Abstract
The human genome is composed of long stretches of DNA with distinct GC contents, called isochores or GC-content domains. A boundary between two GC-content domains in the human NF1 gene region is also a boundary between domains of early- and late-replicating sequences and of regions with high and low recombination frequencies. The perfect conservation of the GC-content distribution in this region between human and mouse demonstrates that GC-content stabilizing forces must act regionally on a fine scale at this locus. To further elucidate the nature of these forces, we report here on the spectrum of human SNPs and base pair substitutions between human and chimpanzee. The results show that the mutation rate changes exactly at the GC-content transition zone from low values in the GC-poor sequences to high values in GC-rich ones. The GC content of the GC-poor sequences can be explained by a bias in favor of GC > AT mutations, whereas the GC content of the GC-rich segment may result from a fixation bias in favor of AT > GC substitutions. This fixation bias may be explained by direct selection by the GC content or by biased gene conversion.
THE mammalian genome is not uniform in its long-range sequence composition, it is a mosaic of sequence stretches of variable length that differ widely in their GC contents. Whether the majority of these stretches really meet the criteria of isochores, as defined by Bernardi (2000), or should be better called GC-content domains, as suggested by Lander et al. (2001), is a matter of debate at the moment (Li 2002; Li et al. 2003; Zhang and Zhang 2004; Cohen et al. 2005; Costantini et al. 2006). Irrespective of this discussion, a correlation exists between the long-range GC content of DNA sequences and the structure of chromosomes on the one hand and a number of other structural and functional genomic features on the other (Holmquist 1992). Regarding chromosome structure, it became apparent that G-bands are composed of mostly GC-poor sequences, whereas R-bands are built up mostly of GC-rich sequences (Saccone et al. 1993). Furthermore, the long-range GC content of genomic regions is correlated with gene density, the distribution of repetitive elements (Lander et al. 2001), replication timing (Watanabe et al. 2002; Woodfine et al. 2004), and recombination frequency (Kong et al. 2002). The evolutionary stability of the GC-content distribution has been demonstrated for a limited number of homologous genes in several species (Bernardi 2000; Federico et al. 2004) and, on a genomewide level, for mice and humans (Waterston et al. 2002). In both cases, GC-rich sequences from one genome were also demonstrated to be GC-rich in the other genome and vice versa. A very fine-scaled conservation of the GC content was demonstrated for the NF1 gene region in humans and mice. In both species, the NF1 gene is located in a GC-poor sequence domain several hundred kilobases in length, whereas the neighboring RAB11FIP4 gene is located in a GC-rich domain of similar length. The boundaries between the two domains are sharp and located at exactly the same place in both species (Schmegner et al. 2005a). The evolutionary stability of the genome structure in mice and humans may be explained by hitherto unknown forces, which exert their effects to stabilize the structure of the mammalian genome. As candidates for these forces three mechanisms are under discussion, i.e., direct selection on the long-range GC content of sequences, regional variation in mutation biases, and biased gene conversion (for review see Eyre-Walker and Hurst 2001). However, results published recently suggest that the mammalian genome is not in a compositional equilibrium; in particular, the GC-rich domains are vanishing (Duret et al. 2002; Arndt et al. 2003; Belle et al. 2004; Meunier and Duret 2004; Comeron 2006; Khelifi et al. 2006). This led to a model according to which the GC-content domains evolved in the common ancestors of reptiles, birds, and mammals and henceforth the GC-content differences have been slowly eroding, at least in the mammalian lineage. In this case, the similarities in the GC-content distribution in the human and mouse genome would not result from an active process, but would simply reflect the common ancestry of the two species. This does not necessarily mean that the GC-content stabilizing forces are no longer active; they may merely be too weak to maintain the GC-rich domains, as suggested by Duret et al. (2002).
On a fine scale, the correlation between DNA-sequence composition and functional genomic features has been demonstrated for a few regions in which the long-range GC content changes abruptly. In humans, GC-content transitions located in the NF1 gene region on chromosome 17 and in the MN1/PITPNB gene region on chromosome 22 are also demonstrated to be boundaries between regions showing high and low recombination frequencies (Eisenbarth et al. 2000, 2001). Boundaries between early- and late-replicating sequences were found to coincide precisely with GC-content transitions in the human MHC locus (Tenzen et al. 1997) and again in the NF1 gene locus (Schmegner et al. 2005a). Interestingly, both the replication timing during S phase and the recombination frequency of a sequence are supposed to influence the composition of DNA sequences (Eyre-Walker and Hurst 2001), the former through changes in the spectrum of base misincorporations during S phase and the latter through two possible mechanisms. The first is based on a postulated correlation between the recombination frequency in a genomic region, the mutation rate, and perhaps the mutation spectrum. The second mechanism derives from the following fact: Recombination and gene conversion are mechanistically connected in a manner by which sequences with different recombination rates ought also to have different conversion rates. Since gene conversion has been shown to be a biased process (Galtier 2003), regional variation of conversion rates probably results in regionally varying patterns of evolutionarily fixed mutations. In summary, it can be assumed that the analysis of genetic variability patterns within a species and of fixed differences between species not only reveals whether GC-rich and GC-poor sequences are in a compositional equilibrium, but also allows for the differentiation between the actions of the stabilizing forces mentioned above. Moreover, the variability patterns, if they result from processes relevant for the local differentiation of the GC content, probably differ according to the GC content of DNA sequences and change at GC-content transitions. For this reason, we analyzed the frequency and the spectrum of polymorphic sites in the human and of fixed mutational differences between human and chimpanzee in DNA sequences located around the GC-content transition in the NF1 gene region.
MATERIALS AND METHODS
Probands and cell lines:
DNA from 29 randomly chosen human probands of self-reported German origin was isolated from peripheral blood. The project was approved by the local ethics committee. DNA from chimpanzee, orangutan, and gorilla was isolated from the Pan troglodytes lymphoblastoid cell line PTR-EB176 (ECACC no. 89072704), the orangutan (Pongo pygmaeus) lymphoblastoid cell line PPY-EB185 (ECACC no. 89072705), and the lymphoblastoid cell line EB(JC) (ECACC no. 89072703) from Gorilla gorilla, which were purchased from the European Collection of Cell Cultures (http://www.ecacc.org.uk).
Resequencing:
Overlapping primers for PCR were designed to span four different, mainly intronic, parts of the NF1 gene, which altogether comprised 24.9 kb, plus two intronic parts of the neighboring RAB11FIP4 gene, which comprised 19.8 kb. The locations of the sequenced regions within the NF1 and the RAB11FIP4 gene are given in Figure 1A. The average amplicon size used was 2.5 kb, and the average overlap between neighboring amplicons was 100 bp. These PCRs were performed using genomic DNA from all 29 human probands, as well as genomic DNA of one chimpanzee, one gorilla, and one orangutan. Samples were sequenced using Big-Dye Terminator chemistry (Applied Biosystems, Foster City, CA) on an ABI 3100 analyzer. Each PCR amplicon was sequenced from both ends and with at least eight additional internal primers to cover the whole 2.5 kb. Sequence data were assembled and compared using the SeqScape software (Applied Biosystems), and the entire sequence chromatograms were visually inspected.
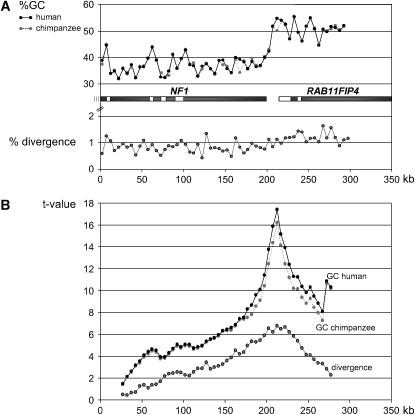
Figure 1.—
(A) GC-content curves for human (solid line) and chimpanzee (dotted line). Below these curves, the position of the NF1 gene (neurofibromatosis type 1) and the neighboring RAB11FIP4 gene (RAB11 family interacting protein 4) is indicated; the open sections represent the genomic regions that were resequenced in 29 human probands, chimpanzee, orangutan, and gorilla. The shaded curve shows the divergence between chimpanzee and human. Remarkable is the sharp GC-content transition in the intergenic region between NF1 and RAB11FIP4, which seems to coincide with a change in the rate of interspecies divergence. Almost all divergence values for the GC-poor region are <1%, whereas the values for the GC-rich region are >1%. (B) Statistical change-point analysis for the GC-content and divergence curves. The position where the t-test statistic takes its maximum value is a guess of a potential change point. The strongest change in both, the GC content and the interspecies divergence, could be assigned to nucleotide position 210–215 kb.
Statistics:
For the change-point analysis of the distributions of the GC contents and the interspecies divergence, a standard approach as described by Hawkins and Qiu (2003) was applied. This approach assumes normally distributed observations and at most one change point in the mean value along the sequence. To this end, all possible two-group comparisons between two adjacent value windows were carried out. The minimum size of each window comprised five values of the respective curve; i.e., the first comparison is between values 1–5 and 6–60 with a partition at nucleotide position 25 kb (Figure 1). The partition for the second comparison takes place at nucleotide position 30 kb, and so on, until the last partition is at nucleotide position 275 kb. For all of the 51 tests, the values of the t-test statistic (allowing for variance heterogeneity) as well as the corresponding P-values were recorded and a Bonferroni correction for 51 tests was carried out. The position where the t-test statistic takes its maximum value (see Figure 1B) is a guess of a potential change point.
One-way chi-square tests were used to compare frequencies of mutations to a model distribution. This model distribution was either 0.5:0.5, when investigating if the frequencies of two types of mutations were equal, or the actual GC:AT content of a sequence, i.e., 0.37:0.63 in the GC-poor domain and 0.51:0.49 in the GC-rich domain. To compare the ratio of AT > GC and GC > AT mutation frequencies (mutations that change an AT into a GC base pair or vice versa) between the domains which is adjusted for the AT contents and the GC contents of the underlying sequences, a test based on the logarithm of the odds ratio for an AT > GC mutation between the domains was used. For adjustment, the model under the null hypothesis claimed an odds ratio of (0.63:0.37)/(0.49:0.51) between the domains. The same procedure was applied to compare SNPs and fixed mutations, with a model odds ratio of 1.
RESULTS
To examine a possible correlation between the GC content of a sequence domain and its substitution rate, we measured the interspecies divergence of altogether 300 kb of homologous noncoding sequences between human and chimpanzee. As only base pair exchanges were taken into account, the term divergence as used here applies only to a part of the overall genomic differences found between human and chimpanzee. For our comparisons, we used the human sequence AC004526 and the homologous chimpanzee sequences, which are located on chimpanzee chromosome 19 between nucleotide position 30.00 Mb and 30.30 Mb (ensemble chimpanzee sequence release 31.2a). This chimpanzee sequence is composed of a number of contigs with just a few small gaps. A sliding window of 5 kb was moved in nonoverlapping 5-kb steps along the human sequence, and thus for each window the GC contents of human and orthologous chimpanzee sequences were determined. The resulting GC-content curves for the two species are given in Figure 1A—they are almost, but not exactly identical. The values obtained for the windows around positions 175.5 and 212.5 kb differ to a larger degree between the species (∼4.5 difference percentage points each). The differences are due to an adenylate kinase 3 pseudogene, present in one window of the human but not found in the chimpanzee genome, in addition to a 1.3-kb gap in the other window of the chimpanzee sequence. In addition to the GC content, the relative frequency of divergent sites was calculated for each window. The values obtained are given graphically in Figure 1A. For this analysis, only intergenic and intronic sequences with a distance of >50 bp to the next coding exon were used. Nevertheless, varying sequence lengths between the windows are supposed to be negligible, since only the homologous sequences of each window were compared. Regarding the GC-content curves in Figure 1A, the sharp GC-content transition located in the intergenic region between NF1 and RAB11FIP4 is clearly visible. In the same region, a transition from low to high values of sequence divergence occurs, with an average divergence of 0.82% in the GC-poor and 1.19% in the GC-rich region. To decide whether the observed distributional changes of GC content and divergence along the sequence are compatible with random variation and, if not, to specify a region where a change takes place, a statistical change-point analysis was performed. The advantage of this method is that, in addition to no prespecified regions being compared, a candidate position for a change between regions is estimated. For the GC content in human and chimpanzee, and for the divergence, change points were observed, which represent actual changes on the 5% level. Visual inspection of the t-value curves given in Figure 1B actually results in the detection of a transition zone of 20 kb where changes of both features, the GC content of the sequences and the interspecies divergence, might occur. The strongest change in the GC content could be assigned to the window around nucleotide position 212.5 kb; this coincides precisely with the strongest change in the interspecies divergence and hence in the substitution rates between GC-poor and GC-rich sequences immediately contacting each other. The boundaries between the GC-content domains and the domains with differing divergences coincide precisely. Under the assumption of a neutral evolution (the reasons for this assumption are discussed below), the observed regional differences in the substitution frequencies can be interpreted as the result of regionally differing mutation rates.
When neutrally evolving sequences are analyzed in a homogeneous population, mutation-rate differences are expected to result in differing SNP densities. To elucidate the SNP densities in both GC-content domains, we resequenced 24.9 kb of intronic sequences from the GC-poor and 19.8 kb from the GC-rich regions in 29 randomly chosen probands of German origin. The sequenced regions (depicted in Figure 1A) were chosen without prior knowledge of GC content or interspecies divergence. The process of resequencing was chosen because reliable SNP densities can hardly be obtained from SNP databases due to varying methods of SNP ascertainment used by the submitters. All told, in our sample of 58 chromosomes, 47 variable sites were detected in the 24.9 kb of NF1 and 71 variable sites in the 19.8 kb of RAB11FIP4. This results in SNP densities of 1.89 SNPs/kb in NF1 and 3.58 SNPs/kb in RAB11FIP4, a difference that also points to mutation-rate differences between the GC-rich and the GC-poor domains.
Next, we wanted to analyze whether or not the spectrum of base pair-changing mutations differs between the two GC-content domains. As it is not possible to directly observe the spectrum of mutations by which a DNA sequence is affected, the spectrum of SNPs found in the human population was used in place of the mutation spectrum. SNPs represent evolutionarily young mutation events. The SNP spectrum is therefore supposed to resemble the mutation spectrum with only minor disturbances. For the analysis we used validated noncoding SNPs, from the dbSNP database, located in a 320-kb region around the GC-content transition at the NF1 locus. The ancestral allele for each SNP was determined through comparisons with the chimpanzee sequence. All SNPs were categorized into four groups: SNPs resulting from mutations that changed an AT base pair into a GC base pair (AT > GC), a GC into an AT (GC > AT), an AT into an TA (AT > TA), and a GC into a CG (GC > CG). GC-rich and GC-poor sequence domains were analyzed separately. As a boundary between the domains, the 5-kb window with the peak in the t-value curve of the GC content (window around nucleotide position 212.5 kb in Figure 1) was used. The results given in Table 1 demonstrate that the SNP pattern is clearly dominated by AT > GC and GC > AT SNPs, which together are called GC-content-changing SNPs in the following text. Henceforth, only this category of SNPs is regarded, because this is the only one that is relevant for the evolution of the GC content of a sequence.
TABLE 1.
Spectrum of noncoding SNPs from dbSNP
| NF1 | RAB11FIP4 | |
|---|---|---|
| AT > GC | 95 (70) | 63 (49) |
| GC > AT | 78 (63) | 111 (69) |
| AT > TA | 12 | 13 |
| GC > CG | 24 (17) | 15 (12) |
Values in parentheses are obtained under exclusion of SNPs at CpG sites.
If the mutational input is responsible for the evolutionary stability of the GC content of a sequence, the number of GC > AT mutations is expected to be equal to the number of AT > GC mutations in this sequence, irrespective of its GC content. Therefore, in both domains a one-way chi-square test was used to determine whether the observed proportions of GC > AT and AT > GC SNPs are compatible with the assumption of equality. The results (given in Table 2) demonstrated that the assumption of equal proportions of GC > AT and AT > GC SNPs cannot be rejected for the GC-poor domain (45% GC > AT:55% AT > GC in GC-content-changing SNPs; P = 0.224), whereas a significant deviation from the assumption of equality was observed when SNPs from the GC-rich domain were analyzed, with an excess of GC > AT over AT > GC SNPs (64% GC > AT:36% AT > GC in GC-content-changing SNPs; P = 0.0003). This excess may be mainly due to a high proportion of GC > AT SNPs resulting from mutations at CpG sites. If this type of SNP is excluded from the analysis, an excess of GC > AT over AT > GC SNPs is still observed in the GC-rich domain, but the differences are no longer statistically significant. (The values obtained under exclusion of SNPs at CpG sites are given in parentheses in Tables 1 and 2.) In the same way, the proportions of GC > AT and AT > GC SNPs were compared to the proportions of GC and AT base pairs in both domains. In this case, a significant excess of GC > AT over AT > GC SNPs was found in both the GC-poor region (45% GC > AT:55% AT > GC in GC-content-changing SNPs in relation to 37% GC:63% AT; P = 0.035) and the GC-rich region (64% GC > AT:36% AT > GC in GC-content-changing SNPs in relation to 51% GC:49% AT; P = 0.0009), irrespective of the in- or exclusion of SNPs at CpG sites. This excess can also be demonstrated by normalizing the proportions of GC > AT SNPs by the GC content of the domains and the proportion of AT > GC SNPs by the AT content. For results see Table 2. Furthermore, the ratios of AT > GC to GC > AT SNP frequencies did not differ significantly between the GC-content domains when the GC-content differences were taken into account (observed odds ratio = 2.15, model odds ratio = 1.77; P = 0.38). Taken together, our results revealed the same mutational bias in favor of GC > AT over AT > GC mutations in both GC-content domains.
TABLE 2.
Statistical analysis of the collected SNP data
| NF1 | RAB11FIP4 | |
|---|---|---|
| Proportion of GC > ATa | 0.45 (0.47) | 0.64 (0.59) |
| Proportion of AT > GCa | 0.55 (0.53) | 0.36 (0.41) |
| χ2 [0.5:0.5]b | P = 0.224 | P = 0.0003 |
| (P = 0.37) | (P = 0.066) | |
| GC content | 0.37 (0.36) | 0.51 (0.49) |
| AT content | 0.63 (0.64) | 0.49 (0.51) |
| χ2 [0.63:0.37]c | P = 0.035 | P = 0.0009 |
| (P = 0.006) | (P = 0.04) | |
| Proportion of GC > ATa/GC content | 1.22 (1.30) | 1.25 (1.20) |
| Proportion of AT > GCa/AT content | 0.87 (0.83) | 0.73 (0.80) |
| OR (AT > GC:GC > AT)d | 2.17 (1.56) | |
| Test whether OR = 1.77 (1.71)e | P = 0.38 (P = 0.73) | |
Values in parentheses are obtained under exclusion of SNPs at CpG sites.
With respect to the GC-content-changing SNPs.
Test whether the true ratio of AT > GC and GC > AT mutations is 0.5:0.5.
Test whether the true ratio of AT > GC and GC > AT mutations is 0.63:0.37.
Odds ratio of AT > GC mutation between NF1 and RAB11FIP4.
Test whether the true odds ratio of AT > GC mutation between NF1 and RAB11FIP4 = (0.63/0.37)/(0.49/0.51) = 1.77 (0.71).
To test whether the sequences of both domains are in a compositional equilibrium, we performed a similar analysis on the basis of fixed base pair substitutions observed between human and chimpanzee. For this purpose, we sequenced the same stretches of DNA as mentioned above from the chimpanzee genome and compared the sequences to homologous GenBank sequences of the human. In addition, homologous DNA fragments from the gorilla and the orangutan were sequenced and used as an outgroup to define the ancestral state of the divergent base pairs found in the human–chimpanzee comparison and, in consequence, to determine the direction of the substitutions. The results are given in Table 3. For both the GC-rich and the GC-poor sequences, a one-way chi-square test was used to analyze whether the proportions of AT > GC and GC > AT substitutions are compatible with the assumption of equality. In this case, in contrast to results obtained with the SNP data, the assumption of equal proportions of AT > GC and GC > AT substitutions could not be rejected for both GC-content domains (49:51% in GC-content-changing substitutions, P = 0.94 for the GC-poor, and 44:56% in GC-content-changing substitutions, P = 0.11 for the GC-rich parts). These results demonstrate that the sequences in both domains are in or close to a compositional equilibrium despite the absolute excess of GC > AT mutations found in the GC-rich domain.
TABLE 3.
Spectrum of fixed base pair substitutions between human and chimpanzee
| NF1 | RAB11FIP4 | |
|---|---|---|
| AT > GC | 77 (55) | 83 (55) |
| GC > AT | 75 (55) | 106 (71) |
| AT > TA | 12 | 8 |
| GC > CG | 23 (16) | 30 (17) |
Values in parentheses are obtained under exclusion of SNPs at CpG sites.
DISCUSSION
Our analysis of the sequence diversity in the NF1 gene region revealed low levels of interspecies divergence and SNP densities in the GC-poor parts, with high levels for both features in the GC-rich parts. A general correlation between GC content and divergence has also been reported for whole-genome comparisons of human and mouse and of human and chimpanzee (Waterston et al. 2002; Mikkelsen et al. 2005). Interspecies divergence and SNP densities are influenced by the mutation rate, by selection on the function of proteins encoded in the region, and possibly by selection on the GC content as well. The potential influence of purifying selection on the obtained values for interspecies divergence and SNP densities due to protein function was minimized by the exclusion of coding and exon-flanking sequences from the analysis. Regarding SNP densities, the low values obtained for the GC-poor NF1 gene may have also resulted from an evolutionarily recent selective sweep, which, due to the linkage disequilibrium pattern in the region, may have influenced the whole NF1 gene but not the neighboring GC-rich sequences. However, the variability pattern of the NF1 gene in the European population (Schmegner et al. 2005b) clearly excludes this possibility. Therefore, the observed pattern of diversity and substitutions in the NF1 gene region most likely reflects differences in the mutation rates between the GC-poor and GC-rich sequence domains. A possible reason for the mutation-rate differences may be a higher density of hypermutable CpG sites in the GC-rich region as compared to the GC-poor region. Reanalysis of the data under elimination of SNPs and substitutions involving CpG sites resulted in a reduction of SNP densities (from 1.89 to 1.24 SNPs/kb in the GC-poor and from 3.58 to 2.47 SNPs/kb in the GC-rich domain) and interspecies divergence (from 0.76 to 0.57% in the GC-poor and from 1.14 to 0.79% in the GC-rich domain), but not in an erosion of the differences between the domains. Hence, mutations of CpG dinucleotides contribute substantially to the numbers of polymorphic and divergent sites in both domains, although they do not explain the mutation-rate differences.
The analysis of the GC content and the human–chimpanzee divergence in a sliding window of 5 kb showed that both characteristics change abruptly within the same narrow transition region. Results published earlier and the inspection of HapMap data (International HapMap Consortium 2005) assigned change points for the recombination frequency (Eisenbarth et al. 2000) and the timing of replication (Schmegner et al. 2005a) to this same transition region. Taken together, these results demonstrate a covariation of all four features—GC content, mutation rate, recombination frequency, and replication timing—on an astonishingly fine scale. On a large scale, various pairwise correlations between these features have been described and a number of causal connections from these correlations have been inferred, especially to explain the mutation-rate variation in the human genome. Covariation of the GC content with the recombination frequency (Kong et al. 2002; Montoya-Burgos et al. 2003; Meunier and Duret 2004), with the mutation rate (Smith et al. 2002), and with the timing of DNA replication (Watanabe et al. 2002; Woodfine et al. 2004) were described for several chromosomes and the human genome as a whole. The direct influence of the GC content on the mutation rate of a sequence due to a higher rate of cytosine deamination in GC-poor DNA was reported (Fryxell and Zuckerkandl 2000; Fryxell and Moon 2005). One hypothesis describes the dependency of both the mutation rate and the mutation spectrum of a sequence on the time in S phase, during which the sequence is replicated. The basis for this hypothesis comes from the observation that the concentrations of free deoxyribonucleotides in a cell fluctuate during S phase and affect the fidelity of DNA replication (Wolfe et al. 1989; Wolfe 1991). A correlation between recombination frequency and the mutation rate of a genomic region has been demonstrated by several groups (Hellmann et al. 2003; Huang et al. 2005). A causal relationship has been suggested in the sense that the process of recombination itself may be mutagenic (Lercher and Hurst 2002b; Filatov and Gerrard 2003; Filatov 2004). Due to the observed covariation of all characters, the results of our work are compatible with all these hypotheses and cannot be used as arguments in favor of or against any of them. Instead, our results suggest that the correlations between structure and function of a genomic region may be more complex than expected from pairwise comparisons of the relevant characters.
To test whether regional variations of the proportions of GC > AT vs. AT > GC mutations were responsible for the creation and the maintenance of the pronounced GC-content differences and the sharp boundary between the sequence domains in the NF1 gene region, the spectrum of base pair exchanges leading to SNPs in the human population was analyzed and taken as a proxy for the mutation spectrum. The results revealed a higher GC > AT than AT > GC mutation rate in both sequence domains, confirming similar observations at the MHC locus, published by Eyre-Walker (1999) and Smith and Eyre-Walker (2001), and for a number of genes on various chromosomes (Alvarez-Valin et al. 2002). Moreover, it turned out that the bias in favor of GC > AT mutations was equally strong in both GC-content domains. These results reject the mutation-bias variation hypothesis as a possible explanation for the compositional heterogeneity of GC-content domains.
A higher rate of GC > AT than AT > GC mutations will lower the GC content of a sequence to <50% over time until an equilibrium is reached, at which both types of mutations occur with the same absolute frequency. This equilibrium appears to be reached for the GC-poor sequences from the NF1 gene locus because the proportions of GC > AT and AT > GC mutations have been found to be equal in this region. As the vast majority of human genomic DNA shows a GC content of <45%, with a peak in the distribution of the GC content in 20-kb windows at 37.5% (Lander et al. 2001), the GC content of the GC-poor NF1 gene region may be taken as representative of the majority of the genomic sequences. In which case, it can be assumed that the sequence composition of the greatest part of the mammalian genome is a direct result of the mutation-rate bias.
The human–chimpanzee comparison revealed equal proportions of GC > AT and AT > GC substitutions for the GC-poor sequences, further demonstrating the compositional equilibrium for this domain. In the GC-rich domain a higher proportion of GC > AT than AT > GC substitutions was observed, although the difference between the two values was statistically not significant. Hence, we cannot categorically state that this domain is also at an equilibrium or whether the GC content of the GC-rich sequences will decrease over time—a fact that would argue in favor of the vanishing isochore theory, which has been discussed controversially during the last few years (Duret et al. 2002; Arndt et al. 2003; Alvarez-Valin et al. 2004; Belle et al. 2004; Meunier and Duret 2004; Khelifi et al. 2006). Irrespective of this discussion, however, our results show a clear discrepancy between the SNP data and the interspecies comparison. This discrepancy may be explained by a change in the mutational biases that occurred after the divergence of the human and the chimpanzee lineages, as suggested by Comeron (2006), or by a fixation bias in favor of AT > GC substitutions, also described earlier (Lercher and Hurst 2002a; Lercher et al. 2002; Webster and Smith 2004). Two forces—direct selection on the GC content and biased gene conversion—can explain the proposed fixation bias. The two forces are not mutually exclusive and it is difficult to distinguish between them. In the NF1 gene region, an abrupt regional change in the action of either of the two forces has to be assumed, because the fixation bias has to be postulated only for the GC-rich but not for the GC-poor domain. Intriguingly, a change in the recombination frequencies of the underlying sequences was observed exactly at the boundary between the two GC-content domains, with a low frequency in the GC-poor and a high recombination frequency in the GC-rich region (Eisenbarth et al. 2000). Moreover, HapMap data (International HapMap Consortium 2005) revealed not only a higher density of recombination hot spots in the GC-rich than in the GC-poor domain, which actually is completely devoid of hot spots, but also substantially elevated recombination rates in the regions outside of the hot spots in the GC-rich compared to the GC-poor domain. Therefore, it can be assumed that the gene-conversion frequency is also higher in the GC-rich domain, a fact that will result in a stronger fixation bias in this domain, if there is a bias in the conversion rates. This postulated correlation was confirmed by data published recently. Kudla et al. (2004) showed that paralogous sequences undergoing gene conversion have a higher GC content than sequences not involved in this process. Webster et al. (2005) demonstrated an effect of the recombination rate of a genomic region on the substitution patterns of Alu sequences but not on their polymorphism pattern and explained the differences in the observations by biased gene conversion. This mechanism can lead to regional differences in the GC content only if regional differences in the conversion rates are stable over long periods of time. Whether this condition has really been met cannot be reported upon at present. The exact location of recombination hot spots and the recombination rates measured over short distances (50 kb) are not well conserved between human and chimpanzee (Ptak et al. 2005), but these results tell us little about the conservation of regional differences of recombination frequencies on a scale that is relevant for the evolution of GC-content domains.
In summary, our results suggest a model according to which the GC content of GC-poor sequence domains, representative of the majority of human genomic DNA, is the result of a mutation bias. The higher GC content of the GC-rich domains cannot be explained by actual regional variation of the mutation bias. For this domain, the action of postmutational processes that lead to a preference in the fixation of AT > GC mutations over GC > AT mutations or an evolutionary very recent change in the mutation bias has to be assumed to explain the data.
References
- Alvarez-Valin, F., G. Lamolle and G. Bernardi, 2002. Isochores, GC3 and mutation biases in the human genome. Gene 300: 161–168. [DOI] [PubMed] [Google Scholar]
- Alvarez-Valin, F., O. Clay, S. Cruveiller and G. Bernardi, 2004. Inaccurate reconstruction of ancestral GC levels creates a “vanishing isochores” effect. Mol. Phylogenet. Evol. 31: 788–793. [DOI] [PubMed] [Google Scholar]
- Arndt, P. F., D. A. Petrov and T. Hwa, 2003. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 20: 1887–1896. [DOI] [PubMed] [Google Scholar]
- Belle, E. M., L. Duret, N. Galtier and A. Eyre-Walker, 2004. The decline of isochores in mammals: an assessment of the GC content variation along the mammalian phylogeny. J. Mol. Evol. 58: 653–660. [DOI] [PubMed] [Google Scholar]
- Bernardi, G., 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241: 3–17. [DOI] [PubMed] [Google Scholar]
- Cohen, N., T. Dagan, L. Stone and D. Graur, 2005. GC composition of the human genome: in search of isochores. Mol. Biol. Evol. 22: 1260–1272. [DOI] [PubMed] [Google Scholar]
- Comeron, J. M., 2006. Weak selection and recent mutational changes influence polymorphic synonymous mutations in humans. Proc. Natl. Acad. Sci. USA 103: 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini, M., O. Clay, F. Auletta and G. Bernardi, 2006. An isochore map of human chromosomes. Genome Res. 16: 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., M. Semon, G. Piganeau, D. Mouchiroud and N. Galtier, 2002. Vanishing GC-rich isochores in mammalian genomes. Genetics 162: 1837–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth, I., G. Vogel, W. Krone, W. Vogel and G. Assum, 2000. An isochore transition in the NF1 gene region coincides with a switch in the extent of linkage disequilibrium. Am. J. Hum. Genet. 67: 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth, I., A. M. Striebel, E. Moschgath, W. Vogel and G. Assum, 2001. Long-range sequence composition mirrors linkage disequilibrium pattern in a 1.13 Mb region of human chromosome 22. Hum. Mol. Genet. 10: 2833–2839. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A., 1999. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics 152: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A., and L. D. Hurst, 2001. The evolution of isochores. Nat. Rev. Genet. 2: 549–555. [DOI] [PubMed] [Google Scholar]
- Federico, C., S. Saccone, L. Andreozzi, S. Motta, V. Russo et al., 2004. The pig genome: compositional analysis and identification of the gene-richest regions in chromosomes and nuclei. Gene 343: 245–251. [DOI] [PubMed] [Google Scholar]
- Filatov, D. A., 2004. A gradient of silent substitution rate in the human pseudoautosomal region. Mol. Biol. Evol. 21: 410–417. [DOI] [PubMed] [Google Scholar]
- Filatov, D. A., and D. T. Gerrard, 2003. High mutation rates in human and ape pseudoautosomal genes. Gene 317: 67–77. [DOI] [PubMed] [Google Scholar]
- Fryxell, K. J., and W. J. Moon, 2005. CpG mutation rates in the human genome are highly dependent on local GC content. Mol. Biol. Evol. 22: 650–658. [DOI] [PubMed] [Google Scholar]
- Fryxell, K. J., and E. Zuckerkandl, 2000. Cytosine deamination plays a primary role in the evolution of mammalian isochores. Mol. Biol. Evol. 17: 1371–1383. [DOI] [PubMed] [Google Scholar]
- Galtier, N., 2003. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 19: 65–68. [DOI] [PubMed] [Google Scholar]
- Hawkins, D. M., and P. Qiu, 2003. The changepoint model for statistical process control. J. Qual. Technol. 35: 355–366. [Google Scholar]
- Hellmann, I., I. Ebersberger, S. E. Ptak, S. Paabo and M. Przeworski, 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist, G. P., 1992. Chromosome bands, their chromatin flavors, and their functional features. Am. J. Hum. Genet. 51: 17–37. [PMC free article] [PubMed] [Google Scholar]
- Huang, S. W., R. Friedman, N. Yu, A. Yu and W. H. Li, 2005. How strong is the mutagenicity of recombination in mammals? Mol. Biol. Evol. 22: 426–431. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium, 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi, A., J. Meunier, L. Duret and D. Mouchiroud, 2006. GC content evolution of the human and mouse genomes: insights from the study of processed pseudogenes in regions of different recombination rates. J. Mol. Evol. 62: 745–752. [DOI] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Kudla, G., A. Helwak and L. Lipinski, 2004. Gene conversion and GC-content evolution in mammalian Hsp70. Mol. Biol. Evol. 21: 1438–1444. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., and L. D. Hurst, 2002. a Can mutation or fixation biases explain the allele frequency distribution of human single nucleotide polymorphisms (SNPs)? Gene 300: 53–58. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., and L. D. Hurst, 2002. b Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337–340. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., N. G. Smith, A. Eyre-Walker and L. D. Hurst, 2002. The evolution of isochores: evidence from SNP frequency distributions. Genetics 162: 1805–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., 2002. Are isochore sequences homogeneous? Gene 300: 129–139. [DOI] [PubMed] [Google Scholar]
- Li, W., P. Bernaola-Galvan, P. Carpena and J. L. Oliver, 2003. Isochores merit the prefix ‘iso’. Comput. Biol. Chem. 27: 5–10. [DOI] [PubMed] [Google Scholar]
- Meunier, J., and L. Duret, 2004. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 21: 984–990. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, T. S., L. W. Hillier, E. E. Eichler, M. C. Zody, D. B. Jaffe et al., 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos, J. I., P. Boursot and N. Galtier, 2003. Recombination explains isochores in mammalian genomes. Trends Genet. 19: 128–130. [DOI] [PubMed] [Google Scholar]
- Ptak, S. E., D. A. Hinds, K. Koehler, B. Nickel, N. Patil et al., 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37: 429–434. [DOI] [PubMed] [Google Scholar]
- Saccone, S., A. De Sario, J. Wiegant, A. K. Raap, V. G. Della et al., 1993. Correlations between isochores and chromosomal bands in the human genome. Proc. Natl. Acad. Sci. USA 90: 11929–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmegner, C., A. Berger, W. Vogel, H. Hameister and G. Assum, 2005. a An isochore transition zone in the NF1 gene region is a conserved landmark of chromosome structure and function. Genomics 86: 439–445. [DOI] [PubMed] [Google Scholar]
- Schmegner, C., J. Hoegel, W. Vogel and G. Assum, 2005. b Genetic variability in a genomic region with long-range linkage disequilibrium reveals traces of a bottleneck in the history of the European population. Hum. Genet. 118: 276–286. [DOI] [PubMed] [Google Scholar]
- Smith, N. G., and A. Eyre-Walker, 2001. Synonymous codon bias is not caused by mutation bias in G+C-rich genes in humans. Mol. Biol. Evol. 18: 982–986. [DOI] [PubMed] [Google Scholar]
- Smith, N. G., M. T. Webster and H. Ellegren, 2002. Deterministic mutation rate variation in the human genome. Genome Res. 12: 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen, T., T. Yamagata, T. Fukagawa, K. Sugaya, A. Ando et al., 1997. Precise switching of DNA replication timing in the GC content transition area in the human major histocompatibility complex. Mol. Cell. Biol. 17: 4043–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., A. Fujiyama, Y. Ichiba, M. Hattori, T. Yada et al., 2002. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: disease-related genes in timing-switch regions. Hum. Mol. Genet. 11: 13–21. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril et al., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Webster, M. T., and N. G. Smith, 2004. Fixation biases affecting human SNPs. Trends Genet. 20: 122–126. [DOI] [PubMed] [Google Scholar]
- Webster, M. T., N. G. Smith, L. Hultin-Rosenberg, P. F. Arndt and H. Ellegren, 2005. Male-driven biased gene conversion governs the evolution of base composition in human alu repeats. Mol. Biol. Evol. 22: 1468–1474. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., 1991. Mammalian DNA replication: mutation biases and the mutation rate. J. Theor. Biol. 149: 441–451. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., P. M. Sharp and W. H. Li, 1989. Mutation rates differ among regions of the mammalian genome. Nature 337: 283–285. [DOI] [PubMed] [Google Scholar]
- Woodfine, K., H. Fiegler, D. M. Beare, J. E. Collins, O. T. McCann et al., 2004. Replication timing of the human genome. Hum. Mol. Genet. 13: 191–202. [DOI] [PubMed] [Google Scholar]
- Zhang, C. T., and R. Zhang, 2004. Isochore structures in the mouse genome. Genomics 83: 384–394. [DOI] [PubMed] [Google Scholar]