Abstract
The accumulation of slightly deleterious mutations in populations leads to the buildup of a genetic load and can cause the extinction of populations of small size. Mutation-accumulation experiments have been used to study this process in a wide variety of organisms, yet the exact mutational underpinnings of genetic loads and their fitness consequences remain poorly characterized. Here, we use an abiotic system of RNA populations evolving continuously in vitro to examine the molecular events that can instigate a genetic load. By tracking the fitness decline of ligase ribozyme populations with bottleneck sizes between 100 and 3000 molecules, we detected the appearance and subsequent fixation of both slightly deleterious mutations and advantageous mutations. Smaller populations went extinct in significantly fewer generations than did larger ones, supporting the notion of a mutational meltdown. These data suggest that mutation accumulation was an important evolutionary force in the prebiotic RNA world and that mechanisms such as recombination to ameliorate genetic loads may have been in place early in the history of life.
AS early as 1937, it was noted by J. B. S. Haldane that mutations with a negative effect on the average fitness of individuals could accumulate in a population (Haldane 1937), leading to what was later called a mutational load by Muller (1950). This prediction has been borne out empirically and experimentally, in a wide variety of wild and laboratory organisms (Wallace 1987; Lynch et al. 1999). In fact, it has led to a vigorous debate over the origins and advantages of sexual reproduction. The argument is often made that sexuality provides an escape from Muller's ratchet because even occasional blending of genotypes can produce offspring with a lowered mutational load—an option not available to strictly asexual lineages. Yet it has been argued that the mutation rate may be too low in many species to explain the advantage of sex, which comes with a high apparent cost compared to asexuality (Keightley and Eyre-Walker 2000). Another issue of great interest is the relationship between mutational load and population size. It has been predicted that these two factors can act synergistically, in that as the load increases, the population size should decrease, leading to a higher probability of fixing new deleterious mutations (Lynch and Gabriel 1990; Gabriel et al. 1993; Lynch et al. 1993). Eventually a threshold is crossed, and the population spirals into extinction via a “mutational meltdown,” as can be seen in ciliated protozoans and fibroblast cultures, for example (Smith and Pereira-Smith 1977; Tagaki and Yoshida 1980).
Mutation-accumulation (MA) experiments have been used effectively for >40 years to address questions related to the buildup of deleterious mutations in populations such as those of Arabidopsis, Caenorhabditis elegans, Daphnia, Drosophila, Escherichia coli, Saccharomyces cerevisiae, and others (reviewed in Mukai 1964; Kibota and Lynch 1996; Lynch and Walsh 1998; Schultz et al. 1999; Pfrender and Lynch 2000; Zeyl et al. 2001; Estes et al. 2004). A species lineage is propagated in a very controlled environment over a large number of generations and is typically forced through a bottleneck in size each generation to exacerbate the effects of random genetic drift. All generations subsequent to the starting, or baseline, generation are monitored for fitness decline and/or for among-line variance in an attempt to interrelate these quantities with genetic parameters such as mutation rate, dominance, and epistasis. The MA approach has led to a deeper understanding of the role of spontaneous mutations in evolution. Nevertheless, it is limited in at least two ways when used with whole organisms. First, the generation time of the organism can be constraining; even the most rapidly growing organisms such as E. coli can be passed only through at most tens of generations in a day. Second, the ultimate causes of fitness declines must be inferred, because it is impractical to determine the complete nucleotide sequence of individuals in the evolving lineages. Often this means that the mutational events that transpire are missed or poorly characterized (Davies et al. 1999), although some recent work with C. elegans has revealed a more thorough examination of the genotypic changes that transpire over the course of a MA experiment (Denver et al. 2004).
One of the goals of the current study was to achieve the first MA experiment with evolving populations of RNA molecules, with the advantage that a detailed genotypic characterization would be within reach. Another goal was to use such a study to observe both the accumulation of slightly deleterious mutations and the mutational meltdown in a very simple and tightly controlled genetic system that was essentially free of confounding factors such as pleiotropy. By using an abiotic milieu of catalytic RNAs (ribozymes) evolving in vitro, we endeavored to test the hypotheses that (i) the mutational load has a clear biochemical origin and (ii) smaller asexual populations are at a greater risk for mutational meltdown than larger ones. At the same time we would be able to examine the influence of the accumulation of deleterious mutations during the origins of life on earth, another subject on which Haldane provided pioneering insight (Haldane 1929). Our intent to track RNA genotypes and phenotypes over time is directly relevant to the RNA world hypothesis, in that life may very well have passed through an RNA stage en route to its current DNA/protein-based existence (Gilbert 1986; Gesteland et al. 2005).
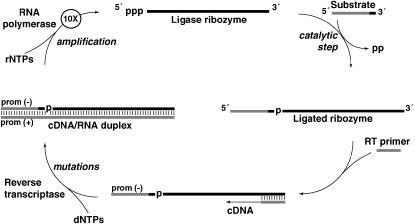
We employed the continuous evolution (CE) system (Wright and Joyce 1997) with ligase ribozymes as a means to observe mutational loads in RNA populations (Figure 1). In this test-tube setting, ribozymes are challenged to perform a catalytic ligation on an exogenous RNA substrate, and only those that succeed can be replicated by the sequential action of two protein enzymes: reverse transcriptase and RNA polymerase. Each completion of the cycle is a generation and leads to an ∼10-fold amplification of the fittest RNA molecules. When the raw materials such as nucleotides and protein enzymes have been exhausted, typically in about three generations, a small fraction of the RNA population can be transferred to a new test tube with fresh reagents for another set of generations. We call each transfer a “burst” because it results in a burst of RNA amplification on the order of 1000-fold (Wright and Joyce 1997; Schmitt and Lehman 1999). This system has highly beneficial features for MA experiments. Each generation is accomplished in ≤10 min, so that in principle hundreds of generations can be completed in a single day. Likewise it is easy to run several lineages in parallel, either in absolute replicate or with variation of single experimental variables (Johns and Joyce 2005). Also, the RNA sequences are short, ∼150 nucleotides, and thus complete sequence analysis of their “genomes” is possible. Last, the bottleneck size is under experimental control yet highly variable, over several orders of magnitude. In the experiments described here, we chose bottleneck population sizes ranging from 100 molecules (167 ymol) to 3000 molecules (5 zmol).
Figure 1.—
The continuous evolution (CE) scheme (Wright and Joyce 1997). RNA strands are solid and DNA strands are shaded. A population of ligase ribozymes (top) is incubated at 37° with an excess of substrate oligonucleotides in a 25-μl volume. Catalytically proficient ribozymes will ligate the substrate to their own 5′ ends (right). Also in the reaction vessel are a primer for reverse transcription that is complementary to the 3′ end of the ribozyme, a pool of dNTPs and rNTPs, and the protein enzymes MMLV reverse transcriptase and T7 RNA polymerase. While all RNAs should be reverse transcribed into double-stranded DNA/RNA hybrids (bottom and left), only those that had successfully performed ligation will be transcribed back into RNA because the substrate contains the necessary T7 promoter sequence. Because ∼10 RNAs are made from each template, and three completions of the cycle as shown occur in each 22-min burst, a 1000-fold amplification of RNA is possible if the initial population contains high-fitness genotypes. After 22 min, the reaction is diluted 1000-fold, fresh protein enzymes, primers, and nucleotides are added, and a new burst is initiated. The cDNA can be amplified via the PCR and genotyped each burst. If the mean fitness of the population falls, the degree of amplification will not keep pace with the dilution factor and eventually the cDNA concentration will drop below the PCR detection threshold.
Mutations in the CE system are generated by the protein enzymes. The MMLV reverse transcriptase used here is particularly error prone in vitro, with mutation rates estimated at 2 × 10−5 mutations/nucleotide/replication pass (De Angioletti et al. 2002). Strictly on the basis of this rate, on average ∼1% of the RNA molecules in each lineage would be expected to suffer a mutation each burst. The T7 RNA polymerase also may contribute to the net mutation rate. Note that although the CE system uses contemporary protein enzymes to accomplish replication—enzymes that would not have been available in a prebiological RNA world (Gilbert 1986)—these enzymes serve as convenient surrogates for RNA replicase ribozymes postulated to have been a crucial feature of the origins of life despite having far higher mutation rates than modern protein polymerases (Johnston et al. 2001). The combined use of these enzymes, a 22-min burst time with three generations per burst, and parallel treatments of lineages, meant that we could accomplish 25 MA lineages of 50–150 generations each with a strong mutational pressure in a few weeks' time.
MATERIALS AND METHODS
RNA preparation:
The starting B16-19 RNA was obtained by run-off transcription of PCR DNA obtained from a cloned genotype arising in a previous in vitro evolution experiment (Schmitt and Lehman 1999) and was gel purified to length homogeneity (152 nucleotides) prior to use. The concentration was measured by UV spectrometry at 260 nm and carefully diluted from 10.0-μm stocks into several separate aliquots of 100 molecules/8.20 μl (2.03 × 10−8 nm), 300 molecules/8.20 μl (6.11 × 10−8 nm), 600 molecules/8.20 μl (1.22 × 10−7 nm), or 3000 molecules/8.20 μl (6.11 × 10−7 nm).
Continuous evolution in vitro:
The CE protocol was followed essentially as described previously (Wright and Joyce 1997; Schmitt and Lehman 1999; Lehman 2004) except that vastly smaller input RNA population sizes were used. Briefly, 8.2 μl of a diluted RNA stock was incubated with 64 pmol S-163 DNA/RNA substrate (5′-CTTGACGTCAGCCTGGACTAATACGACTCACUAUA-3′, with the T7 promoter sequence underlined and the ribonucleotides shown in boldface type), 50 pmol RT primer (5′-GCTGAGCCTGCGATTGG-3′), 250 units M-MLV reverse transcriptase (United States Biochemicals, Cleveland), 50 units T7 RNA polymerase (Ambion, Austin, TX), 5 nmol each dNTP, 50 nmol each rNTP, and 25 mm MgCl2 in reaction buffer [50 mm KCl, 30 mm 4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid (EPPS), pH 8.3] in a total volume of 25 μl for 22 min at 37°. At the end of the incubation period, 3 μl were removed and diluted into 981 μl of water. An 8.2-μl aliquot of this dilution was used to seed the next 25-μl burst, resulting in an overall 1000-fold dilution from one burst to the next. In the second and all subsequent bursts, the diluted mixture from the previous burst was incubated with fresh amounts of substrate, primer, protein enzymes, nucleotides, and buffer in the quantities described above. To ensure that the dilution factor was matching the amplification factor each burst, some lineages were run as above but additionally in the presence of 3.75 μCi [α-32P]ATP. In these cases, an additional 3 μl were removed after 22 min, quenched in acrylamide gel-loading buffer (0.05% bromphenol blue, 40% sucrose), and subjected to electrophoresis through 6% polyacrylamide/8 m urea gels and phosphorimaging. After overnight exposure to the phosphor screen, failure to detect the appearance of a 152-nt RNA species after >10 bursts, despite the appearance of strong 187-bp PCR products (see below), was indicative that the RNA population was not growing because the net dilution over this time would be as high as 1030-fold (Wright and Joyce 1997; Johns and Joyce 2005).
Genotypic monitoring:
A total of 25 lineages were maintained, 6 each of 100-, 300-, and 3000-molecule sizes, and 7 of a 600-molecule size. The status of each lineage was monitored by amplification of 2.75 μl of the 981-μl postburst dilutions using the RT primer and a second primer (5′-CTTGACGTCAGCCTGGA-3′) matching a portion of the S-163 sequence. Amplification of the B16-19 genotype, or of point mutations of this genotype, generates a 187-bp product. These products were digested with TaqαI to detect the CUGAACCUUA(123–132) → AAUCG mutation (which shortens the PCR product to 182 bp, generating a TaqαI restriction site and fragments of 160 and 22 bp) and with XmnI to detect the U62 → A mutation (which destroys a XmnI restriction site in the B16-19 sequence that would cut the PCR product essentially in half). Products from selected bursts were also cloned via the TOPO-TA cloning kit (Invitrogen, San Diego). DNA extracts from single bacterial colonies from these clones were amplified with the same primers as above. Selected burst PCR pools and individual cloned amplification products were both subjected to sequence analysis on an ABI 3100 Prism using BigDye (v.3) chemistry.
Phenotypic analyses:
RNA transcripts from cloned DNA amplicons from selected genotypes were prepared and diluted to various concentrations. For kinetic assays, the kcat parameter was estimated for each genotype by the y-intercept of modified Eadie–Hofstee plots of the observed rate of reaction (kobs) as a function of kobs/[ribozyme]. The kobs-values of ligation were determined at 22° under ribozyme excess conditions of 5 nm 5′-γ-32P-labeled substrate and 0.1–1.0 μm ribozyme. Six time points ranging between 5 sec and 2 min were taken when the ribozymes were incubated in CE buffer. The extent of reaction was quantified as a fraction of total RNA in the ligated form in 5% denaturing polyacrylamide gels. Single-turnover self-ligation kobs-values were obtained for each ribozyme concentration by an exponential curve fit to the equation f = A(1 − exp(−kobs × t)), where f is the fraction of substrate reacted at time t, and A is the asymptote (projected maximum f). For analysis of energetics of putative base-paired regions in genotypes with the insurance mutation, the mfold program (Zuker 2003) was employed.
RESULTS AND DISCUSSION
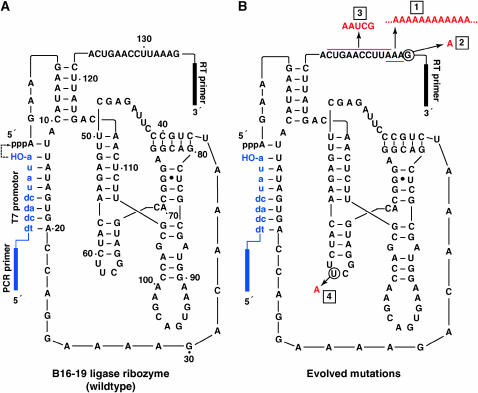
We began each MA lineage with a genotypically pure population of a high-fitness ligase ribozyme genotype, denoted B16-19 (Figure 2A). This sequence is a strong competitor in the CE environment and has been selected repeatedly from randomized populations under a variety of experimental conditions (Schmitt and Lehman 1999; Lehman 2004). Therefore mutations that arise in this sequence are likely to be deleterious; B16-19 sits atop a high fitness peak. Of course many mutations would be lethal, either destroying the ligase activity of the ribozyme or rendering it unable to fold properly in the 22-min burst time. But those types of mutations are not assayed by MA experiments, only those with small effect that can be fixed in small populations through the sampling error of genetic drift. In CE, this drift is manifest because one one-thousandth of the population is transferred to a new reaction vessel each burst, resulting in effective population sizes small enough to allow less-fit genotypes to increase in frequency by chance.
Figure 2.—
Ribozymes used in this study. (A) The B16-19 ligase ribozyme. This ribozyme catalyzed the ligation of the substrate (blue, lowercase letters) to its 5′ end (dashed arrow). (B) Mutations (red) detected in various continuous evolution lineages: 1, 3′ polyadenylation (mutational load); 2, G135 → A (floodgate); 3, CUGAACCUUA at positions 123–132 → AAUCG (immunity); and 4, U62 → A (insurance).
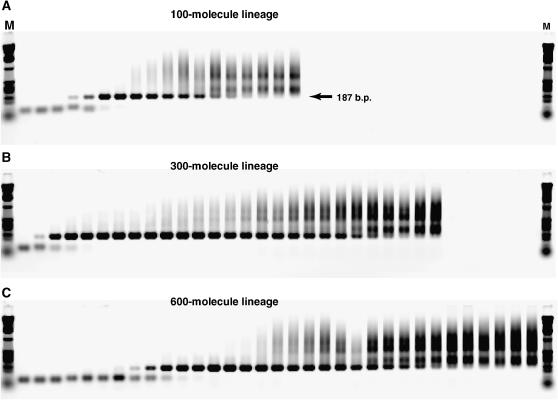
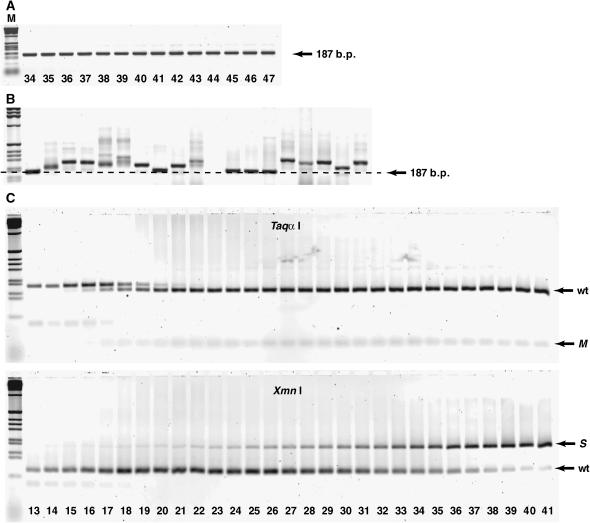
We monitored the progress of CE via PCR amplification of the cDNA that is made during the reaction cycle. Samples of a CE lineage were taken every burst and amplified with primers specific to the ligation substrate and to the reverse transcriptase primer-binding site such that all fit ligase ribozymes should be amplified. While some lineages produced robust PCR products of the expected size (187 bp), others faded out over time, typically developing a high-molecular-weight smear and eventually losing the 187-bp band (Figure 3). We equated complete loss of the main band with lineage death, as this meant that the amplification of the wild-type length sequences was not strong enough to keep up with the 1/1000-fold dilution each burst. Note that the loss of a PCR product should trail a few bursts behind the loss of the actual RNA population until the residual cDNA from the last RNA survivors gets diluted below the PCR detection threshold. We also monitored the RNA population itself, in a few cases, by the use of [α-32P]ATP nucleotides in the reaction milieu and polyacrylamide gel electrophoresis to ensure that the RNA population was not substantially growing and outpacing the dilution factor. In fact, the 22-min burst time was chosen to maintain this balance in the sampled lineages (data not shown).
Figure 3.—
Tracking of lineage progress and death. Agarose gel electrophoretic images are of PCR amplifications of samples taken from consecutive bursts from three example lineages. (A) Bursts 1–18 of 100-molecule lineage 4V declared dead (i.e., complete loss of 187-bp band) at burst 18. (B) Bursts 1–27 of 300-molecule lineage 3J declared dead at burst 27. (C) Bursts 1–33 of 600-molecule lineage 3X declared dead at burst 37 (not shown on gel).
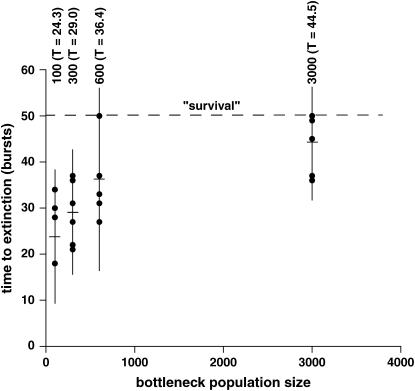
We continued each lineage until it died or survived to 50 bursts (∼150 generations), whichever came first, and we ran six or seven replicates of each bottleneck population size (Table 1). Strikingly, the average time to extinction is negatively correlated with population size (Figure 4). The 100-molecule (bottleneck population size) lineages never survived past 34 bursts, while two each of the 600- and 3000-molecule lineages survived strongly to 50 bursts, and a third 3000-molecule lineage died at burst 49. The average times to extinction were calculated using a value of 50 for those four lineages that survived to burst 50, even though they could have persisted for much longer (Figure 5A). Thus the averages for the 600- and 3000-molecule populations are conservative underestimates. Nevertheless, using these averages, there is a statistical difference between time to extinction when the 100-molecule lineages are compared to the 600- or 3000-molecule lineages (P = 0.050 and 0.0060 respectively) and when the 300-molecule lineages are compared to the 3000-molecule lineages (P = 0.010). All tests were made using a model I ANOVA with multiple (k = 6) planned comparisons of means (T′ method).
TABLE 1.
Fates of the 25 continuous evolution lineages
| Lineage designation | Bottleneck population size | Extinction at burst no. | Mutations observeda (burst appeared, burst fixed) |
|---|---|---|---|
| 4Vb | 100 | 18 | A (10, 18), F |
| 4R | 100 | 18 | A (11, 18), F |
| 4Z | 100 | 18 | A (13, 18), F |
| 4Y | 100 | 28 | A (18, 28) |
| 4W | 100 | 30 | A (22, 30) |
| 4U | 100 | 34 | A (21, 34), S (ND, ND) |
| 3K | 300 | 21 | A (15, 21), F |
| 3O | 300 | 22 | A (16, 22), F |
| 3J | 300 | 27 | A (22, 27), F |
| 3N | 300 | 31 | A (21, 31) |
| 3W | 300 | 36 | A (30, 36), S (ND, 28) |
| 3X | 300 | 37 | A (29, 37), S (ND, ND) |
| 3A | 600 | 27 | A (20, 27), F |
| 3V | 600 | 27 | A (20, 27), F |
| 3S | 600 | 31 | A (19, 31), F |
| 3E | 600 | 33 | A (25, 33) |
| 3Q | 600 | 37 | A (26, 37) |
| 5A | 600 | 50+ | M (15, 21); S (20, 43) |
| 3D | 600 | 50+ | M (ND, NF); S (ND, 32) |
| 2I | 3000 | 36 | A (18); S (7, gone by 25) |
| 2K | 3000 | 37 | A (22); S (3, 30) |
| 2A | 3000 | 45 | A (31); S (8, gone by 10) |
| 2J | 3000 | 49 | A (23, NF); M (40, NF); S (6, 30) |
| 1A | 3000 | 50+ | M (5, 10); S (6, 10) |
| 1C | 3000 | 50+ | M (5, 6); S (6, 6) |
A, polyadenylations; F, floodgate; M, immunity; S, insurance; NF, never fixed (always polymorphic); ND, not determined. Not all bursts were surveyed for the presence of the floodgate mutation.
Lineages in italics were subjected to direct sequence analyses, as well as to RFLP.
Figure 4.—
Mutational meltdown in RNA populations. Each dot represents a lineage, with the mean time to extinction (T) indicated by a crosshair in a vertical line spanning the mean ± 2 SD of trials for each of the four population sizes. Values of T are significantly different for the 100- vs. 600-, 100- vs. 3000-, and 300- vs. 3000-molecule comparisons (see text). The dashed line indicates the a priori designation of 50 bursts as being the threshold for a surviving lineage, at which point the lineages were discontinued.
Figure 5.—
Tracking genotypic changes over time. (A) PCR amplification of bursts 34–47 of 600-molecule lineage 5A, which survived at least to burst 50. There is no development of a smear above the 187-bp band, which are the amplicons from wild-type-length RNAs (152 nt). This lineage did not suffer a mutational load imposed by 3′ polyadenylations, while it did evolve both the immunity and the insurance mutations (see C). M denotes size marker (1-kb ladder). (B) PCR amplification of 20 cloned molecules from burst 13 of a 100-molecule lineage that went extinct in burst 18. Size differences are a consequence of 3′ polyadenylations. (C) RFLP analysis of bursts 13–41 of 600-molecule lineage 5A, which survived to burst 50 (shown also in A). The TaqαI digest distinguishes between the wild-type (wt) B16-19 genotype and the immunity (M) mutant; the XmnI digest distinguishes between the B16-19 genotype and the insurance (S) mutant. In this lineage, the immunity mutation arises in about burst 15 and is rapidly fixed in the population by burst 21. The insurance mutation arises later, around burst 20, and slowly accumulates to fixation around burst 43 (not shown on gel).
Thus we observed clear evidence of mutational meltdown in these abiotic populations. To determine the underlying mutational events leading to these lineage deaths, we genotyped the PCR populations resulting from selected bursts. This was done both by RFLP analysis of the PCR DNA from each and every burst in our study and by direct nucleotide sequence analysis of selected bursts. For the direct sequence analysis, we performed batch sequence analysis from at least 1 burst from all lineages, typically within 10 bursts of extinction. Primarily, however, we focused on two lineages, one 100-molecule lineage that died the earliest (4V; Table 1) and one 600-molecule lineage that survived to burst 50 (5A; Table 1). We performed batch sequence analysis of the burst PCR products on 10 bursts of lineage 4V (6–16 except 11) and on two bursts of lineage 5A (6 and 9). In addition, we cloned bursts 5, 9, 13, and 14 from lineage 4V and bursts 22, 34, and 47 from lineage 5A. From each of these cloned populations, we obtained complete bidirectional sequence data from between 6 and 10 individual molecules.
From these genotypic data, four types of mutation were evident (Figure 2B):
The load (polyadenylation): First, the lineages that developed smears above the 187-bp PCR product became increasingly dominated over time by molecules possessing poly(A) tracts near the 3′ end of the RNA. The mutated RNAs in these populations contained between 1 and ≥1000 additional adenosine residues in the region immediately preceding the primer-binding site for reverse transcriptase. In the starting B16-19 RNA, the last nucleotides prior to the primer are AAAG, and this set of three A's is where the poly(A) expansion takes place. These mutations appear in lineages that are destined for extinction, which often, but not always, evolved by the following sequence of events: appearance of a smear above the 187-bp band, intensifying and lengthening of the smear, loss of the 187-bp band entirely, and then eventual fading away of the population as a whole. In some lineages the smear appeared quite early. For example, the smear appeared by the 10th burst of one of the 100-molecule lineages (4V), and the lineage was declared extinct by burst 18. Cloned individuals from burst 13 of this population revealed that most molecules contained between 0 and 150 additional A's, although cloning could be biased against recovery of the longest mutants (Figure 5B).
The floodgate (G135 → A): In some mutants, the terminal guanosine prior to the reverse transcriptase primer-binding site was missing. All of these mutants possessed long poly(A) tracts as well. Although a few poly(A) mutated RNAs were found not to contain the G135 → A mutation, its presence was so clearly associated with the existence of long poly(A) tracts that we termed this mutation the “floodgate.” When it occurs, the mechanism to produce polyadenylation is apparently greatly enhanced.
The immunity: While the first two classes of mutations were associated with lineages destined for extinction, two types of advantageous mutations were occasionally observed. The first is the conversion of the 10-nt sequence CUGAACCUUA (from positions 123 to 132) to the 5-nt sequence AAUCG. This mutation appeared in all four lineages that survived to 50 bursts. Because this mutation results in the creation of a unique TaqαI restriction site in the PCR DNA, it was possible to survey its frequency in any given burst, and we did so in all 25 lineages at all bursts prior either to death or to the appearance of a poly(A) smear. It was detected only once in the 21 lineages that died prior to burst 50, and that was in the last few bursts of the 3000-molecule lineage that died at burst 49. On the other hand, it became fixed between bursts 15 and 20 in the two surviving 600-molecule lineages and fixed between bursts 5 and 10 in the two surviving 3000-molecule lineages. Thus this mutation appears to provide immunity against poly(A) tract formation, by an unknown mechanism.
The insurance (U62 → A): The final common mutation that we encountered was U62 → A. This mutation was found in all six 3000-molecule lineages, in three 600-molecule lineages, in two 300-molecule lineages, and in one 100-molecule lineage. Its appearance was related to that of the immunity mutation. It became fixed in all four surviving lineages and in the one 3000-molecule lineage that survived to burst 49, but in only one other lineage. In the other six lineages where it appeared, it was present at a low frequency in a given population (i.e., <10%) and often persisted for only a few bursts before disappearing. When it appeared in the four lineages that survived to 50 bursts, the U62 → A mutation appeared after the establishment of the immunity mutation (Figure 5C). While the beneficial mechanism of the immunity mutation is uncertain, that of the insurance mutation is likely related to RNA folding. In RNAs with the immunity mutation, a misfold is possible in the secondary structure if nucleotides 63–70 make an 8-bp pseudoknot interaction with nucleotides 124–131. The U62 → A mutation interrupts this 8-bp stretch in the middle, greatly diminishing its contribution to the free energy of the overall fold. Without the insurance mutation, the base-paired section contributes ∼ −4.5 kcal/mol of free energy to the overall folded conformation at 37°, but with the mutation the contribution drops to −0.1 kcal/mol, or practically nothing.
The mutations observed here are examples, at the raw molecular level, of mutational loads and of epistatic responses to counteract them. The polyadenylation happens gradually, as the lengths of the smears increase with generational time (Figure 3). The cause of this process is most likely slippage by the reverse transcriptase as it attempts to copy the three adenosines immediately past its primer. As each additional adenosine is incorporated, the chances for further slippage increase, and the accumulation of adenosines accelerates. The poly(A) tracts do not completely inhibit ligase activity, but they can contribute to a genetic load. As the molecule gets longer, more time is needed for both reverse and forward transcription, and even small increments in these times can lead to lower fitness in the CE environment. Moreover, should the number of adenosines in the poly(A) tract greatly exceed the size of the rest of the molecule, two additional negative fitness consequences could arise. First, the enlarged 3′ end of the molecule may interfere with proper folding. Second, the large numbers of adenines in the RNAs can deplete the dTTP pool for reverse transcription and the ATP pool for forward transcription, lowering the reproduction rates for all members of the population. This scenario bears resemblance to genetic deteriorations such as triplet repeat expansions that accompany fragile-X syndrome and Huntington's chorea. In our system, the load-producing process seems hastened by the appearance of floodgate mutations, but it is retarded or prevented by the appearance of the immunity and insurance mutations, events more probable in populations with a larger effective size. Note that the effective population sizes of fluctuating populations such as these are determined by the harmonic mean of the bottleneck sizes, meaning that the smaller lineages such as the 100-molecule lineage indeed have less genetic variability from which to draw.
As a control for the advantageous nature of the immunity mutation, we carried for 40 bursts two 100-molecule lineages initiated with a pure population of RNAs containing this mutation but no other. Both lineages survived to burst 40 with no poly(A) tract formation evident, while all 100-molecule lineages initiated with B16-19 developed poly(A) smears by burst 22 and went extinct by burst 34 (Table 1). We also initiated two lineages with pure populations of B16-19 ribozymes with 23-nt poly(A) tails in place, and these both developed fatal smears within 6 bursts (data not shown).
The four classes of mutations described above were by far the most frequent that we observed in our lineages. The batch sequence analyses of later bursts from all lineages did not reveal any other mutation at a frequency detectable by such methodology (i.e., >10%). The sequence analysis of >50 clones from several bursts of these two lineages also failed to detect any other common mutations. Aside from a few examples of a C123 → A mutation in lineage 4V clones from burst 13 and a single example of a U124 → C mutation in a 4V clone from burst 14, we did not observe any other mutations. One explanation for this is that the starting B16-19 ligase was highly adapted and is likely in a strong local fitness optimum (Lehman 2004). In fact, if the secondary structure in Figure 2 is correct, then less than half of the ligase's nucleotides are not base paired (62 of 135), and some of these are involved in tertiary contacts (Bergman et al. 2004). Thus relatively little of the molecule is actually free to tolerate mildly deleterious mutations; all of the ones that we did see lay in non-base-paired regions (Figure 2B). The high frequencies of the four major classes of mutations imply that sequencing errors (e.g., Taq polymerase infidelity) were not a concern in our study.
An important consideration in this study was to guard against the possibility that the results were not simply the result of stochastic sampling failures. If sheer numbers of RNA molecules were being lost by sampling dilute solutions each burst regardless of genotype, then one might expect premature deaths for the smaller populations. We have at least two strong pieces of evidence that this was not happening. First, the lineages do not go extinct suddenly, as would be predicted if RNA was simply not being transferred from burst to burst. The lineages tend to die out gradually, as the cDNA in their composite populations becomes less concentrated and has a less robust input for the PCR, akin to the process of qPCR. Second, and more importantly, the specific mutations we observe in the lineages are correlated either to lineage survival or to extinction. If transfer failures were the ultimate cause of lineage extinction, then the appearance of, say, the immunity mutation in all four surviving lineages (and only once elsewhere) would not be expected.
To analyze more critically the fitness of some of the specific RNAs, we performed comparative kinetic assays on four important RNA genotypes in isolation. We assayed the pseudo-first-order catalytic rate constant kcat for these ribozymes as a measure of their relative reproductive rates. This parameter estimates the speed at which substrate oligomers are covalently ligated to the ligase ribozymes once bound, the critical step in the reproductive cycle (Figure 1). Because these ligases are among the fastest known non-self-cleaving catalytic RNAs (Wright and Joyce 1997; Schmitt and Lehman 1999; Bergman et al. 2000), these rate assays needed to be performed outside the CE milieu and at room temperature to slow down the reaction enough to get accurate measurements. We did, however, assay them in the salt and pH conditions that they experienced during CE. For a ribozyme with no mutations (B16-19), for one with just the immunity mutation, for one with both the immunity and the insurance mutations, and for one with 23 additional 3′ A's (but no other mutations), we measured rate constants of 13 min−1, 11 min−1, 18 min−1, and 6.6 min−1, respectively. These data indicate that the immunity mutation does not engender a faster ribozyme, but that a poly(A) tail of 23 A's slows the ribozyme to half of its wild-type rate. The insurance mutation, which we propose lessens the probability of misfolding of RNAs with the immunity mutation, may slightly enhance the catalytic rate. In any event, the rate constants provide corroboration of the notion that 3′ polyadenylation is a slightly deleterious mutation for these ribozymes. To our knowledge, such direct assessment of the fitness implications of specific genotypes that arise during intense selection regimes has rarely been reported, with the exception perhaps of those seen in RNA viruses (e.g., Sanjuan et al. 2004; Heineman et al. 2005).
Our data demonstrate that RNA populations can accumulate a genetic load in an abiotic environment. The observed mutations can be ascribed to concrete biochemical events that affect fitness. These data reiterate empirical evidence that small population sizes and population bottlenecks magnify the efficacy of random genetic drift, mechanisms proposed to spark significant evolutionary transitions within lineages (Lynch and Conery 2003). New genotypes arise via high mutation rates imposed in a strictly “asexual” (here, nonrecombining) mode of reproduction. There is little chance of recombination by template jumping by reverse transcriptase (Hu and Temin 1990; Negroni and Buc 2001) because RNA concentrations are so low in these CE experiments (e.g., 5 zmol in 25 μl = 2 × 10−10 μm). It remains to be seen whether the loads in these populations can be ameliorated by recombination, which could in theory be deliberately introduced in vitro (Lehman and Unrau 2005).
While mutation accumulation and mutational meltdown have been documented for extant populations of organisms, the conditions that promote genetic loads and mutational meltdowns would have been especially prevalent during an RNA world, before the advent of cellular life. Naked RNA molecules evolving on the primitive earth would have suffered high mutation rates for replication, initially low population sizes, and extremely rugged fitness landscapes, all of which would have exacerbated the accumulation of deleterious mutations. Although computer simulations show that compensatory and phenotypically neutral mutations can relax the error threshold for larger populations (tens of thousands) of catalytic RNAs (Kun et al. 2005), our experimental results show that small populations would still be at risk for accumulation of sublethal mutations. It is likely, then, that recombination would have been needed early in the history of life—even before the advent of cellular life and true sexual reproduction—as a means to maintain high-fitness genotypes.
Acknowledgments
We thank B. Lehman, A. Krummel, R. I. Barr, and K. Rusterholtz for technical support; A. Burton, S. Estes, and E. Hayden for useful discussions; and the National Science Foundation for funding (DEB-0315286 to N.L.).
References
- Bergman, N. H., W. K. Johnston and D. P. Bartel, 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123. [DOI] [PubMed] [Google Scholar]
- Bergman, N. H., N. C. Lau, V. Lehnert, E. Westhof and D. P. Bartel, 2004. The three-dimensional architecture of the class I ligase ribozyme. RNA 10: 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, E. K., A. D. Peters and P. D. Keightley, 1999. High frequency of cryptic deleterious mutations in Caenorhabditis elegans. Science 285: 1748–1751. [DOI] [PubMed] [Google Scholar]
- De Angioletti, M., A. Rovira, M. Sadelain, L. Luzzatto and R. Notaro, 2002. Frequency of missense mutation in the coding region of a eukaryotic gene transferred by retroviral vectors. J. Virol. 76: 1991–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, M. Lynch and W. K. Thomas, 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. [DOI] [PubMed] [Google Scholar]
- Estes, S., P. C. Phillips, D. R. Denver, W. K. Thomas and M. Lynch, 2004. Mutation accumulation in populations of varying size: the distribution of mutational effects for fitness correlates in Caenorhabditis elegans. Genetics 166: 1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, W., M. Lynch and R. Bürger, 1993. Muller's ratchet and mutational meltdowns. Evolution 47: 1744–1757. [DOI] [PubMed] [Google Scholar]
- Gesteland, R. F., T. R. Cech and J. F. Atkins, 2005. The RNA World, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Gilbert, W., 1986. The RNA world. Nature 319: 618. [Google Scholar]
- Haldane, J. B. S., 1929. The origin of life. Ration. Ann. 3: 3–10. [Google Scholar]
- Haldane, J. B. S., 1937. The effect of variation on fitness. Am. Nat. 71: 337–349. [Google Scholar]
- Heineman, R. H., I. J. Molineux and J. J. Bull, 2005. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J. Mol. Evol. 61: 181–191. [DOI] [PubMed] [Google Scholar]
- Hu, W. S., and H. M. Temin, 1990. Retroviral recombination and reverse transcription. Science 250: 1227–1233. [DOI] [PubMed] [Google Scholar]
- Johns, G. C., and G. F. Joyce, 2005. The promise and peril of continuous in vitro evolution. J. Mol. Evol. 61: 253–263. [DOI] [PubMed] [Google Scholar]
- Johnston, W. K., P. J. Unrau, M. S. Lawrence, M. E. Glasner and D. P. Bartel, 2001. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292: 1319–1325. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and A. Eyre-Walker, 2000. Deleterious mutations and the evolution of sex. Science 290: 331–333. [DOI] [PubMed] [Google Scholar]
- Kibota, T. T., and M. Lynch, 1996. Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature 381: 694–696. [DOI] [PubMed] [Google Scholar]
- Kun, A., M. Santos and E. Szathmáry, 2005. Real ribozymes suggest a relaxed error threshold. Nat. Genet. 37: 1008–1011. [DOI] [PubMed] [Google Scholar]
- Lehman, N., 2004. Assessing the likelihood of recurrence during RNA evolution in vitro. Artif. Life 10: 1–22. [DOI] [PubMed] [Google Scholar]
- Lehman, N., and P. J. Unrau, 2005. Recombination during in vitro evolution. J. Mol. Evol. 61: 245–254. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2003. The origins of genome complexity. Science 302: 1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and W. Gabriel, 1990. Mutation load and the survival of small populations. Evolution 44: 1725–1737. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA.
- Lynch, M., R. Burger, D. Butcher and W. Gabriel, 1993. The mutational meltdown in asexual populations. J. Hered. 84: 339–344. [DOI] [PubMed] [Google Scholar]
- Lynch, M., J. Blanchard, D. Houle, T. Kibota, S. Schultz et al., 1999. Perspective: spontaneous deleterious mutation. Evolution 53: 645–663. [DOI] [PubMed] [Google Scholar]
- Mukai, T., 1964. The genetic structure of populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 72: 335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1950. Our load of mutations. Am. J. Hum. Genet. 2: 111–176. [PMC free article] [PubMed] [Google Scholar]
- Negroni, M., and H. Buc, 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35: 275–302. [DOI] [PubMed] [Google Scholar]
- Pfrender, M. E., and M. Lynch, 2000. Quantitative genetic variation in Daphnia: temporal changes in genetic architecture. Evolution 54: 1502–1509. [DOI] [PubMed] [Google Scholar]
- Sanjuan, R., A. Moya and S. F. Elena, 2004. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc. Natl. Acad. Sci. USA 101: 15376–15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, T., and N. Lehman, 1999. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem. Biol. 6: 857–869. [DOI] [PubMed] [Google Scholar]
- Schultz, S. T., M. Lynch and J. H. Willis, 1999. Spontaneous deleterious mutation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 11393–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. R., and O. Pereira-Smith, 1977. Colony size distribution as a measure of age in cultured human cells. A brief note. Mech. Ageing Dev. 6: 283–286. [DOI] [PubMed] [Google Scholar]
- Tagaki, Y., and M. Yoshida, 1980. Clonal death associated with the number of fissions in Paramecium caudatum. J. Cell Sci. 41: 177–191. [DOI] [PubMed] [Google Scholar]
- Wallace, B., 1987. Fifty years of genetic load. J. Hered. 78: 134–142. [DOI] [PubMed] [Google Scholar]
- Wright, M. C., and G. F. Joyce, 1997. Continuous in vitro evolution of catalytic function. Science 276: 614–617. [DOI] [PubMed] [Google Scholar]
- Zeyl, C., M. Mizesko and J. A. G. M. deVisser, 2001. Mutational meltdown in laboratory yeast populations. Evolution 55: 909–917. [DOI] [PubMed] [Google Scholar]
- Zuker, M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]