated fatty acid could therefore lead to an increase in porosity of the plasma membrane.
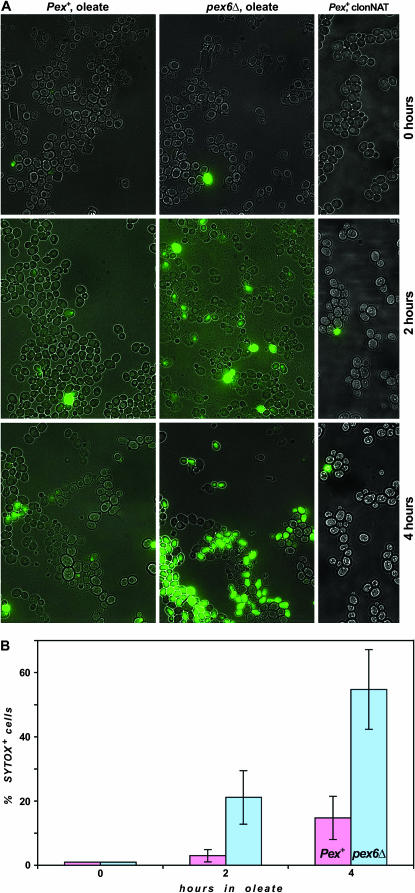
The effect of oleate on the integrity of the plasma membrane was tested by examining the ability of Sytox green, a vital dye, to enter cells. The fluorescence enhancement of Sytox upon interaction with nucleic acid has previously been used to probe membrane permeability of Aspergillus (Theis et al. 2003), Neurospora, and Saccharomyces (Thevissen et al. 1999). Figure 8A shows Sytox-stained Pex+ and pex6Δ strains after 0, 2, and 4 hr of exposure to oleate. Prior to oleate treatment, Sytox entered a similar small fraction (<2%) of cells of the two strains. With increased time of exposure to oleate, a significantly greater fraction of pex6Δ cells than of Pex+ cells were capable of taking up the dye. Quantitation of these results in Figure 8B shows that by 4 hr of oleate treatment, three- to fourfold more pex6Δ cells than Pex+ cells were Sytox positive.
Figure 8.—
Oleate increases the ability of Sytox to enter a peroxisome-deficient strain. (A) Pex+ (BY4741) and pex6Δ (YD1115) cultures were grown in STY + 2% glucose to midlog and then either pelleted and resuspended in STY + oleate (0.1%) or supplemented with clonNAT (0.1 mg/ml). Samples taken at 0, 2, and 4 hr were stained with Sytox and fluorescence microscopic images were acquired. (B) Quantitation of the percentage of Sytox-positive Pex+ and pex6Δ at the three indicated times of oleate treatment. The bar heights are the mean values from five independent experiments and standard deviations are also provided.
Exposure to oleate allows entry of Sytox into cells either because oleate directly influences processes at the plasma membrane to allow dye to enter the cell or because oleate kills the cells by some other means after which the dead cells become permeable to Sytox. To distinguish between these two possibilities, yeast were killed by poisoning with clonNAT, a drug that is known to kill by mistranslation (Haupt et al. 1978). Although <5% of either Pex+ or pex6Δ cells were viable after 2 hr of clonNAT treatment, there was no increase in Sytox-positive cells of either the Pex+ (Figure 8A) or the pex6Δ (data not shown) strain even after 4 hr of clonNAT treatment. Thus oleate-induced but not clonNAT-induced cell death allowed entry of Sytox into cells within the time course of this experiment. In addition to PEX6, we also found that deletion of SAP190, ILM1, or PEX19 led to increases in the fraction of Sytox-positive cells comparable to those seen for the pex6Δ strain in Figure 8 (data not shown).
DISCUSSION
We have identified genes whose products enable optimal growth of Saccharomyces in the presence of oleate. This strategy was used by Erdmann et al. (1989) because of the peroxisomal location of β-oxidation: impairment of peroxisome function could block fatty acid catabolism, thereby preventing oleate from serving as a carbon and energy source. Our results demonstrate, however, that the growth impairment of the yeast deletion strains isolated in our screen is mainly caused by the inhibition of their growth by oleate not by their inability to utilize oleate. Erdmann et al. (1989) reported that wild-type yeast are completely incapable of growth in the absence of oleate or any other added carbon source. It was therefore not possible, presumably because of differences in either strain background or media components in the two studies, for the earlier study to observe oleate inhibition. In addition to inhibition of the growth of mutants, our data also show that oleate slightly stimulates growth of wild-type yeast. Oleate thus appears to be a carbon/energy source preferable to the unknown component(s) of STY medium and/or agar that permits growth in the absence of any added carbon source.
This study had its origins in our search for additional proteins that are required for the maintenance of the peroxisome. At the project's inception, there was evidence that the peroxisome might be an autonomous organelle, i.e., that a new peroxisome could form only from a preexisting peroxisome that would serve as a structural template. The mitochondrion, for example, is undoubtedly such an autonomous organelle (Lockshon 2002), yet, unlike the peroxisome, it performs at least one essential function (Kispal et al. 2005) and must therefore continue to be maintained. Pex3 and Pex19 were known to be required for the maintenance of the peroxisome (Hohfeld et al. 1991; Gotte et al. 1998), yet the ability of peroxisomes to be reformed in pex3Δ and pex19Δ strains after being resupplied with PEX3 and PEX19, respectively, argues against the structural templating of this organelle. However, structures termed “protoperoxisomes” were reported in pex19Δ (Snyder et al. 1999) and pex3Δ (Hazra et al. 2002) mutants of Pichia pastoris and in pex3 strains of Hansenula polymorpha (Faber et al. 2002) and S. cerevisiae (K. Huang and P. B. Lazarow, unpublished data cited in Lazarow 2003). Protoperoxisomes, in principle, would be capable of preserving the putative peroxisomal structural information, which could then serve as a template for the reestablishment of peroxisomes upon genetic complementation. Our oleate screen was therefore initiated to identify proteins that are necessary for the maintenance of both the peroxisome and the protoperoxisome. None of the 68 deletions examined had any effect on peroxisome integrity. In light of more recent evidence, it is clear why this search was unsuccessful: Hoepfner et al. (2005) have clearly shown that peroxisomes of S. cerevisiae, rather than being autonomous, can be derived from the endoplasmic reticulum.
While much is known about the activities of the proteins that are responsible for peroxisome function and about the biochemical pathways that occur there, the biological role of this organelle is still poorly understood. There is extraordinary variation in metabolic processes carried out in the peroxisomes of the different eukaryotic species, yet all peroxisomes carry out β-oxidation (Moyersoen et al. 2004). The mitochondrial β-oxidation pathway found in animals, on the other hand, has thus far been found neither in plants nor in fungi (Kunau et al. 1988). Perhaps the catabolism of dietary fatty acid is also a relatively specialized process and is carried out mainly or solely in mitochondria. We propose that peroxisomal β-oxidation instead fulfills a different function: catabolism of fatty acids that have been removed from membrane phospholipids.
This hypothesis is consistent with the requirement of a functional peroxisome to prevent growth inhibition by oleate. We have also presented data that further suggest that oleate exerts its toxic effect on oleate-sensitive mutants by permeabilizing the plasma membrane. This is the anticipated effect of a decrease in the packing density of the membrane bilayer, which would result from the over-incorporation of this unsaturated fatty acid into phospholipids. Although confirmation of an effect of supplemental oleate on phospholipid fatty acid content in pex strains awaits biochemical analysis of membrane fractions from such cultures, previous studies have indeed demonstrated the incorporation of fed fatty acids into Saccharomyces phospholipids (Bossie and Martin 1989; Stukey et al. 1989). While it is economical to propose that oleate exerts its toxicity on mutants in all the functional categories listed in Table 1 by its effect on the plasma membrane, this is not necessarily the case since the mitochondrial entries in Table 1, for example, imply a toxic effect of oleate on mitochondrial membrane. The nuclear-localized proteins listed in Table 1 perhaps cause oleate sensitivity indirectly by adversely influencing the ability of PEX genes to be induced by oleate.
Two large-scale screens have identified proteins required for proper cell-wall function in yeast (Lussier et al. 1997; Page et al. 2003). Twenty of these proteins, among them those listed in Table 1 in the “cell wall” functional category, were found in our screen: Bud14, Bud22, Dfg5, Fox2, Gal11, Ilm1, Med2, Nbp2, Pex6, Pex12, Ptc1, Rps11A, Sac1, Sap190, Sod1, Srb2, Srb5, Ste11, Thp1, and Tif3. This overlap implies that, for the “cell wall” functional category, oleate might exert its toxic effects directly on the plasma membrane. It is possible that this subset of mutants is deficient primarily in the ability to adequately control plasma membrane composition, a defect that in turn could influence the wall. Indeed, the cell wall is emerging as a dynamic structure that interacts in complex ways with the plasma membrane (Firon et al. 2004).
The sap190Δ and ilm1Δ mutants are of interest because they are particularly sensitive to oleate (C18:1) yet unaffected by palmitoleate (C16:1). We hypothesize that Sap190 and Ilm1 function in the control of the ratio of the palmitoleate-to-oleate content of phospholipids. The ratio of these two phospholipid components, the predominant unsaturated fatty acids in S. cerevisiae (Daum et al. 1999), decreases ∼1.7-fold when the growth temperature of S. cerevisiae increases from 10° to 35° (Suutari et al. 1990), implying an effect of the C16/C18 ratio on membrane fluidity. Moreover, acyl chain length is known to influence bacterial membrane fluidity (Cronan 1996). Although the mechanisms by which several prokaryotic species control membrane fluidity is understood in some detail (Mansilla et al. 2004), the mechanisms by which eukaryotes control this process are completely unknown.
No Sit4-associated proteins other than Sap190 are needed to prevent oleate sensitivity. To date, the Saps of Saccharomyces have been shown to be involved in sensitivity to Kluyveromyces lactis zymocin (Jablonowski et al. 2001, 2004), regulating the level of the Npr1 kinase (Jacinto et al. 2001), influencing the toxic effects of rapamycin (Rohde et al. 2004), and modulating K+ efflux (Manlandro et al. 2005). These previously reported functions, and perhaps oleate sensitivity as well, all concern events at the plasma membrane. None of these previous studies, however, have convincingly demonstrated that Sap190 plays a unique role. The sensitivity of the sit4Δ strain to oleate also suggests that a Sap190/Sit4 complex is the entity that prevents oleate inhibition. The ability of 0.03% oleate in YPD to inhibit the sap190Δ strain but not the sit4Δ strain (Figure 6B), however, suggests that Sap190 may be capable of functioning in concert with (an) additional PP2A-type phosphatase(s) (Duvel and Broach 2004) to form an alternative functional unit with Sap190.
Deletion of Ilm1, a 203-residue membrane protein of unknown function that resides in the ER, was first reported to increase the loss of mitochondrial (ilm) DNA (Entian et al. 1999) although we are unable to reproduce this phenotype (data not shown). Our screen showed ilm1Δ to cause oleate sensitivity; further examination showed that this deletion, like sap190Δ, has no effect on sensitivity to palmitoleate. This suggests that Ilm1 may also participate in the control of the C16/C18 ratio. A yeast two-hybrid screen carried out by The Yeast Resource Center (http://www.yeastrc.org/pdr/pages/front.jsp) has identified Mga2 as one of seven yeast proteins that strongly interact with Ilm1. Mga2p is a key transcription factor that controls expression of Ole1, the sole fatty acyl desaturase in S. cerevisiae responsible for conversion of the saturated fatty acids stearate (C18) and palmitate (C16) to oleate and palmitoleate, respectively (Zhang et al. 1999; Chellappa et al. 2001). The ratio of saturated to unsaturated fatty acids (UFA/SFA) is known from studies of Bacillus subtilis, for example, to be a key determinant of membrane fluidity (Grau and de Mendoza 1993; Weber et al. 2001). Suutari et al. (1990), however, showed that the UFA/SFA ratio of Saccharomyces is unaffected by growth temperature. On the other hand, the strong induction of OLE1 expression upon shifting yeast from 30° to 10° (Nakagawa et al. 2002) strongly implicates the UFA/SFA ratio in yeast membrane fluidity control. Although yeast are poikilotherms, as are all microbes, an understanding of the contributions of C16/C18 and UFA/SFA ratios to yeast membrane fluidity is likely to be relevant to homeotherms as well.
As judged by Sytox staining, pex6Δ and other deletions augment the ability of oleate to cause permeabilization of the yeast plasma membrane. Thus, Pex6 and other proteins may participate in the maintenance of plasma membrane integrity. Of several possible mechanisms that account for this role, we favor the participation of the peroxisome in preventing the alteration of phospholipid fatty acid content by oleate. This model is consistent with the role of the peroxisome in fatty acid catabolism, one strategy by which yeast can remove excess oleate. However, the complete absence of β-oxidation in the pox1Δ and fox2Δ strains caused only slight oleate sensitivity; deletion of POT1, which encodes the third enzyme essential for β-oxidation, caused a more severe oleate phenotype. Thus, prevention of oleate toxicity by the peroxisome is apparently more complex than merely the organelle's role in fatty acid catabolism. Yeast can also sequester added fatty acid in the lipid particle, a membrane-bound organelle (Athenstaedt et al. 1999). Indeed, yeast that lack a lipid particle (Sandager et al. 2002) are also highly sensitive to oleate (D. Lockshon, unpublished results). An intimate association between the lipid particle and the peroxisome of Saccharomyces has recently been reported (Binns et al. 2006), suggesting that perhaps the peroxisome may also participate in lipid storage. Other possible models, which implicate the peroxisome in membrane resealing (Jedd and Chua 2000; McNeil and Steinhardt 2003) or in multi-drug efflux pump function (Ernst et al. 2005), have not been ruled out.
A cellular role for the peroxisome has yet to be determined. This work suggests that it could have an important role in governing yeast fatty acid levels and perhaps in governing the composition of membrane phospholipids. The relative simplicity of the metabolic processes that occur in the yeast peroxisome does not readily suggest a cellular function for the organelle. The human peroxisome, on the other hand, houses multiple pathways, perhaps all of which involve membrane component metabolism (Wanders and Tager 1998), thus implying a close relationship between the organelle and cellular membrane composition.
Acknowledgments
Stan Fields and members of his lab graciously allowed us to use their Biomek 2000. Thanks to Alex Merz and Margo Murphy for a critical reading of the manuscript. The work was supported by a Congressionally Directed Medical Research Program Grant (DAMD17-03-0497) to B.K.K.
References
- Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein and G. Daum, 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181: 6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, K., and S. Eaton, 2004. Mitochondrial beta-oxidation. Eur. J. Biochem. 271: 462–469. [DOI] [PubMed] [Google Scholar]
- Binns, D., T. Januszewski, Y. Chen, J. Hill, V. S. Markin et al., 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund, S., and C. M. Gustafsson, 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30: 240–244. [DOI] [PubMed] [Google Scholar]
- Bossie, M. A., and C. E. Martin, 1989. Nutritional regulation of yeast delta-9 fatty acid desaturase activity. J. Bacteriol. 171: 6409–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Chellappa, R., P. Kandasamy, C. S. Oh, Y. Jiang, M. Vemula et al., 2001. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 276: 43548–43556. [DOI] [PubMed] [Google Scholar]
- Ciriacy, M., 1975. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol. Gen. Genet. 138: 157–164. [DOI] [PubMed] [Google Scholar]
- Cronan, J. E., and C. O. Rock, 1996. Biosynthesis of membrane lipids, pp. 612–636 in Escherichia coli and Salmonella: Cellular and Molecular Biology, edited by F. C. Neidhardt. American Society of Microbiology, Washington, DC.
- Cronin, S. R., R. Rao and R. Y. Hampton, 2002. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 157: 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, G., G. Tuller, T. Nemec, C. Hrastnik, G. Balliano et al., 1999. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast 15: 601–614. [DOI] [PubMed] [Google Scholar]
- Duvel, K., and J. R. Broach, 2004. The role of phosphatases in TOR signaling in yeast. Curr. Top. Microbiol. Immunol. 279: 19–38. [DOI] [PubMed] [Google Scholar]
- Eckert, J. H., and R. Erdmann, 2003. Peroxisome biogenesis. Rev. Physiol. Biochem. Pharmacol. 147: 75–121. [DOI] [PubMed] [Google Scholar]
- el-Sherbeini, M., and J. A. Clemas, 1995. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-beta-glucan in vitro. J. Bacteriol. 177: 3227–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian, K. D., T. Schuster, J. H. Hegemann, D. Becher, H. Feldmann et al., 1999. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262: 683–702. [DOI] [PubMed] [Google Scholar]
- Erdmann, R., M. Veenhuis, D. Mertens and W. H. Kunau, 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86: 5419–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, R., F. F. Wiebel, A. Flessau, J. Rytka, A. Beyer et al., 1991. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell 64: 499–510. [DOI] [PubMed] [Google Scholar]
- Ernst, R., R. Klemm, L. Schmitt and K. Kuchler, 2005. Yeast ATP-binding cassette transporters: cellular cleaning pumps. Methods Enzymol. 400: 460–484. [DOI] [PubMed] [Google Scholar]
- Faber, K. N., G. J. Haan, R. J. Baerends, A. M. Kram and M. Veenhuis, 2002. Normal peroxisome development from vesicles induced by truncated Hansenula polymorpha Pex3p. J. Biol. Chem. 277: 11026–11033. [DOI] [PubMed] [Google Scholar]
- Firon, A., G. Lesage and H. Bussey, 2004. Integrative studies put cell wall synthesis on the yeast functional map. Curr. Opin. Microbiol. 7: 617–623. [DOI] [PubMed] [Google Scholar]
- Foss, E. J., 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte, K., W. Girzalsky, M. Linkert, E. Baumgart, S. Kammerer et al., 1998. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol. 18: 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau, R., and D. de Mendoza, 1993. Regulation of the synthesis of unsaturated fatty acids by growth temperature in Bacillus subtilis. Mol. Microbiol. 8: 535–542. [DOI] [PubMed] [Google Scholar]
- Gurvitz, A., J. K. Hiltunen, R. Erdmann, B. Hamilton, A. Hartig et al., 2001. Saccharomyces cerevisiae Adr1p governs fatty acid beta-oxidation and peroxisome proliferation by regulating POX1 and PEX11. J. Biol. Chem. 276: 31825–31830. [DOI] [PubMed] [Google Scholar]
- Haupt, I., R. Hubener and H. Thrum, 1978. Streptothricin F, an inhibitor of protein synthesis with miscoding activity. J. Antibiot. 31: 1137–1142. [DOI] [PubMed] [Google Scholar]
- Hazel, J. R., 1995. Thermal adaptation in biological membranes: Is homeoviscous adaptation the explanation? Annu. Rev. Physiol. 57: 19–42. [DOI] [PubMed] [Google Scholar]
- Hazra, P. P., I. Suriapranata, W. B. Snyder and S. Subramani, 2002. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3: 560–574. [DOI] [PubMed] [Google Scholar]
- Hettema, E. H., W. Girzalsky, M. van Den Berg, R. Erdmann and B. Distel, 2000. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19: 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen, J. K., A. M. Mursula, H. Rottensteiner, R. K. Wieranga, A. J. Kastaniotis et al., 2003. The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27: 35–64. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen and H. F. Tabak, 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122: 85–95. [DOI] [PubMed] [Google Scholar]
- Hohfeld, J., M. Veenhuis and W. H. Kunau, 1991. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 114: 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jablonowski, D., A. R. Butler, L. Fichtner, D. Gardiner, R. Schaffrath et al., 2001. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 159: 1479–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski, D., L. Fichtner, M. J. Stark and R. Schaffrath, 2004. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell 15: 1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle and M. N. Hall, 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8: 1017–1026. [DOI] [PubMed] [Google Scholar]
- Jedd, G., and N. H. Chua, 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2: 226–231. [DOI] [PubMed] [Google Scholar]
- Johnston, G. C., J. R. Pringle and L. H. Hartwell, 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105: 79–98. [DOI] [PubMed] [Google Scholar]
- Jones, D. L., J. Petty, D. C. Hoyle, A. Hayes, E. Ragni et al., 2003. Transcriptome profiling of a Saccharomyces cerevisiae mutant with a constitutively activated Ras/cAMP pathway. Physiol. Genomics 16: 107–118. [DOI] [PubMed] [Google Scholar]
- Karpichev, I. V., and G. M. Small, 1998. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6560–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal, G., K. Sipos, H. Lange, Z. Fekete, T. Bedekovics et al., 2005. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2: E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer, D., 2002. Monitoring protein degradation. Methods Enzymol. 351: 639–647. [DOI] [PubMed] [Google Scholar]
- Kunau, W. H., S. Buhne, M. de la Garza, C. Kionka, M. Mateblowski et al., 1988. Comparative enzymology of beta-oxidation. Biochem. Soc. Trans. 16: 418–420. [DOI] [PubMed] [Google Scholar]
- Lazarow, P. B., 2003. Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 15: 489–497. [DOI] [PubMed] [Google Scholar]
- Lockshon, D., 2002. A heritable structural alteration of the yeast mitochondrion. Genetics 161: 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke, M. M., F. Della Seta, C. J. Di Como, H. Sugimoto, R. Kobayashi et al., 1996. The SAPs, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16: 2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y., I. V. Karpichev, R. A. Kohanski and G. M. Small, 1996. Purification, identification, and properties of a Saccharomyces cerevisiae oleate-activated upstream activating sequence-binding protein that is involved in the activation of POX1. J. Biol. Chem. 271: 12068–12075. [DOI] [PubMed] [Google Scholar]
- Lussier, M., A. M. White, J. Sheraton, T. di Paolo, J. Treadwell et al., 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manlandro, C. M., D. H. Haydon and A. G. Rosenwald, 2005. Ability of Sit4p to promote K+ efflux via Nha1p is modulated by Sap155p and Sap185p. Eukaryot. Cell 4: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla, M. C., L. E. Cybulski, D. Albanesi and D. de Mendoza, 2004. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 2004: 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., J. J. Smith, S. Jung, E. Yi, A. I. Nesvizhskii et al., 2004. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 167: 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch, M., R. Erdmann, M. Veenhuis and W. H. Kunau, 1994. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13: 4908–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, P. L., and R. A. Steinhardt, 2003. Plasma membrane disruption: repair, prevention, adaptation. Annu. Rev. Cell Dev. Biol. 19: 697–731. [DOI] [PubMed] [Google Scholar]
- Moyersoen, J., J. Choe, E. Fan, W. G. Hol and P. A. Michaels, 2004. Biogenesis of peroxisomes and glycosomes: trypanosomatid glycosome assembly is a promising new drug target. FEMS Microbiol. Rev. 28: 603–643. [DOI] [PubMed] [Google Scholar]
- Nakagawa, Y., N. Sakumoto, Y. Kaneko and S. Harashima, 2002. Mga2p is a putative sensor for low temperature and oxygen to induce OLE1 transcription in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 291: 707–713. [DOI] [PubMed] [Google Scholar]
- Oh, C. S., D. A. Toke, S. Mandala and C. E. Martin, 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272: 17376–17384. [DOI] [PubMed] [Google Scholar]
- Page, N., M. Gerard-Vincent, P. Menard, M. Beaulieu, M. Azuma et al., 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163: 875–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue, P. E., and P. B. Lazarow, 2001. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17: 701–752. [DOI] [PubMed] [Google Scholar]
- Rohde, J. R., S. Campbell, S. A. Zurita-Martinez, N. S. Cutler, M. Ashe et al., 2004. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol. Cell. Biol. 24: 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner, H., A. J. Kal, M. Filipits, M. Binder, B. Hamilton et al., 1996. Pip2p: a transcriptional regulator of peroxisome proliferation in the yeast Saccharomyces cerevisiae. EMBO J. 15: 2924–2934. [PMC free article] [PubMed] [Google Scholar]
- Sandager, L., M. H. Gustavsson, U. Stahl, A. Dahlqvist, E. Wiberg et al., 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277: 6478–6482. [DOI] [PubMed] [Google Scholar]
- Schorr, M., A. Then, S. Tahirovic, N. Hug and P. Mayinger, 2001. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11: 1421–1426. [DOI] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Simon, M., G. Adam, W. Rapatz, W. Spevak and H. Ruis, 1991. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol. Cell. Biol. 11: 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, W. B., K. N. Faber, T. J. Wenzel, A. Koller, G. H. Luers et al., 1999. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol. Biol. Cell 10: 1745–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, D., and G. Daum, 2003. Triacylglycerol biosynthesis in yeast. Appl. Microbiol. Biotechnol. 61: 289–299. [DOI] [PubMed] [Google Scholar]
- Stukey, J. E., V. M. McDonough and C. E. Martin, 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264: 16537–16544. [PubMed] [Google Scholar]
- Sutton, A., D. Immanuel and K. T. Arndt, 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11: 2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suutari, M., K. Liukkonen and S. Laakso, 1990. Temperature adaptation in yeasts: the role of fatty acids. J. Gen. Microbiol. 136: 1469–1474. [DOI] [PubMed] [Google Scholar]
- Theis, T., M. Wedde, V. Meyer and U. Stahl, 2003. The antifungal protein from Aspergillus giganteus causes membrane permeabilization. Antimicrob. Agents Chemother. 47: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen, K., F. R. Terras and W. F. Broekaert, 1999. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl. Environ. Microbiol. 65: 5451–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipper, D. J., and C. A. Harley, 2002. Yeast genes controlling responses to topogenic signals in a model transmembrane protein. Mol. Biol. Cell 13: 1158–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toke, D. A., and C. E. Martin, 1996. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271: 18413–18422. [DOI] [PubMed] [Google Scholar]
- van den Bosch, H., R. B. Schutgens, R. J. Wanders and J. M. Tager, 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61: 157–197. [DOI] [PubMed] [Google Scholar]
- Veenhuis, M., M. Mateblowski, W. H. Kunau and W. Harder, 1987. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast 3: 77–84. [DOI] [PubMed] [Google Scholar]
- Veenhuis, M., J. A. Kiel and I. J. Van Der Klei, 2003. Peroxisome assembly in yeast. Microsc. Res. Tech. 61: 139–150. [DOI] [PubMed] [Google Scholar]
- Wanders, R. J., and J. M. Tager, 1998. Lipid metabolism in peroxisomes in relation to human disease. Mol. Aspects Med. 19: 69–154. [DOI] [PubMed] [Google Scholar]
- Weber, M. H., W. Klein, L. Muller, U. M. Niess and M. A. Marahiel, 2001. Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol. Microbiol. 39: 1321–1329. [DOI] [PubMed] [Google Scholar]
- Wiebel, F. F., and W. H. Kunau, 1992. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature 359: 73–76. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Zaremberg, V., and C. R. McMaster, 2002. Differential partitioning of lipids metabolized by separate yeast glycerol-3-phosphate acyltransferases reveals that phospholipase D generation of phosphatidic acid mediates sensitivity to choline-containing lysolipids and drugs. J. Biol. Chem. 277: 39035–39044. [DOI] [PubMed] [Google Scholar]
- Zhang, S., Y. Skalsky and D. J. Garfinkel, 1999. MGA2 or SPT23 is required for transcription of the Δ9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z., and J. Zou, 2001. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J. Biol. Chem. 276: 41710–41716. [DOI] [PubMed] [Google Scholar]