Abstract
The symbiotic relationship between the mitochondrial and nuclear genomes coordinates metabolic energy production and is fundamental to life among eukaryotes. Consequently, there is potential for strong selection to shape interactions between these two genomes. Substantial research attention has focused on the possibility that within-population sequence polymorphism in mitochondrial DNA (mtDNA) is maintained by mitonuclear fitness interactions. Early theory predicted that selection will often eliminate mitochondrial polymorphisms. However, recent models demonstrate that intergenomic interactions can promote the maintenance of polymorphism, especially if the nuclear genes involved are linked to the X chromosome. Most empirical studies to date that have assessed cytonuclear fitness interactions have studied variation across populations and it is still unclear how general and strong such interactions are within populations. We experimentally tested for cytonuclear interactions within a laboratory population of Drosophila melanogaster using 25 randomly sampled cytoplasmic genomes, expressed in three different haploid nuclear genetic backgrounds, while eliminating confounding effects of intracellular bacteria (e.g., Wolbachia). We found sizable cytonuclear fitness interactions within this population and present limited evidence suggesting that these effects were sex specific. Moreover, the relative fitness of cytonuclear genotypes was environment specific. Sequencing of mtDNA (2752 bp) revealed polymorphism within the population, suggesting that the observed cytoplasmic genetic effects may be mitochondrial in origin.
DESPITE an increasing interest among evolutionary biologists in the role of the mitochondrial genome in adaptive evolution, only a limited number of studies has tested the effect of mitochondrial polymorphism on life-history traits and fitness (reviewed in Ballard and Whitlock 2004; Gemmell et al. 2004; Rand et al. 2004). In fact, variation in mitochondrial genes by tradition has been assumed to be selectively neutral (see Rand 2001; Ballard and Whitlock 2004; Ballard and Rand 2005). This is perhaps surprising, because the interplay between the mitochondrial and nuclear genomes of eukaryotes undoubtedly represents one of the most important symbioses among living organisms. Together, the two genomes coordinate the cellular function fundamental to energy production, and there is thus large potential for selection to shape their integration (Ballard and Rand 2005).
Early theoretical work realized that simple, additive genetic fitness variation will be low among mitochondrial haplotypes within a population because detrimental and beneficial mutations are rapidly purged and fixed, respectively (Takahata 1984). However, the fact that the mitochondrial genome exhibits sequence polymorphism within populations of some species (Clark 1984), in conjunction with the observation that different mitochondrial DNA (mtDNA) haplotypes can have a significant influence on organismal fitness (reviewed in Gerber et al. 2001; Ballard and Rand 2005), has motivated researchers to explore the underlying processes maintaining such polymorphisms. Substantial research attention has focused on the possibility that this mtDNA sequence polymorphism can be upheld by cytonuclear fitness interactions. Initial theoretical studies suggested that such interactions probably cannot maintain polymorphism in mitochondrial genes within panmictic populations, except under restrictive conditions of frequency-dependent selection or strong sex-specific selection (Clark 1984; Gregorius and Ross 1984; Babcock and Asmussen 1996, 1998). A few empirical studies also confirmed these predictions. For example, Clark and Lyckegaard (1988) found no evidence of cytonuclear fitness interactions among lines of Drosophila melanogaster from within panmictic populations. However, such cytonuclear interactions have been demonstrated among lines of flies created from distinct geographic origins (e.g., Clark 1985; Hiraizumi 1985; Clark and Lyckegaard 1988; James and Ballard 2003). These findings would seem to collectively suggest that polymorphism in cytonuclear genotypes is removed quickly within panmictic populations, but that mitochondrial and nuclear genes do coevolve, such that intergenomic epistatic variation is found when different populations are crossed.
The early theoretical models that examined possibilities for cytonuclear fitness polymorphisms were based on autosomal loci (Clark 1984; Gregorius and Ross 1984; Babcock and Asmussen 1996, 1998). In diploid species where females are the homogametic sex, the patterns of cytoplasm–nuclear chromosome transmission are different for the X chromosome than for the autosomes. On average, X chromosomes cosegregate with the cytoplasm in two-thirds of cases, whereas autosomes cosegregate with the cytoplasm in only half of cases. This increased rate of cotransmission of X chromosomes with cytoplasms may facilitate the transmission of favorable allelic combinations with the cytoplasms. Furthermore, the fact that X chromosomes spend two-thirds of their time in females makes them a hotspot for sex-specific fitness variation (Rice 1984; Gibson et al. 2002). These points motivated Rand et al. (2001) to develop a model of cytonuclear fitness polymorphism, in which the interacting nuclear locus is located on the X chromosome. This model demonstrated that multiple mitochondrial haplotypes can indeed be maintained within populations when interacting nuclear genes are X-linked. Additionally, they provided empirical support that such interactions may occur both among lines of D. melanogaster derived from diverse geographic localities and from lines derived from the same geographic populations. Specifically, these interactions were sex specific and the nuclear genes involved were X-linked.
The above studies that tested for within-population, cytonuclear interactions used chromosome segregation assays (Clark and Lyckegaard 1988; Rand et al. 2001). With this genetic design, females produced two types of offspring: homozygous and heterozygous for the nuclear chromosome copy examined (males were hemizygous when X chromosomes were tested). The heterozygous offspring carried one copy of a genetically marked balancer chromosome. The proportion of hatching homozygous to heterozygous offspring, within each sex, was then used as a joint estimate of segregation and viability (egg-to-adult survival: i.e., juvenile fitness) to test for differences among the assayed chromosomes in each cytoplasm. An advantage of this experimental design is that it is possible to examine differences in offspring sex ratio between crosses and thus test for sex-specific viability. However, the design has two drawbacks. First, it is difficult to evaluate how representative cytonuclear interactions (or the lack thereof) are in outbred populations because results reflect the difference in viability of chromosomes when expressed in their inbred vs. outbred (the homologous chromosome was a genetically marked cytogenetic construct derived from a different population) states in different cytoplasms. Second, although juvenile viability is an important component of fitness, estimates suggest that it composes only ∼15% of the nuclear genetic variation for fitness in D. melanogaster and, furthermore, that sex-specific effects on fitness of particular alleles do not seem to be expressed until the adult stage (see Chippindale et al. 2001; Gibson et al. 2002). Thus, cytonuclear fitness interactions may be more ubiquitous within populations than currently assumed, and there are good reasons to focus on the adult part of the life cycle when studying the effects of mtDNA polymorphisms within populations. A final limitation of previous studies is that they have not attempted to remove potentially confounding effects of cytoplasmic, maternally inherited bacteria, such as Wolbachia, using tetracycline treatment (Clark and Lyckegaard 1988; Rand et al. 2001).
Here, we experimentally test whether there is genetic variation in cytoplasmic genes for female adult fitness within a panmictic population of D. melanogaster. In particular, we examine cytonuclear interactions for fitness. This was achieved by establishing 25 randomly sampled cytoplasmic lines, using backcrossing to disassociate each line from its original nuclear background. These lines were tetracycline treated to remove bacteria such as Wolbachia. The cytoplasm of each line was then expressed in three different haploid nuclear genetic backgrounds derived from the same population, enabling cytonuclear interactions on adult female fitness to be determined following straightforward assays.
MATERIALS AND METHODS
Laboratory flies:
Flies used in this experiment were sampled from a large, outbred laboratory population (LHM) of D. melanogaster. This population is cultured on a 14-day discrete generation cycle. Each generation is started by 56 “juvenile competition vials” (each 10-dram vial contains 10 ml of cornmeal–molasses killed-yeast medium) trimmed to contain 150–200 eggs in each. Larval, pupal, and early adult stages reside in these vials for 11.25 days, at which point a thoroughly mixed sample of 1792 adults from the 56 “juvenile competition vials” are transferred to “adult competition vials” (16 pairs/vial with 10 mg of live yeast added on top of the medium). Eighteen hours before the end of the 14-day generation cycle, the flies are transferred to 56 fresh vials where eggs are laid that will propagate the next generation, so that fecundity during these 18 hr represents lifetime fecundity in this population. At the start of these experiments, this population had adapted to this specific laboratory environment for >300 generations. Flies were reared at 25°, on a 12-hr light:12-hr dark cycle (see Chippindale and Rice 2001 and Friberg et al. 2005 for further details on the LHM population).
Construction of mt lines:
Cytoplasms were sampled from the laboratory population by randomly collecting 25 mated females. Each female was effectively a “mitochondrial Eve” that was used to found a separate “mt line” fixed for her cytoplasmic (cyto)/mitochondrial (mito) type. To accurately discern variation in cytoplasms in relation to fitness, it was necessary to disassociate each sampled cytoplasm from the nuclear background with which it was originally associated. This was achieved via 27 successive generations of backcrossing with males from the outbred LHM base population. For the first 20 generations of backcrossing, 8 daughters were collected from each mt line and mated to 8 random males from the laboratory population. To ensure that sampling error would not create differences in nuclear DNA among mt lines, in generation 21–25, the number of daughters used for each backcross was increased to an average of 70 (range: 42–100) and the number of random base males to 50. At generation 26, each line was increased to contain 150 daughters and 100 random LHM males, distributed equally across five vials. According to diagnostic polymerase chain reaction (PCR) conducted at generation 15, all lines were free from Wolbachia infection. Nonetheless, at generation 26, all larvae were treated with tetracycline hydrochloride to ensure that they were uninfected with cytoplasmic bacteria, such as Wolbachia. From generation 28, the lines were closed and each subsequent generation was propagated in five replicate vials, by 32 pairs/vial, and subsequent egg densities were counted and trimmed to 150–200 eggs/vial.
During the backcrossing, daughters from the mt lines were not collected immediately upon hatching. Most females were therefore already mated (some with their brothers) before they were introduced to random LHM males. This slowed down the rate at which the nuclear DNA, originally associated with the sampled cytoplasms, was replaced. Remating is, however, frequent in this population, ranging from 55 to 98% (W. R. Rice, personal communication), and sperm displacement is high. The average sperm displacement rate reported for D. melanogaster is 93% (Simmons 2001) and 85% for the LHM population (W. R. Rice, personal communication). Sperm displacement was also measured in this population in another experiment that was conducted concurrently with this one and found to be 81% (U. Friberg and D. K. Dowling, unpublished data). To calculate the percentage of the original nuclear background that had been replaced after the 27 generations of backcrossing, we conservatively assumed that all females mated first to a brother, then remated to a random LHM male in 55% of cases, and, when remating, fertilized 80% of the ova with sperm from the second male. This reduces the effective replacement rate of nuclear DNA to 22%/generation, rather than 50%. Assuming that all “mitochondrial Eves” were initially mated to nonrelatives in the first generation, a highly probable scenario, this results in 99.92% of the original nuclear background being replaced at generation 27, which is equivalent to 10 generations of backcrossing using virgin females.
Production of inbred lines:
Inbred lines were created so that the 25 mt lines could be crossed with distinct and controlled nuclear genetic backgrounds. Thirty inbred lines were initiated by collecting and isolating 30 mated females from the LHM population. One virgin daughter and one son were collected from each mated female and crossed. In each generation, a virgin daughter was mated with a full-sib brother. Of the 30 lines, 6 persisted over nine generations of such inbreeding, and 3 of these were randomly chosen to be used in the experimental assays. At this point, the inbreeding coefficient was 0.859 and the probability of fixation of a single allele at any one locus, under the conservative assumption that four alleles were present in the initial mating, was 0.736 (Falconer and Mackay 1996). These three lines were then maintained as populations in which each generation was propagated in 10 replicate vials, by 14 pairs/vial, and subsequent egg densities were counted and trimmed to 150–200 eggs/vial.
Experimental design:
The experiment was conducted in three blocks that were separated in time. To start each block, each mt line and each inbred line was further replicated in two vials of 32 pairs (using 7- to 8-day-old adults) and in 10 vials of 5 pairs (using 1- to 2-day-old adults), respectively. These pairs were provided with fresh vials with yeast each day for 6 days and subsequent egg densities were trimmed to 150–200 eggs in each vial. These eggs were thus propagated by mothers aged between 7 and 13 days for the mt lines and between 1 and 7 days for the inbred lines. Thus, the mt lines to be used in the experiment were propagated by moderate-aged females and the inbred lines by young females.
In each block, 90 virgin adult females were collected from each mt line and divided into three groups of 30. For practical reasons, the first group of 30 females/mt line was collected from mothers aged 7–9 days, the second group from mothers aged 9–11 days, and the third group aged 11–13 days. Each of these groups in turn was subdivided into three vials such that 10 females/vial were stored. At the same time, 750 males were collected from each inbred line and divided into 25 groups of 30 males. Each of these groups was also subdivided so that 10 males/vial were stored. Again for practical reasons, the males of each inbred line were collected from different aged mothers. Specifically, in blocks 1 and 3, inbred line 1 was collected from mothers aged 1–3 days, inbred line 2 from mothers aged 3–5 days, and line 3 from mothers aged 5–7 days. In block 2, inbred line 2 was collected from mothers aged 1–3 days, line 1 from mothers aged 3–5 days, and line 3 from mothers aged 5–7 days.
When 1–2 days old, each group of 30 females (three vials/group) was crossed to a corresponding group of 30, 1- to 2-day-old males from an inbred line (three vials/group) in each possible combination so that there were 75 crosses in total. As indicated above, the 30 pairs for each mt line × inbred line combination were divided into three subgroups of 10 pairs each, and each such subgroup received a separate vial within which they mated and the females laid eggs. Twenty-four hours later, the flies were discarded and egg density was trimmed to 150–200 eggs/vial. Nine days after egg laying, within the first 7 hr of hatching (flies remain virgins for ∼8 hr) but no earlier than 1 hr after hatching, 23 virgin daughters were collected from each cross under light CO2 anesthesia. These females, then, had inherited their cyto/mito type along with a random haploid set of nuclear chromosomes from their mother. The second haploid set of nuclear chromosomes was inherited from their father and can be assumed to be distinct across the three inbred lines (Figure 1). These 23 females/cross were the focal females for this experiment. Their parents were all of identical age (1–2 days), which minimizes the possibility of parental effects confounding the experiment. The grandparents to these focal females, however, were of variable age. Nonetheless, it is unlikely that grandparental effects will confound this experiment and we explain why within the discussion.
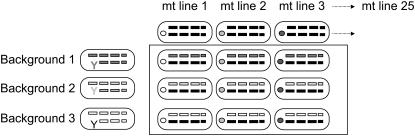
Figure 1.—
Schematic of karyotypes for offspring of the 75 mt line × nuclear background crosses. Small circles, of different shades, within each eclipse indicate particular cyto/mito types. The first set of rectangles on the left side of the eclipse indicates the sex chromosomes: Y denotes the Y chromosome, and a rectangle the X chromosome. The three pairs of rectangles to the right of this are autosomes. The shading of all chromosomes indicates their origin: solid indicates random wild-type chromosomes from the base population and paler shades of three different intensities indicate chromosomes inherited from males of each of the three nuclear backgrounds. Thus, each offspring receives its cyto/mito type from its mt line of origin; 50% of its nuclear DNA is inherited from a specific, controlled nuclear background, while 50% represents random, wild-type nuclear DNA from the base population.
When 4 days old, the 23 focal females collected from each of the 75 crosses were mated (without CO2 anesthesia) to 35 randomly selected 3-day-old males from the LHM base population for exactly 2 hr. Twenty of these mated females were then collected under light anesthesia and placed in individual vials with medium, upon which a standardized amount of live yeast was added (1.6 g dry yeast was thoroughly mixed with 10 ml H2O and then 5 μl was added to each vial before drying overnight, resulting in 0.8 mg live yeast/female). An incision was made in the medium of each vial to further entice females to oviposit. Females were allowed exactly 18 hr to oviposit before they were discarded. This protocol mimicked the rearing conditions for the base population in terms of the average amount of live yeast consumed by females [which is linearly related to the number of eggs laid by females in this population (Orteiza et al. 2005)] and the timing and length of the oviposition period. However, two deviations were made from the standard culturing procedure to avoid interdependencies between females from the same line and block. First, live yeast was provided at the time of oviposition rather then before oviposition and, thus, females did not have to compete over this resource. Second, females were exposed to males for a shorter period of time. Ten days later, offspring emerging from each vial were counted and sex was assigned. All vials were rechecked on day 11 to include any late-hatching offspring in our fitness assays. Thus, the measure of female adult fitness used here is the total number of hatchlings emerged per female by day 11, which is in concurrence with the regular culturing protocol since flies hatching later than this will not contribute to the subsequent generation. Overall, ∼60 broods were scored for each of the 75 mt line × nuclear background crosses, divided into three blocks. Some females (n = 50) did not produce any offspring and were subsequently removed from the final analyses reported below. However, their removal did not qualitatively alter the results. Data were accidentally lost from four mt line × nuclear combinations during block 2. In total, 98,725 offspring from 4370 broods were included in the analyses.
Screening for mtDNA variation:
To probe for mtDNA sequence polymorphism among the 25 mt lines, we sequenced four protein-coding mtDNA gene fragments in (initially) one individual for each mt line. The four fragments consisted of a fragment within CytB, Cox2, and two nonoverlapping fragments within ND5. Primers were designed from the published complete mtDNA sequence of D. melanogaster (Embl entry U37541) using the software PRIMER3 (Rozen and Skaletsky 2000). Primer sequences (all given in 5′–3′ direction) were CytB-F ACCTTTACGAAATTCCCATCC, CytB-R GGGTCTCCCAATAAATTTGGTC, Cox2-F TGGCAGATTAGTGCAATAGATTTAAG, Cox2-R GACCAGTACTTGCTTTCAGTCATC, ND5-I-F TTTGTTCTTATAATTTCTTCTTTAGTGA, ND5-I-R GCCCCAGCACATATAAACAA, ND5-II-F TTTAAAGCATTATTGTTTATATGTGC, and ND5-II-R GACCTCCAAAATATTCTGATCAAC. DNA was extracted from single flies using the Puregene tissue extraction kit (Gentra, Research Triangle Park, NC). PCR was carried out in 20-μl volumes comprising 2 μl of extract, 0.5 units of AmpliTaq Gold (Applied Biosystems, Foster City, CA), 1× GeneAmp PCR buffer II (Applied Biosystems), 0.2 mm of each dNTP, and 0.4 μm of each primer at a final concentration of 2.5 mm MgCl2 (3.75 mm for fragment ND5I). We used the following thermal profile on a Mastercycler gradient instrument (Eppendorf, Madison, WI): 38 cycles at 95° for 30 sec, at 56° for 30 sec (55° for fragment ND5I), and at 72° for 45 sec. Before the first cycle, a prolonged denaturation step (95° for 7 min) was included and the last cycle was followed by an additional annealing step at 55°–56° for 1 min and a final extension step at 72° for 10 min. PCR products were cleaned using ExoSAP-IT (Amersham Biosciences) and sequenced on both strands with the original primers by Macrogen. Forward and reverse electropherograms were checked manually. We used the software MEGA3 (Kumar et al. 2004) for sequence alignment, comparison, and amino acid characterization.
Statistical analysis:
Cytoplasmic and nuclear effects, as well as the interaction between them, on fitness were estimated using the restricted maximum likelihood (REML) algorithm (PROC MIXED statement) in SAS version 9.1. The mt line, nuclear genetic background, and block were modeled as random-effects variables. Cytonuclear effects on female fitness were also examined separately within each block. To test for sex specificity of the effects of the mt line, nuclear background, as well as the interaction between them, on juvenile viability, we conducted a repeated measures general linear model (GLM) in which mt line, background, and block were “between-subjects” factors and sex the “within-subjects” factor. Sex-specific effects were also examined separately within each block.
RESULTS
Cytonuclear fitness effects:
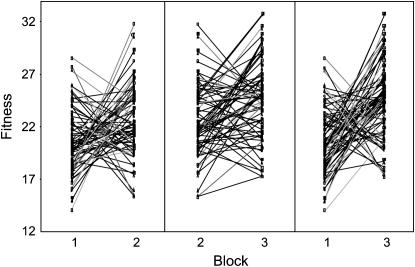
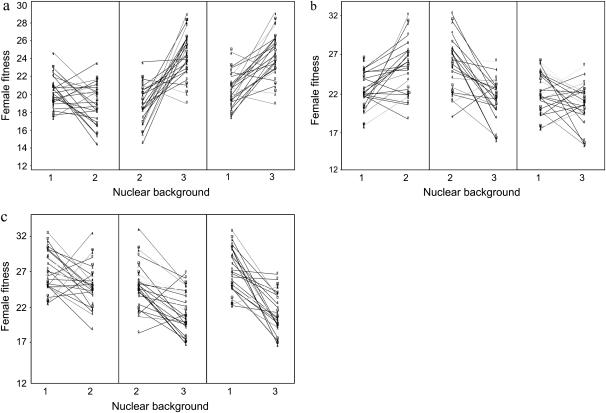
We found no significant main effects of mt line or nuclear background on adult female fitness (Table 1). However, there was a highly significant interaction among mt line, nuclear background, and block on adult female fitness that accounted for ∼8% of the observed phenotypic variance in adult female fitness (95% C.I: 6.1–11.2%) (Table 1). This demonstrates that the relative fitness of specific mt line × nuclear crosses was contingent upon the block in which the assay was conducted (Figure 2). We note that negative variance component estimates were obtained for some model parameters, as indicated in Table 1. Although the REML algorithm is normally considered robust against the potentially biasing effects of negative variance component estimates (e.g., Shaw 1987), we conducted separate analyses within each block to verify the results revealed by the main model. Again, there were no main effects of mt line or nuclear background within each of the three blocks, but highly significant mt line × nuclear interactions that accounted for 4.5% (C.I.: 2.6–9.5%), 12.4% (C.I.: 8.3–20.5%), and 8.5% (C.I.: 5.1–17.0%) of the phenotypic variance in female adult fitness in blocks 1, 2, and 3, respectively (see Table 2). Thus, within each block, some mt lines had high fitness when expressed in some nuclear backgrounds but low fitness when expressed in others (Figure 3). To help elucidate whether there were any consistent patterns in mt line × nuclear effects on fitness across blocks, we reanalyzed the data, including block only as a main effect. In this analysis, we controlled for variation attributable to block while pooling the variation attributable to the mt line × nuclear × block effect with the residual error. This revealed a significant mt line × nuclear background effect on female fitness that accounted for ∼1.6% (C.I.: 0.8–4.3%) of the variation in female fitness (z = 2.49, P = 0.006).
TABLE 1.
Variance component estimates for cytonuclear effects on adult female fitness
| Source | Estimate | Standard error | Z | P | Scaled estimate % total variance |
|---|---|---|---|---|---|
| mt line | 0.175 | 0.284 | 0.62 | 0.269 | 0.31 |
| Background | 0 | — | — | — | — |
| Block | 1.203 | 3.687 | 0.33 | 0.372 | 2.11 |
| mt line × background | 0 | — | — | — | — |
| mt line × block | 0 | — | — | — | — |
| Background × block | 6.470 | 3.897 | 1.66 | 0.048 | 11.37 |
| mt line × background × block | 4.602 | 0.708 | 6.50 | <0.0001 | 8.09 |
| Residual | 44.440 | 0.976 | 45.55 | <0.0001 | 78.12 |
Figure 2.—
Interaction plot of adult female fitness for the 75 mt line × nuclear background combinations (cytonuclear genotypes) for each block. The substantial crossing over of the reaction norms illustrates that the relative order of fitness per cytonuclear genotype differs across blocks.
TABLE 2.
Variance component estimates for cytonuclear effects on female fitness within each block
| Source | Estimate | Standard error | Z | P | Scaled estimate % total variance |
|---|---|---|---|---|---|
| Block 1 | |||||
| mt line | 0 | — | — | — | 0 |
| Background | 7.254 | 7.444 | 0.97 | 0.165 | 13.15 |
| mt line × background | 2.474 | 0.798 | 3.10 | 0.001 | 4.48 |
| Residual | 45.442 | 1.709 | 26.59 | <0.0001 | 82.37 |
| Block 2 | |||||
| mt line | 0 | — | — | — | 0 |
| Background | 4.439 | 4.793 | 0.93 | 0.177 | 8.92 |
| mt line × background | 6.170 | 1.398 | 4.41 | <0.0001 | 12.40 |
| Residual | 39.149 | 1.517 | 25.81 | <0.0001 | 78.68 |
| Block 3 | |||||
| mt line | 0.503 | 1.034 | 0.49 | 0.313 | 0.81 |
| Background | 7.658 | 7.967 | 0.96 | 0.168 | 12.38 |
| mt line × background | 5.263 | 1.584 | 3.32 | 0.0004 | 8.51 |
| Residual | 48.456 | 1.830 | 26.48 | <.0001 | 78.30 |
Figure 3.—
Cytonuclear effects on adult female fitness within each block. Interaction plots for each block of female fitness for the 25 mt lines expressed in three different nuclear genetic backgrounds. The substantial crossing over of reaction norms indicates that the relative order of fitness per mt line changes according to nuclear background: (a) block 1, (b) block 2, and (c) block 3.
Sex-specific effects:
Theory suggests that the maintenance of cytonuclear polymorphisms is promoted by differences between the sexes in cytonuclear fitness ranks (see Introduction). We could not definitively test this possibility here because male adult fitness was not measured. However, following the approach used previously (Clark 1985; Rand et al. 2001), we measured sex-specific viability (i.e., number of eclosing offspring of each sex) of offspring to the focal females. In an overall analysis (across blocks), we found no evidence for sex specificity of the effects of mt lines, nuclear genotypes, or their interaction on viability (see Table 3). However, in separate analyses within each block, sex-specific effects of mt line × nuclear background (block 1) or mt line (block 2) on viability were revealed within two of the blocks (Table 4).
TABLE 3.
Repeated measures GLM testing specifically for sex-specific differences in fitness among cytonuclear genotypes
| Source | d.f. | Sum of squares | F | P |
|---|---|---|---|---|
| Sex | 1 | 108.84 | 9.889 | 0.0017 |
| Sex × block | 2 | 3.44 | 0.156 | 0.8555 |
| Sex × mt line | 24 | 259.39 | 0.982 | 0.4868 |
| Sex × background | 2 | 27.53 | 1.251 | 0.2864 |
| Sex × mt line × block | 48 | 691.91 | 1.310 | 0.0750 |
| Sex × background × block | 4 | 31.94 | 0.726 | 0.5744 |
| Sex × mt line × background | 48 | 456.04 | 0.863 | 0.7363 |
| Error | 4242 | 46,688.67 |
Block, background, and mt line are “between-subjects” factors and offspring sex (Sex) is the “within-subjects” factor. Only interactions involving Sex (the “within-subjects” results) are included in the table because the model was constructed explicitly to test for sex specificity of effects. The “Sex × mt line × background × block” interaction did not resolve and was thus omitted from the analysis.
TABLE 4.
Repeated measures GLM testing specifically for sex-specific differences in fitness between cytonuclear genotypes within each block
| Source | d.f. | Sum of squares | F | P |
|---|---|---|---|---|
| Block 1 | ||||
| Sex | 1 | 40.93 | 4.24 | 0.0398 |
| Sex × mt line | 24 | 189.47 | 0.82 | 0.7180 |
| Sex × background | 2 | 36.98 | 1.91 | 0.1479 |
| Sex × mt line × background | 48 | 634.27 | 1.37 | 0.0498 |
| Error | 1414 | 13,663.65 | ||
| Block 2 | ||||
| Sex | 1 | 42.17 | 3.83 | 0.0505 |
| Sex × mt line | 24 | 428.01 | 1.62 | 0.0296 |
| Sex × background | 2 | 24.78 | 1.13 | 0.3246 |
| Error | 1376 | 15,138.51 | ||
| Block 3 | ||||
| Sex | 1 | 22.45 | 1.83 | 0.1758 |
| Sex × mt line | 24 | 339.81 | 1.16 | 0.2728 |
| Sex × background | 2 | 10.00 | 0.41 | 0.6649 |
| Sex × mt line × background | 48 | 520.79 | 0.89 | 0.6933 |
| Error | 1404 | 17,187.49 | ||
Only the “within-subjects” results are presented. The “Sex × mt line × background” interaction did not resolve in block 2 because the term “background” was unbalanced in the subject stratum (due to missing data in block 2; see materials and methods)
Sequence polymorphism in mtDNA:
We obtained DNA sequences from a total of 2752 bp of mtDNA (701 bp of CytB, 715 bp of Cox2, plus 698 and 638 bp, respectively, for the two ND5 fragments). Sequences have been submitted to the Embl database (accession nos. AM403327–AM403330). The Cox2 and ND5 fragments were monomorphic across all 25 mt lines. However, the CytB fragment harbored variation: site 204 showed a transition (A in mt lines 6 and 11, G in the remaining 23 lines). The variable site was unambiguous in forward and reverse electropherograms and was confirmed by sequencing an additional two individuals from lines 6 and 11. The DNA sequence variation corresponds to an amino acid replacement: Tyr in lines 6 and 11, Cys in the remaining lines.
To assess whether the particular polymorphism detected might have influenced our results, we recoded the 25 mt lines into the two distinct mtDNA variants found and then explored whether there were mtDNA × nuclear effects on female adult fitness, using the same models as described above. These analyses did not reveal any significant additive mtDNA or nuclear effects, or any mitonuclear effects, either across or within blocks. We note, however, that this is a weak test because our classification was based on sequence data from only 14% of the total amount of coding mtDNA of D. melanogaster. Genetic variation outside our sequenced mtDNA domains thus would have been misclassified in these analyses.
DISCUSSION
The cytonuclear interactions that we document support the contention that multiple cyto/mito types can be maintained within a panmictic population. Our results suggest that this maintenance is caused by epistatic interactions between cytoplasmic and nuclear genes, since the fitness of any particular mt line depends upon the nuclear genetic background with which it is coexpressed. Furthermore, the relative fitness of specific cytonuclear genotype combinations is apparently dependent upon the environment in which they persist, as revealed by substantial crossing over of cytonuclear fitness reaction norms across our three blocks. Following standard convention, we ascribe variation attributable to the effect of blocks in our experiment to minor unpredictable environmental heterogeneity across blocks. Moreover, we found limited evidence that the cytonuclear interactions may be sex specific. That is, specific cytonuclear combinations may encode high fitness in one sex but low fitness in the other. However, we note that our measure of sex-specific effects was restricted to juvenile viability and that these effects were inconsistent across blocks. It is also worth noting that we failed to find any main effects of cyto type or nuclear genetic type on fitness. Taken together, these results are consistent with, and extend upon, earlier work that has explored the possible role that cytonuclear fitness interactions play in maintaining mitochondrial haplotype polymorphism within populations (Clark 1984; Gregorius and Ross 1984; Clark and Lyckegaard 1988; Babcock and Asmussen 1996, 1998; Rand et al. 2001).
Multiple studies have demonstrated cytonuclear effects on fitness when using cytonuclear genotypes constructed from strains of different geographic origins (Clark 1985; Hiraizumi 1985; Clark and Lyckegaard 1988; James and Ballard 2003; Zeyl et al. 2005; Dowling et al. 2007). However, very few studies have tested whether fitness variation occurs among cytonuclear genotypes within a population. Clark and Lyckegaard (1988) assessed the potential for interactions between cytoplasms and the second autosomal chromosome in D. melanogaster, but found no evidence of such interactions. This result was consistent with theoretical models between cytoplasms and a polymorphic autosomal locus (Gregorius and Ross 1984; Babcock and Asmussen 1996, 1998). In another study, Rand et al. (2001) assessed interactions between cytoplasms and the X chromosome, motivated by the fact that the X chromosome cotransmits with the cytoplasm more frequently than do the autosomal chromosomes. This extension upon previous studies was logical because theoretical and empirical study has shown that the X chromosome is enriched with sexually antagonistic fitness variation (Rice 1984; Gibson et al. 2002). Rand et al. (2001) found sex-specific, cytonuclear fitness interactions in two of three populations assayed (however, one of these two populations was infected with Wolbachia). The nuclear genes involved in these interactions were X-linked and, consistent with theory, there was some evidence of sex-specific fitness effects according to cytoplasmic and nuclear genes. Thus, from these studies it appears that cytonuclear fitness interactions may be maintained in specific circumstances, namely when the nuclear genes involved are X-linked and consequently under sex-specific viability selection.
In this study, we used a different approach to assess the potential for intrapopulation cytonuclear fitness interactions and addressed whether such interactions are more ubiquitous and stronger than currently assumed. First, we studied a more comprehensive measure of fitness (adult fitness instead of segregation and viability, i.e., juvenile fitness). Estimates suggest that the adult stage accounts for more than five times the amount of genetic variation for fitness than does the juvenile stage in D. melanogaster (Gibson et al. 2002). Furthermore, there is no detectable ontogenetic sexual conflict involving the X chromosome at the juvenile stage, but there is a clear conflict at the adult stage [the X chromosome accounts for 97% of genomewide sexually antagonistic variation (Chippindale et al. 2001; Gibson et al. 2002)]. Thus, our approach should have provided higher power in terms of detecting cytonuclear fitness interactions in general. Second, our method of using distinct haploid genomes as genetic backgrounds provided increased power over previous studies, because genetic interactions were assessed over the whole genome compared to only a particular autosome or X chromosome (cf. Clark and Lyckegaard 1988; Rand et al. 2001). Furthermore, we avoided the usage of inbred chromosomes and thus tested for cytonuclear interactions across cytonuclear variants that actually occur “naturally” within our laboratory population. Our approach also avoided problems associated with using cytogenetic constructs, like balancer chromosomes, in the flies scored for fitness. The main limitation of our approach, of course, is that we were unable to identify the location(s) of nuclear genes involved in these interactions (i.e., autosomal or X-linked).
Our results also revealed that the relative fitness of a joint cytonuclear genotype is dependent upon the environment in which it is expressed, as shown by differences in cytonuclear fitness ranks across the three blocks. The D. melanogaster population (LHM) that we studied has been cultured under controlled environmental conditions for >10 years. Laboratory conditions were also controlled during our experiments and average adult female fitness was indeed similar across the three blocks. Nonetheless, the minor environmental heterogeneity that always occurs across blocks in terms of temperature, humidity, and/or food quality was apparently sufficient to generate differences in the relative fitness ranks of the 75 cytonuclear genotypes studied here. Genotype by environment interactions for fitness are common (Remold and Lenski 2001), but their general ability to maintain genetic variation when variation is temporal is disputed (reviewed in Hedrick 1986). Theory predicts that a temporally fluctuating environment can maintain genetic variation only under special conditions and that these conditions are particularly unlikely when generations are nonoverlapping (see Roff 1997), such as in the LHM population studied here. A possible exception is when expression of the genes in question is sex limited (Sasaki and Ellner 1997; Reinhold 2000). In contrast to temporal variation, spatial variation in selection and division of populations into linked subgroups may play a more important role for the maintenance of genetic variation when migration between demes is limited (see Hedrick 1986), although we are unaware of any theory directly addressing the maintenance of mitochondrial genetic variation in fitness by cytonuclear interactions under spatially varying selection. Because the LHM population is subdivided during both larval and adult competition (see above), it is possible that spatial variation in selection may have contributed at least to the maintenance of the cytonuclear interactions seen in our assays. However, the fact that environmental differences across competition vials should be relatively minor, combined with the fact that mixing of individuals between “competitive demes” is extensive between generations (i.e., very high “migration” between vials), casts doubt on this possibility.
We also assessed whether the cytonuclear fitness effects documented in this study may be upheld by sex-specific selection and found some evidence of sex-specific viability attributable to mt line or the mt line × nuclear background interaction in two of three blocks. Thus, cytonuclear effects were seemingly sex specific in two of three blocks. This is consistent with the theory postulating that sex-specific viability selection is a key mechanism behind cytonuclear fitness interactions. It is important to note that our test for sex specificity was conservative and not ideal for two reasons. First, we did not test for sex-specific interactions in adult flies and, second, the cytonuclear genotype of the offspring (being tested for sex-specific viability) differed partially from that of their mother's (the focal female's) genotype. The offspring (both sons and daughters) had inherited their mother's mt haplotype, but only half of the controlled haploid genome derived from their maternal grandfather. There was thus a 50% reduction in controlled nuclear background in the offspring compared to the focal females. Although this should not create any directional biases among crosses, it will reduce the power by which sex-specific cytonuclear interactions for viability can be detected. Thus, although the sex-specific effects that we found were inconsistent, they suggest that the X chromosome may be more likely to be involved in these cytonuclear interactions than the autosomes, and this is consistent with recent theory and empirical research (Rand et al. 2001).
Although 25 cytoplasms were sampled from the population, only three haploid nuclear genetic backgrounds were used, which compose only a minute fraction of all possible nuclear backgrounds. In addition, we controlled for only 50% of the nuclear genome in the focal females. Consequently, it is difficult to estimate the absolute magnitude of intergenomic fitness interactions. Despite these limitations, the interaction between mt line and nuclear background was significant, accounting for at least 3–8% (lower 95% C.I.'s) of total variation in fitness within blocks. Although these values are highly relevant in terms of the genetic interactions occurring in this population, it is important to recognize that they do not provide exact quantitative estimates of naturally occurring variation for fitness explained by such interactions.
We found no difference in fitness among mt lines per se. Moreover, no evidence was found for additive differences among the nuclear backgrounds, despite extensive testing of these haploid genomes (1460–1500 females were tested for each). This was somewhat unexpected, because previous research has shown that there is a substantial amount of additive genetic variation for female fitness in this population (Gibson et al. 2002). One possibility for the apparent lack of additive differences among the nuclear backgrounds is, of course, that the set of three haploid genomes that we employed, by chance, had similar breeding values for female fitness.
We acknowledge a possible alternative explanation for these results. Although we have clearly identified cytoplasmic effects, the cyto × nuclear interactions can also be explained by cyto × grandparental age effects interactions. However, we believe that it is unlikely that such grandparental effects confounded our interpretation of the results. As explained in materials and methods, the flies sampled from the mt and nuclear backgrounds (inbred lines) at the outset of the experiment had been propagated by parents of different ages (the age of which covaried with the nuclear background being tested). Notably, the parents propagating the inbred lines were all relatively young (1–7 days old) and those propagating the mt lines were all of moderate age (7–13 days). Moreover, they were not the parents of the focal females, but rather the grandparents. Although Hercus and Hoffmann (2000) provided evidence for grandmaternal effects in Drosophila, their experiment compared differences in egg hatching when the grandmothers differed in age by >20 days (compared to an average of 4 days over the first to third nuclear backgrounds and 4 days per set of mt lines over the three nuclear backgrounds in our experiment). Furthermore, if female fitness was to decrease with increasing grandparental age, we would expect both a general decrease in fitness from nuclear backgrounds 1–3, as well as a decrease in fitness per mt line over the three backgrounds. Yet these effects were not observed over the whole data set or within blocks. There was no consistent tendency for fitness to decline over the three backgrounds. In sum, we can confidently conclude that the grandparental age is unlikely to have confounded the results.
Although we cannot completely rule out the existence of some other factor varying among our mt lines, there are very good reasons to assume that the effects revealed here reflect variation in mitochondrial genes. First, careful attempts were made to control for potential confounding effects of the nuclear genome by extensive backcrossing to the base population. Second, screening via diagnostic PCR followed by precautionary tetracycline treatment of all lines eliminated the possibility of cytoplasmic bacteria, such as Wolbachia, confounding the results. Finally, although we sequenced only ∼14% of the total amount of coding mtDNA, we found nucleotide variation with phenotypic effects among the 25 mt lines, with no signs of heteroplasmy [commonly found for length differences of the mtDNA control region in laboratory populations of D. melanogaster; (Townsend and Rand 2004)]. This confirms the presence of mtDNA polymorphism in our population, which is a prerequisite for mitochondrial genetic effects. Nevertheless, the potential exists that the cytonuclear fitness effects demonstrated in this study may be caused or confounded by variation among lines in infection by a host of cytoplasmically transmitted but unknown viruses, such as sigma and C viruses (Clark 1985).
Overall, this study provides some of the first evidence that the maintenance of cytoplasmic (most likely mitochondrial) polymorphism within a panmictic population is likely to be promoted by cytonuclear fitness interactions. This result is especially notable, considering that North American populations of D. melanogaster (and by implication, our study population) apparently possess lower levels of polymorphism in nucleotide diversity of mtDNA than populations from other geographic regions (see Solignac 2004). Thus, the tests presented here may actually be conservative, with the implication that cytonuclear fitness interactions may be both ubiquitous and sizable within panmictic populations.
Acknowledgments
We thank William R. Rice for kindly providing the LHM population and Fleur Champion de Crespigny for conducting diagnostic PCR for Wolbachia on the fly lines. We thank Alexei Maklakov for helpful discussions, Magdalena Nystrand for statistical assistance, the editor and referees for their valuable comments on the manuscript, and the subdepartment of Animal Ecology, Uppsala University, for supporting this work. D.K.D. was funded by a Wenner-Gren Foundations postdoctoral scholarship and a Swedish Institute Guest Scholarship. The study was supported by grants from Magn. Bergvalls Stiftelse and Stiftelsen Lars Hiertas Minne to U.F., from Stiftelsen för zoologisk forskning, Uppsala University, to D.K.D., and from the Swedish Research Council to G.A.
References
- Babcock, C. S., and M. A. Asmussen, 1996. Effects of differential selection in the sexes on cytonuclear polymorphism and disequilibria. Genetics 144: 839–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock, C. S., and M. A. Asmussen, 1998. Effects of differential selection in the sexes on cytonuclear dynamics: life stages with sex differences. Genetics 149: 2063–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard, J. W. O., and D. M. Rand, 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36: 621–642. [Google Scholar]
- Ballard, J. W. O., and M. C. Whitlock, 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13: 729–744. [DOI] [PubMed] [Google Scholar]
- Chippindale, A. K., and W. R. Rice, 2001. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98: 5677–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale, A. K., J. R. Gibson and W. R. Rice, 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98: 1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., 1984. Natural selection with nuclear and cytoplasmic transmission. I. A deterministic model. Genetics 107: 679–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., 1985. Natural selection with nuclear and cytoplasmic transmission. II. tests with Drosophila from diverse populations. Genetics 111: 97–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., and E. M. S. Lyckegaard, 1988. Natural selection with nuclear and cytoplasmic transmission. III. Joint analysis of segregation and mtDNA in Drosophila melanogaster. Genetics 118: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, D. K., K. Chávez Abiega and G. Arnqvist, 2007. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution (in press). [DOI] [PubMed]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman Harlow, Essex, UK.
- Friberg, U., T. A. Lew, P. G. Byrne and W. R. Rice, 2005. Assessing the potential for an ongoing arms race within and between the sexes: selection and heritable variation. Evolution 59: 1540–1551. [PubMed] [Google Scholar]
- Gemmell, N. J., V. J. Metcalf and F. W. Allendorf, 2004. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol. 19: 238–244. [DOI] [PubMed] [Google Scholar]
- Gerber, A. S., R. Loggins, S. Kumar and T. E. Dowling, 2001. Does nonneutral evolution shape observed patterns of DNA variation in animal mitochondrial genomes? Annu. Rev. Genet. 35: 539–566. [DOI] [PubMed] [Google Scholar]
- Gibson, J. R., A. K. Chippindale and W. R. Rice, 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. R. Soc. Lond. Ser. B 269: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorius, H.-R., and M. D. Ross, 1984. Selection with gene-cytoplasm interactions. I. Maintenance of cytoplasm polymorphisms. Genetics 107: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W., 1986. Genetic polymorphism in heterogenous environments: a decade later. Annu. Rev. Ecol. Syst. 17: 535–566. [Google Scholar]
- Hercus, M. J., and A. A. Hoffmann, 2000. Maternal and grandmaternal age influence offspring fitness in Drosophila. Proc. R. Soc. Lond. Ser. B 267: 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraizumi, Y., 1985. Genetics of factors affecting the life history of Drosophila melanogaster. I. Female productivity. Genetics 110: 452–464. [PMC free article] [PubMed] [Google Scholar]
- James, A. C., and J. W. O. Ballard, 2003. Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 164: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Orteiza, N., J. E. Linder and W. R. Rice, 2005. Sexy sons from re-mating do not recoup the direct costs of harmful male interactions in the Drosophila melanogaster laboratory model system. J. Evol. Biol. 18: 1315–1323. [DOI] [PubMed] [Google Scholar]
- Rand, D. M., 2001. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 32: 415–448. [Google Scholar]
- Rand, D. M., A. G. Clark and L. M. Kann, 2001. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics 159: 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, D. M., R. A. Haney and A. J. Fry, 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19: 645–653. [DOI] [PubMed] [Google Scholar]
- Reinhold, K., 2000. Maintenance of a genetic polymorphism by fluctuating selection on sex-limited traits. J. Evol. Biol. 13: 1009–1014. [Google Scholar]
- Remold, S. K., and R. E. Lenski, 2001. Contribution of individual random mutations to genotype-by-environment interactions in Escherichia coli. Proc. Natl. Acad. Sci. USA 98: 11388–11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Roff, D. A., 1997. Evolutionary Quantitative Genetics. Chapman & Hall, New York.
- Rozen, S., and H. J. Skaletsky, 2000. PRIMER3, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Clifton, NJ (http://www-genome.wi.mit.edu/genome_software/other/primer3.html). [DOI] [PubMed]
- Sasaki, A., and S. Ellner, 1997. Quantitative genetic variance maintained by fluctuating selection with overlapping generations: variance components and covariances. Evolution 51: 682–696. [DOI] [PubMed] [Google Scholar]
- Shaw, R. G., 1987. Maximum-likelihood approaches applied to quantitative genetics of natural populations. Evolution 41: 812–826. [DOI] [PubMed] [Google Scholar]
- Simmons, L. W., 2001. Sperm Competition and Its Evolutionary Consequences in the Insects. Princeton University Press, Princeton, NJ.
- Solignac, M., 2004. Mitochondrial DNA in the Drosophila melanogaster complex. Genetica 120: 41–50. [DOI] [PubMed] [Google Scholar]
- Takahata, N., 1984. A model of extranuclear genomes and the substitution rate under within-generation selection. Genet. Res. 44: 109–116. [Google Scholar]
- Townsend, J. P., and D. M. Rand, 2004. Mitochondrial genome size variation in New World and Old World populations of Drosophila melanogaster. Heredity 93: 98–103. [DOI] [PubMed] [Google Scholar]
- Zeyl, C., B. Andreson and E. Weninck, 2005. Nuclear-mitochondrial epistasis for fitness in Saccharomyces cerevisiae. Evolution 59: 910–914. [PubMed] [Google Scholar]