Abstract
A combination of cytogenetic and bioinformatic procedures was used to test the chromosomal phylogeny relating Drosophila buzzatii with D. repleta. Chromosomes X and 2, harboring most of the inversions fixed between these two species, were analyzed. First, chromosomal segments conserved during the divergence of the two species were identified by comparative in situ hybridization to the D. repleta chromosomes of 180 BAC clones from a BAC-based physical map of the D. buzzatii genome. These conserved segments were precisely delimited with the aid of clones containing inversion breakpoints. Then GRIMM software was used to estimate the minimum number of rearrangements necessary to transform one genome into the other and identify all possible rearrangement scenarios. Finally, the most plausible inversion trajectory was tested by hybridizing 12 breakpoint-bearing BAC clones to the chromosomes of seven other species in the repleta group. The results show that chromosomes X and 2 of D. buzzatii and D. repleta differ by 12 paracentric inversions. Nine of them are fixed in chromosome 2 and entail two breakpoint reuses. Our results also show that the cytological relationship between D. repleta and D. mercatorum is closer than that between D. repleta and D. peninsularis, and we propose that the phylogenetic relationships in this lineage of the repleta group be reconsidered. We also estimated the rate of rearrangement between D. repleta and D. buzzatii and conclude that rates within the genus Drosophila vary substantially between lineages, even within a single species group.
THE cytological analysis of animal and plant chromosomes has a long tradition in the study of evolution (White 1973). Recently, the availability of physical maps and whole-genome sequences from many species has provided an unparalleled opportunity to investigate further the structural changes in the eukaryotic genome. The studies carried out so far have led to three main conclusions about genome evolution (Eichler and Sankoff 2003; Sankoff 2003; Kazazian 2004; Coghlan et al. 2005; Shapiro 2005): (1) the eukaryotic genome is exceptionally malleable, (2) rates and patterns of chromosomal rearrangement vary significantly between different evolutionary lineages, and (3) repetitive DNA sequences are ubiquitous in eukaryotic genomes and responsible for most of the structural dynamism.
Paracentric inversions are the most frequent type of rearrangement within the Drosophila and Anopheles genera with a similar average rate of rearrangement fixation, ∼0.05 disruptions/Mb/MY (González et al. 2002; Sharakhov et al. 2002). However, in the more distant comparison between Drosophila and Anopheles, extensive interchromosomal exchange has been observed (Zdobnov et al. 2002). In nematodes, the rearrangement rate is four times that in Drosophila and the ratio of translocations to inversions to transpositions is 1:1:2 (Coghlan and Wolfe 2002). Whole-genome duplications as well as various types of rearrangements characterize plant genome evolution, the evolution rate of the most dynamic plant genomes (∼0.03 disruptions/Mb/MY) being half the rate reported for insects (Lagercrantz 1998; Yogeeswaran et al. 2005). In mammals, the X chromosome shows extensive syntenic conservation whereas the autosomes show a variety of intra- and interchromosomal rearrangements and the Y chromosome is an example of rapid and unconstrained evolution. In addition, rates of mammalian chromosome evolution vary radically among lineages (Murphy et al. 2005).
In Drosophila, one of the most striking features of chromosomal evolution is the nonuniform distribution of inversions (Sperlich and Pfriem 1986; Powell 1997). Some species have dozens of inversions segregating in all chromosomes, other species carry them in only one or two chromosomes, and there are species with no polymorphic inversions. Another remarkable observation is the nonrandom distribution of inversion breakpoints within a given chromosome, with many sites where two or more inversion breakpoints seem to coincide (Tonzetich et al. 1988; Wasserman 1992; Cáceres et al. 1997). Similar observations have been made in the mosquito Anopheles (Coluzzi et al. 2002). These results are based on the cytological comparison of polytene chromosomes and may suffer from the limited resolution of this method. Breakpoint clustering has also been described as an outstanding feature of mammal genome evolution (Pevzner and Tesler 2003; Bourque et al. 2004; Murphy et al. 2005). Pevzner and Tesler (2003) coined the word “reuse” to describe this phenomenon. We use this term here as a shorthand for “close clustering” but emphasize, following their proponents, that it does not imply the use of exactly the same genomic position (nucleotide) as an endpoint of different rearrangements.
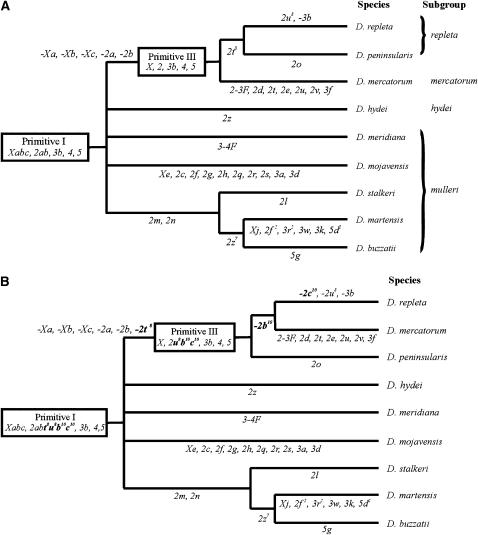
The repleta species group belongs to the Drosophila subgenus, which diverged from the Sophophora subgenus, the other main lineage in the Drosophila genus, 40–62 MYA (Powell 1997; Tamura et al. 2004). This species group is one of the largest in the genus and includes >100 species, many of them cactophilic species living in the deserts and arid zones of the American continent. In an outstanding cytological effort, Wasserman (1982, 1992) determined the inversion relationships between 70 species and divided the repleta group into five subgroups: repleta, mercatorum, hydei, mulleri, and fasciola. He used Drosophila repleta chromosomes (Wharton 1942) as a reference and inferred that the chromosomal arrangement of the ancestor of the repleta species group, named Primitive I, was one differing from that of D. repleta by six inversions and that can be represented as Xabc, 2ab, 3b, 4, 5 (Figure 1A).
Figure 1.—
Phylogenetic relationships between the species analyzed in this study. Chromosomal inversions fixed in each branch of the tree are indicated. Inversions fixed in the lineage from Primitive I to D. repleta are represented with the minus sign to indicate that they must be subtracted from the formula of Primitive I to get the formula for D. repleta. (A) Current phylogeny based on comparisons of polytene chromosome-banding patterns (Wasserman 1992; Ruiz and Wasserman 1993). (B) Phylogeny inferred from the results of in situ hybridizations of DNA probes to the polytene chromosomes (Ranz et al. 2003; this work). Inversion changes in the phylogenetic tree are shown in boldface type.
The chromosomes of D. buzzatii, a member of the mulleri subgroup that diverged from D. repleta 15–22 MYA (Spicer 1988; Russo et al. 1995), have been particularly difficult to disentangle. This was one of the first repleta group species to be cytologically analyzed (Wasserman 1954) and in the subsequent 50 years its inversion phylogeny has suffered many alterations (Wasserman 1962, 1982, 1992; Ruiz et al. 1982). Ruiz and Wasserman (1993) proposed that the D. buzzatii chromosomes derive from the putative ancestral karyotype of the repleta group, Primitive I, by the fixation of four inversions: 2m, 2n, 2z7, and 5g (Figure 1A). However, they also discovered that the D. buzzatii chromosome 2 differs from that of D. repleta by two small additional inversions, 2t8 and 2u8. Being fixed in the lineage leading from Primitive I to D. repleta (Figure 1A), these two small inversions were not incorporated to the D. buzzatii chromosome maps (Ruiz and Wasserman 1993).
Inversion phylogenies are usually considered very reliable and have been used as a benchmark for comparison with allozyme and DNA sequence phylogenies and assessment of congruence (MacIntyre and Collier 1986; O'Grady et al. 2001). There are, however, several sources of error, one of them being observational mistakes (Wasserman 1963, 1992). As a way to overcome the limitations of cytological studies and get a deeper insight into the molecular organization and evolution of D. buzzatii chromosomes, we undertook a decade ago a project to map DNA clones to its salivary gland chromosomes by in situ hybridization (Ranz et al. 1997). The map location of nearly 300 molecular markers from D. melanogaster was compared between D. buzzatii and D. repleta (Ranz et al. 2003). The results were consistent with some of the previous cytological results but uncovered also another two inversions fixed in chromosome 2, 2b10 and 2c10, overlooked in previous studies. These two inversions are apparently fixed in the lineage leading from Primitive I to D. repleta, yet no attempt was made to determine their distribution in the phylogenetic tree (Ranz et al. 2003). In a further effort to determine the precise number and extent of structural changes between D. buzzatii and D. repleta, we have turned to clones from the recently produced BAC library and BAC-based physical map of the D. buzzatii genome (González et al. 2005). We have mapped to D. repleta chromosomes 180 BAC clones from D. buzzatii chromosomes X and 2 that harbor most of the rearrangements fixed between these two species (Figure 1A). Specifically the aims of this study are: (i) to determine the number and orientation of conserved chromosomal segments between D. buzzatii and D. repleta, (ii) to test the inversion phylogeny proposed for D. buzzatii, (iii) to ascertain whether the coincidence of inversion breakpoints described by cytological inspection of chromosomes still holds at an enhanced resolution level, and (iv) to estimate the rate of chromosomal rearrangement between D. repleta and D. buzzatii for comparison with that of other lineages.
MATERIALS AND METHODS
Flies:
Stocks of nine Drosophila species representing four of the five subgroups in the repleta species group were used (see Figure 1A). All stocks are homokaryotypic for the standard arrangement in all chromosomes except the D. mercatorum stock that is homokaryotypic for the polymorphic inversion 2v3. The stocks of D. repleta (15084-1611.06), D. peninsularis (15081-1401.00), D. meridiana (15081-1341.03), D. stalkeri (15081-1451.00), and D. mojavensis (15081-1352.10) come from the Tucson Drosophila Stock Center; the remaining stocks belong to the collection of the Departament de Genètica i de Microbiologia (Universitat Autònoma de Barcelona).
Probes:
A total of 180 clones from the D. buzzatii BAC library CHORI-225 (available from BACPAC Resources at http://bacpac.chori.org) were successfully hybridized to the polytene chromosomes of D. repleta. Most clones were hybridized individually but 42 clones were hybridized as pools of 2 or 3 nearby clones and each pool was considered a single marker. Pools were designed to close the gaps between clones already hybridized. BAC clones were chosen according to their localization in the D. buzzatii genome map (González et al. 2005) to optimize coverage of chromosomes X (57 clones) and 2 (123 clones). Particular attention was paid to those regions that have been rearranged during the divergence between D. repleta and D. buzzatii. When two clones mapping relatively close in D. buzzatii hybridized to two different chromosomal sites in D. repleta, clones spanning the region between them were also hybridized. This allowed us to determine accurately the boundaries of chromosomal segments conserved throughout evolutionary time and also to infer the minimum number of inversion breakpoints fixed in that particular genomic region. Clones giving one signal in D. buzzatii and two signals in D. repleta contain an inversion breakpoint fixed between the two species. When this result was obtained with a pool of clones, further hybridizations were carried out with each clone separately until we identified the clone containing the breakpoint. Twelve clones containing inversion breakpoints were then hybridized to the polytene chromosomes of other species of the repleta group to test for the presence of the inversions fixed between D. buzzatii and D. repleta in the other species.
In situ hybridization and chromosomal maps:
Only female larvae were used for hybridizing the BAC clones mapping on the X chromosome because the single X of the male shows a somewhat reduced level of hybridization while the efficiency of hybridization on the female X is equivalent to that on the autosomes (Pardue et al. 1987). Polytene chromosome squashes, hybridization, and detection were carried out as in Montgomery et al. (1987). When possible, the same DNA previously used for fingerprinting and in situ hybridization of BAC clones in D. buzzatii (González et al. 2005) was used as a probe to hybridize to D. repleta chromosomes. Otherwise DNA from BAC clones was extracted following the alkaline lysis miniprep protocol available from http://bacpac.chori.org/protocols.htm. Probes were labeled with biotin-16-dUTP by random primer. Hybridization results were recorded as digital images captured with a phase contrast Nikon Optiphot-2 microscope at 600× magnification and a Nikon Coolpix 4500 camera. Hybridization signals were localized using the cytological map of D. repleta (Wharton 1942). For the other species of the repleta group cut-and-paste reconstructions of chromosome 2 according to the inversions putatively fixed during their divergence were produced and used to locate the signals (Wasserman 1992; Ruiz and Wasserman 1993).
Bioinformatic analysis:
GRIMM software (Tesler 2002) implements the Hannenhalli and Pevzner algorithms for computing unichromosomal and multichromosomal genomic distances and was used here to calculate the minimum number of rearrangement events required to transform one genome into another. This algorithm also finds optimal scenarios for the transformation of one genome into another via these rearrangement events. The program was run online at the publicly available server http://www-cse.ucsd.edu/groups/bioinformatics/GRIMM/, using the linear unichromosomal genome and signed conserved segments options. Each chromosome (X or 2) or chromosomal region (distal or proximal half of chromosome 2) was separately analyzed. The program gives one of the multiple optimal scenarios to transform one genome into the other. To find out all the possible scenarios we ran the program several times, introducing each time one of the inversions in the source genome until all the possible combinations of inversions were found.
RESULTS
Identification of conserved segments:
Detailed results for the 180 D. buzzatii BAC clones hybridized to the polytene chromosomes of D. repleta are given in supplemental Tables S1 and S2 (at http://www.genetics.org/supplemental/). The X chromosome clones amount to 53 markers and represent 23 contigs of the D. buzzatii physical map of chromosome X (González et al. 2005). Another 69 markers (gene clones, cosmids, and P1 phages) previously mapped to the X chromosome of both species (Ranz et al. 2003) were also included in our analysis, raising the total number of available markers to 122. The chromosome 2 clones amount to 106 markers, representing 22 contigs of the D. buzzatii physical map of chromosome 2 (González et al. 2005). We included in the analysis another 143 markers previously mapped to this chromosome in the two species (Ranz et al. 2003) for a total number of 249 markers. The markers are distributed all along chromosomes X and 2, effectively covering the entire chromosome length in both cases.
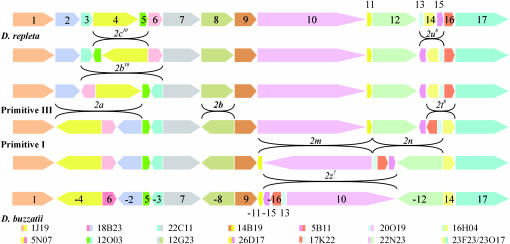
The chromosomal localization of all markers was compared in the two species to identify the number and orientation of conserved segments. A conserved segment is defined here as a set of markers that are consecutive (show the same relative order) in D. buzzatii and D. repleta. Seven conserved segments were identified in chromosome X and 17 in chromosome 2 (Figures 2 and 3). Most conserved segments are quite big, made up of at least 10 markers (supplemental Tables S1 and S2 at http://www.genetics.org/supplemental/). However, a few smaller conserved segments were also identified. Despite their small size, these conserved segments span at least three chromosomal bands with different markers hybridizing to different bands, allowing us to ascertain their relative orientation in both species. Most segments were precisely delimited with the aid of BAC clones that yield two hybridization signals in D. repleta and thus contain an inversion breakpoint fixed between the two species. Two such clones were identified in chromosome X, whereas 15 such clones were found in chromosome 2 (Table 1).
Figure 2.—
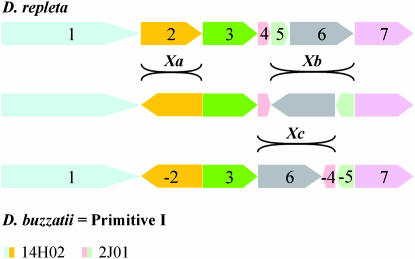
Relative position and orientation of the seven chromosome X segments conserved during the divergence between D. repleta and D. buzzatii. Conserved segments are numbered consecutively (from telomere to centromere) in D. repleta. The three rearrangements necessary to transform the X chromosome of one species into that of the other are indicated.
Figure 3.—
Relative position and orientation of the 17 chromosome 2 segments conserved during the divergence between D. repleta and D. buzzatii. Conserved segments are numbered consecutively (from telomere to centromere) in D. repleta. Nine rearrangements are required to transform chromosome 2 of one species into that of the other. The particular inversion trajectory depicted here is the one subject to testing and corroborated by further evidence.
TABLE 1.
Paracentric inversions fixed between D. repleta and D. buzzatii on chromosomes X and 2
| Distal breakpoint
|
Proximal breakpoint
|
||||
|---|---|---|---|---|---|
| Chromosome | Inversion | Banda | Clone | Banda | Clone |
| X | Xa | C4d | 14H02-D | D4d | 14H02-D |
| Xb | F1d | — | G1g | — | |
| Xc | E4d | 2J01-P | F2h | 2J01-P | |
| 2 | 2a | A3c | 1J19-D | C1e | 1J19-D |
| 2b10 | B1b | 18B23-D | C3a | 18B23-D | |
| 22C11-P | 22C11-P | ||||
| 2c10 | B1i | 12O03-D | C1k | 12O03-D | |
| 5N07-P | 5N07-P | ||||
| 2b | C6a | 12G23-D | D1g | 12G23-D | |
| 2t8 | G1g | 5B11-D | G2g | 5B11-D | |
| 23F23-P | 23F23-P | ||||
| 23O17-P | 23O17-P | ||||
| 2u8 | F6e | 17K22-P | G2a | 17K22-P | |
| 22N23-D | |||||
| 2m | D3e | 14B19-D | F2a | 14B19-D | |
| 20O19-P | |||||
| 2n | F2a | 16H04-P | F6h | 16H04-P | |
| 20O19-D | |||||
| 2z7 | F1h | 26D17-D | G2a | 26D17-D | |
| 22N23-P | |||||
D. buzzatii BAC clones giving two hybridization signals in the D. repleta chromosomes and containing inversion breakpoints are also indicated (P or D after clone name indicates that the clone contains the proximal or distal inversion breakpoint in the D. buzzatii chromosome).
Breakpoint coordinates refer to the cytological map of D. repleta (Wharton 1942).
Estimation of the number of rearrangements:
GRIMM software (Tesler 2002) was used to estimate the minimum number of rearrangement events and find optimal scenarios for the transformation of one genome into another. A minimum of three rearrangement events are needed to transform the X chromosome of one species into the other. The extension and relative position of these three rearrangements (Figure 2) agree well with the three inversions, Xa, Xb, and Xc, fixed in the lineage from Primitive I to D. repleta. However, the breakpoints of these inversions (given in Table 1) differ slightly from those previously reported (Ranz et al. 2003). Inversions Xb and Xc are overlapping whereas inversion Xa is independent from the other two (Figure 2). GRIMM software provides three possible scenarios for the occurrence of these three inversions that differ in the temporal position of Xa relative to the other two inversions. The order of fixation of these three inversions was not investigated further as they appear to co-occur in all those species where they are present (Wasserman 1992). However, we can conclude that it must be D. buzzatii = Primitive I → Xc → Xb → D. repleta with inversion Xa taking place before Xc, after Xc, or after inversion Xb.
To explain the present organization of chromosome 2 in both species the minimum number of rearrangements is nine (Figure 3). Rearrangements taking place in the distal region of the chromosome, involving chromosomal segments 1–9, are independent of those occurring in the proximal region of the chromosome, segments 9–17. Thus, both regions can be separately analyzed. In the distal region, the minimum possible number of rearrangements is four (Figure 3) and there are eight different pathways transforming one chromosome into the other, implying the same four inversions and differing only in the order in which they took place. The extension and relative position of these four rearrangements agree well with the inversions 2a, 2b, 2b10, and 2c10 fixed in the lineage from Primitive I to D. repleta (Ranz et al. 2003), yet some of the breakpoint coordinates must be slightly modified (Table 1). In the proximal region of chromosome 2 at least five inversions are needed to transform the D. repleta chromosome into that of D. buzzatii. There are 18 possible scenarios with five inversions and different scenarios imply different inversions. This is so because there are only 9 conserved segments and eight breakpoints for five inversions, implying a minimum of two breakpoint reuses (see discussion). We have explored all possible scenarios and 2 of them approximately match the inversions proposed to have been fixed in this region between these species: 2m, 2n, 2z7, 2u8, and 2t8 (Figure 3). The 2 scenarios differ only in the order of occurrence of 2m and 2n, two tandemly arranged inversions that seem to share the middle breakpoint.
Test of the inversion phylogeny:
Twelve clones containing inversion breakpoints were hybridized to seven other Drosophila species, besides D. buzzatii and D. repleta, to test for the presence of the inversions in these species. Each clone produces a single hybridization signal in D. buzzatii chromosome 2 (the source of the BAC clones) and two hybridization signals in D. repleta chromosome 2 (implying a breakpoint). If the clone produces a single hybridization signal in the chromosome of a third species, this implies a similar arrangement to that of D. buzzatii. If the clone yields two hybridization signals, then an arrangement similar to D. repleta can be inferred. The important point is that the two clones that represent the distal and proximal breakpoints of an inversion should behave similarly. That is, if the proposed inversion trajectory is correct, the results produced by the two breakpoints of an inversion should be congruent. This is not expected if the proposed inversion pathway is wrong.
Sixty-two out of 64 hybridizations attempted with the 12 breakpoint-bearing BAC clones were successful and the results were generally congruent (Table 2). For inversion 2u8 a clone containing the proximal breakpoint was available and this clone failed to produce a detectable hybridization signal in D. hydei and D. meridiana. In two other cases, one clone containing the breakpoint of an inversion gave one signal whereas a second clone containing the other breakpoint yielded two signals. This occurred with inversion 2m in D. meridiana and inversion 2z7 in D. peninsularis (Table 2). These two exceptional cases were interpreted as meaning that the same arrangement as in D. repleta was present (but one of the hybridization signals could not be detected). The localization of the signals in chromosome 2 of these species (not shown) supports this interpretation.
TABLE 2.
Number of hybridization signals produced by 12 BAC clones containing inversion breakpoints in nine Drosophila species of the repleta group
| Clone | 18B23 | 22C11 | 12O03 | 5N07 | 17K22 | 23F23 | 5B11 | 14B19 | 20O19 | 16H04 | 22N23 | 26D17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inversion | 2b10 | 2b10 | 2c10 | 2c10 | 2u8 | 2t8 | 2t8 | 2m | 2m/2n | 2n | 2u8/2z7 | 2z7 |
| Breakpoint | D | P | D | P | P | P | D | D | P/D | P | D/P | D |
| D. repleta | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D. mercatorum | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D. peninsularis | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| D. hydei | ND | ND | ND | ND | 0 | 1 | 1 | 2 | 2 | 2 | 2 | 2 |
| D. meridiana | ND | ND | ND | ND | 0 | 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| D. mojavensis | ND | ND | ND | ND | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 |
| D. stalkeri | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| D. martensis | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D. buzzatii | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
D, distal breakpoint; P, proximal breakpoint; ND, not determined.
Four clones containing the breakpoints of inversions 2b10 and 2c10 were hybridized to the chromosomes of D. peninsularis and D. mercatorum. These clones were tested only in these two species because inversions 2b10 and 2c10 were seemingly fixed in the lineage leading from Primitive I to the D. repleta and D. mercatorum subgroups. In D. peninsularis, the four clones gave a single signal, indicating that this species does not contain any of the two inversions (Table 2). In D. mercatorum, clones containing the breakpoints of inversion 2c10 gave a single signal while clones containing the breakpoints of inversion 2b10 gave two signals (Table 2). Thus D. mercatorum contains inversion 2b10 but does not contain inversion 2c10. We cannot conclude when inversion 2b took place but the order of the other inversions in the distal region of the chromosome is as follows: D. buzzatii = Primitive I → 2a → 2b10 → 2c10 → D. repleta.
Eight clones containing the breakpoints of the five inversions fixed in the proximal half of chromosome 2 were hybridized to the chromosomes of seven other species of the repleta group (Table 2). The two clones from inversion 2t8 gave two signals only in D. mercatorum and D. peninsularis, implying that this inversion is fixed in both species (and absent in all the other species). In contrast, the clone containing the proximal breakpoint of inversion 2u8 produced a single signal in all species but D. repleta, implying that 2u8 is fixed only in the latter species. The three clones containing the breakpoints of inversions 2m and 2n gave a single signal in D. stalkeri and D. martensis, which means they are fixed in these two species (as in D. buzzatii), and two signals in the remaining species (that lack these two inversions). Finally, the two clones from inversion 2z7 gave a single signal in D. martensis that must be fixed for the inversion (as D. buzzatii) and two signals in the rest of the species (that must lack this inversion as in D. repleta). The order of inversions fixed in the proximal region of the chromosome is either D. buzzatii → 2z7 → 2m → 2n → 2t8 → 2u8 → D. repleta or D. buzzatii → 2z7 → 2n → 2m → 2t8 → 2u8 → D. repleta.
DISCUSSION
Inversion phylogeny of the repleta group:
A number of bioinformatic approaches and methods are available for inferring the number and trajectory of rearrangements fixed during the divergence of two genomes from comparative mapping data (see Sankoff 2004 for a review). However, bioinformatic analyses of genome sequences often deviate from more traditional cytogenetic views of chromosomal evolution (Bourque et al. 2006; Froenicke et al. 2006). Here, a combination of bioinformatic and cytological approaches is used to determine the correct rearrangement phylogeny of D. buzzatii. We have used in situ hybridization of 180 clones from a D. buzzatii genomic BAC library to the chromosomes of D. repleta and previous mapping data for another 212 markers (Ranz et al. 2003) to determine the number and orientation of conserved segments in chromosomes X and 2 between D. buzzatii and D. repleta. The previous markers were spread approximately at random over chromosomes X and 2, thus leaving gaps or uncovered regions of variable size (Ranz et al. 2003). In contrast, we selected our BAC clones to optimize coverage and ascertain the extent and orientation of conserved segments. In most cases BAC clones were identified, encompassing the conserved segments boundaries (i.e., bearing inversion breakpoints). Thus, we are reasonably confident that all conserved segments >150 kb have been identified. Seven conserved segments were found in chromosome X and 17 in chromosome 2. The extent and orientation of all conserved segments was determined (Figures 2 and 3). In addition we used GRIMM (Tesler 2002) to estimate the minimum number of rearrangements required to transform the chromosomes of D. repleta into those of D. buzzatii and to explore all possible scenarios or pathways.
The hybridization of 12 breakpoint-bearing clones representing seven different inversions in seven other Drosophila species of the repleta group was used to ascertain the correct inversion phylogeny. The results are summarized in Figure 1B. D. buzzatii differs from D. repleta by 14 chromosomal inversions and its chromosomal arrangement can be represented by the formula: Xabc, 2abt8u8b10c10mnz7, 3b, 4, 5g. The arrangement of the conservative chromosomes 3, 4, and 5 has not been analyzed here but inferred from previous work (Wasserman 1992; Ruiz and Wasserman 1993). The cytological map of D. buzzatii corresponding to its chromosomal arrangement can be seen in Figure 2 of González et al. (2005).
Ten inversions became fixed in the lineage from Primitive I to D. repleta: Xa, Xb, Xc, 2a, 2b, 2t8, 2u8, 2b10, 2c10, and 3b (Figure 1B). Inversion 2t8 was found by Ruiz and Wasserman (1993) in D. repleta and D. peninsularis and led Wasserman (1992) to include the latter species in the repleta complex of the repleta subgroup. However, Ruiz and Wasserman (1993) did not analyze the chromosomes of D. mercatorum. The results presented here show that in fact this species also has 2t8 fixed and thus this inversion does not indicate a close relationship between D. peninsularis and D. repleta. Wasserman (1992) reported that only two inversions, 2a and 2b, have been fixed in the distal region of chromosome 2. Our results support the observations of Ranz et al. (2003), which proposed that besides 2a and 2b another two inversions, 2b10 and 2c10, have been fixed in this chromosomal region. These two inversions were overlooked in previous analyses because 2c10 is included within 2b10 and the combination of the two inversions nearly restores the original banding pattern. We also show that inversion 2b10 is found in D. repleta and D. mercatorum but not in D. peninsularis whereas 2c10 is exclusive of D. repleta. The closer relationship between D. mercatorum and D. repleta than between D. peninsularis and D. repleta contradicts the current inversion phylogeny (Figure 1A) but is consistent with diverse lines of evidence. Wharton (1944) classified D. peninsularis as a member of the D. mercatorum subgroup. Vilela (1983) noted that D. peninsularis is morphologically closer to D. carcinophila (a member of the D. mercatorum subgroup) than to any other described species and accordingly placed D. peninsularis within the D. mercatorum subgroup. Finally, Durando et al. (2000), using sequences from four mitochondrial genes and one nuclear gene, observed a close relationship between the repleta and the mercatorum subgroups. Their data and our cytological results suggest that the classification of these lineages as two separate subgroups is probably not warranted and should be reconsidered.
Breakpoint reuse and genome evolution model:
In the repleta species group the cytological coincidences between breakpoints of different inversions are quite common (Wasserman 1992). For instance, 96 breakpoint reuses can be inferred for the 208 inversions described in chromosome 2. This rate of coincidence still holds true within subgroups or even within complexes (Cáceres et al. 1997). Obviously, some of these apparent coincidences can be the consequence of the limited resolution of the cytological technique. The increased resolution provided by the in situ hybridization technique allows us to map more accurately inversion breakpoints and test for breakpoint reuses.
In the species analyzed here three breakpoint reuses have been described, one in chromosome X and two in chromosome 2. Wasserman (1992) stated that inversions Xb and Xc were arranged in tandem and shared the middle breakpoint. However, our results support the claim of Ranz et al. (2003) that these two inversions are in fact overlapping (Figure 2). In chromosome 2, Ruiz and Wasserman (1993) proposed that inversions 2m and 2n are also tandemly arranged and share the middle breakpoint and also that inversion 2z7 shares its distal breakpoint with that of inversion 2m. The identification of clone 20O19 containing the proximal breakpoint of 2m and the distal breakpoint of 2n confirms the first coincidence. The fact that this clone produces two hybridization signals on the D. repleta chromosomes instead of three suggests that the two breakpoints must be very close to each other or even at the same molecular site. However, our results show that inversion 2z7 does not share its distal breakpoint with that of inversion 2m although it is relatively close (see Figure 3). On the other hand, our results suggest that inversion 2z7 shares one breakpoint with inversion 2u8 (Figure 3). Again, the fact that clone 22N23 gives two hybridization signals instead of three suggests a close proximity of the two breakpoints. In summary, of three breakpoint reuses previously described (Ruiz and Wasserman 1993), only one still holds at the resolution level of the clone size used in this study (∼150 kb), but a new one has been found, leaving the total at two. This coincidence of inversion breakpoints is intriguing and deserves further scrutiny at the DNA sequence level.
Breakpoint reuse is a quite common phenomenon in diverse organisms. Pevzner and Tesler (2003) compared the human and mouse genomes and observed clumps of closely located breakpoints that could not be explained by the “random breakage model” (Nadeau and Taylor 1984). They proposed an alternative model that envisages mammalian genomes as a mosaic of relatively short fragile regions with a high propensity for rearrangements and solid regions with a low propensity for rearrangements. These fragile regions may correspond to segmental duplications or regions with an unusually high concentration of transposable elements (TEs) or with a palindromic structure (Eichler and Sankoff 2003; Murphy et al. 2005). Segmental duplications represent ∼5% of the human genome (Bailey et al. 2002) and ∼2% of the mouse genome (Bailey et al. 2004a). They induce rearrangements by unequal crossing over (Shaffer and Lupski 2000) and are hotspots for mammalian chromosomal evolution (Bailey et al. 2004b; Zody et al. 2006).
The Drosophila genome is quite different from that of mammals: the amount of repetitive DNA is much lower (5 vs. 44%) and the fraction of segmental duplications is negligible (Lander et al. 2001; Celniker and Rubin 2003). Accordingly, inversion breakpoint reuse in Drosophila is likely to have a different cause. Molecular studies of breakpoint regions in natural Drosophila inversions have revealed the presence of TEs in some cases (Cáceres et al. 1999, 2001; Casals et al. 2003). By contrast no clear evidence for the implication of TEs was found in other studies (Wesley and Eanes 1994; Cirera et al. 1995; Andolfatto et al. 1999; Matzkin et al. 2005; Richards et al. 2005). The conclusion is that at least in some species or species groups, TEs are responsible for the origin of chromosomal inversions.
A plausible hypothesis for breakpoint reuse in Drosophila can be set forth. When an inversion has been generated by a TE, copies of this element will be flanking the inverted segment in the chromosome with the inversion (Lim and Simmons 1994; Gray 2000). If the inversion succeeds and goes to fixation, these TE insertions will be brought to fixation as well. These fixed TE insertions will have a much higher probability to be involved in further chromosome breakages than the rest of the TE insertions (that usually have a rather low population frequency because of the equilibrium between transposition and selection; Charlesworth et al. 1994). Although one expects that these insertions will be removed by deletion in the long run (Petrov et al. 1996; Singh and Petrov 2004), they may last in the genome several million years (average time to loss of 50% nonfunctional DNA is ∼12 MY). Furthermore, because of the reduction of recombination in the heterokaryotypes, inversion breakpoint regions often accumulate additional TE insertions besides the one that originated the inversion (Cáceres et al. 2001, 2003; Sharakhov et al. 2006). When the inversion goes to fixation, some of these TE insertions may become fixed with the inversion while other may remain polymorphic. In any case, the unusual high density of TE insertions at inversion breakpoints will increase the chances of further chromosome breakages at these sites, i.e., the chances of breakpoint reuse. One prediction of this model is that inversions sharing breakpoints are expected to arise in a temporal succession within the same lineage, something that seems to be frequent in the repleta species group (Wasserman 1992). In our study, the model would apply to the breakpoint coincidence between inversions 2m and 2n (which occurred in the same lineage) but not to that between inversions 2z7 and 2u8 (which occurred in different lineages).
Rates of chromosomal rearrangement fixation in the genus Drosophila:
Our results show that three paracentric inversions in chromosome X and nine in chromosome 2 have been fixed during the divergence between D. repleta and D. buzzatii. These numbers agree with the general pattern in the repleta group where chromosome 2 has been found to be the most dynamic, harboring ∼70% of all inversions (Wasserman 1992). The size of the genome of the repleta group species is ∼220 Mb with 70% in the euchromatin (Schulze and Lee 1986). The total number of inversions fixed between these two species is 14 and thus we can estimate an average rate of rearrangement fixation of 0.004 disruptions/Mb/MY.
Rates of rearrangement fixation have been estimated in the genus Drosophila using different species pairs. We have normalized these estimates as the number of disruptions per megabase and per million years to make them comparable (Table 3). These estimates must be taken with caution because different estimation methods have been used for different comparisons and some of the studies did not include all chromosomal elements. However, they are likely to be accurate enough for a broad overview of chromosomal evolution in the genus. Two conclusions can be drawn. First, rates of rearrangement vary between chromosomal elements as proposed by González et al. (2002), although the element exhibiting the highest rate can vary. Second, rates of rearrangement differ between lineages, the rate within the Sophophora subgenus being generally higher that that within the Drosophila subgenus (Papaceit et al. 2006). This agrees well with the distribution of polymorphic inversions in these subgenera (Sperlich and Pfriem 1986; Powell 1997). The lowest rate for the entire genus is that observed here for the comparison D. buzzatii–D. repleta. The highest rate is probably that corresponding to the comparison D. miranda–D. pseudoobscura (Bartolomé and Charlesworth 2006) that is 24–58 times higher. Twenty-fold differences in rearrangement rate have been reported between different vertebrate lineages (Coghlan et al. 2005; Murphy et al. 2005). It is remarkable that similar differences can be found within a single genus of flies. Substantial variation in rearrangement rate is also evident even within the repleta group (Figure 1B). Some lineages, such as those leading to D. mercatorum or D. mojavensis, are highly dynamic, whereas other lineages, such as that of D. hydei, are much more conservative. Four factors may help to explain the great variation in rearrangement rates between Drosophila lineages: generation time, population size, mutation rate, and environmental conditions (i.e., selection regime). While all of them probably make a contribution, we believe that mutation rate may play a crucial role. If chromosomal inversions are generated by TEs, their generation rate is likely to vary greatly between lineages because so does the activity of TEs. It is intriguing, however, that the rate of rearrangement seems to be highest in the subgenus Sophophora where until now clear evidence for the implication of TEs in the generation of chromosomal inversions has not been found.
TABLE 3.
Rates of chromosomal rearrangement fixation (number of disruptions per megabase and per million years) in the genus Drosophila
| Muller's chromosomal element
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Subgenus | Species paira | A | B | C | D | E | A–F | Referenceb |
| Sophophora | D. pseudoobscura–D. melanogaster | 0.088 | 0.144 | 0.153 | — | — | 0.128 | 1, 2 |
| D. subobscura–D. pseudoobscura | 0.067 | 0.183 | 0.125 | 2 | ||||
| D. subobscura–D. melanogaster | 0.143 | 0.133 | 0.138 | 2 | ||||
| D. pseudoobscura–D. melanogaster | 0.087 | 0.055 | 0.092 | 0.055 | 0.073 | 0.072 | 3 | |
| D. miranda–D. pseudoobscura | 0.130–0.204 | — | 0.126–0.379 | — | 0.047–0.158 | 0.096–0.233 | 4 | |
| D. miranda–D. melanogaster | 0.011–0.026 | — | 0.021–0.048 | — | 0.007–0.032 | 0.012–0.030 | 4 | |
| D. pseudoobscura–D. melanogaster | 0.011–0.064 | — | 0.023–0.048 | — | 0.007–0.016 | 0.013–0.040 | 4 | |
| Drosophila | D. virilis–D. novamexicana | 0.053 | — | — | 0.018 | — | 0.018 | 5 |
| D. virilis–D. montana | 0.041 | — | — | 0.07 | — | 0.012 | 5 | |
| D. montana–D. novamexicana | 0.041 | — | — | 0.011 | — | 0.013 | 5 | |
| D. virilis–D. buzzatii | — | — | — | — | 0.021 | 0.021 | 6 | |
| D. virilis–D. repleta | — | — | — | — | 0.029 | 0.029 | 6 | |
| D. repleta–D. buzzatii | 0.005 | 0.001 | 0.001 | 0 | 0.006 | 0.004 | 7 | |
| Sophophora– Drosophila | D. melanogaster–D. repleta | 0.087 | 0.021 | — | 0.045 | 0.079 | 0.058 | 8 |
For all species pairs including D. melanogaster, we used the genome size and chromosome size in Celniker and Rubin (2003). For comparisons with D. pseudoobscura, the genome and chromosome sizes reported by Richards et al. (2005) were used. For comparisons between repleta group species, we used 154 Mb as the euchromatic genome size (Schulze and Lee 1986) and the relative sizes of each chromosome given by Wasserman (1992). Finally, for the virilis species group 150 Mb was used as the euchromatic genome size (Hartl and Lozovskaya 1995) and  for each chromosome.
for each chromosome.
References: (1) Segarra et al. (1995); (2) Papaceit et al. (2006); (3) Richards et al. (2005); (4) Bartolomé and Charlesworth (2006); (5) Vieira et al. (1997); (6) Ranz et al. (1999); (7) Wasserman (1992), Ruiz and Wasserman (1993), and this work; (8) Ranz et al. (2001), González et al. (2002).
Acknowledgments
We thank Alejandra Delprat, Oriol Calvete, Marta Puig, and two anonymous reviewers for useful comments on the manuscript and Tomás Morán for the D. hydei stock. This work was supported by grants BMC2002-01708 and BFU2005-02237 from the Secretaría de Estado de Universidades e Investigación (Ministerio de Educación y Ciencia, Spain) awarded to A.R.
References
- Andolfatto, P., J. D. Wall and M. Kreitman, 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153: 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. A., Z. Gu, R. A. Clark, K. Reinert, R. V. Samonte et al., 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Bailey, J. A., D. M. Church, M. Ventura, M. Rocchi and E. E. Eichler, 2004. a Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 14: 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. A., R. Baertsch, W. J. Kent, D. Haussler and E. E. Eichler, 2004. b Hotspots of mammalian chromosomal evolution. Genome Biol. 5: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé, C., and B. Charlesworth, 2006. Rates and patterns of chromosomal evolution in Drosophila pseudoobscura and D. miranda. Genetics 173: 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., P. A. Pevzner and G. Tesler, 2004. Reconstructing the genome architecture of ancestral mammals: lessons from human, mouse and rat genomes. Genome Res. 14: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., G. Tesler and P. A. Pevzner, 2006. The convergence of cytogenetics and rearrangement-based models of ancestral genome reconstruction. Genome Res. 16: 311–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres, M., A. Barbadilla and A. Ruiz, 1997. Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: Is inversion length a selected trait? Evolution 51: 1149–1155. [DOI] [PubMed] [Google Scholar]
- Cáceres, M., J. M. Ranz, A. Barbadilla, M. Long and A. Ruiz, 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415–418. [DOI] [PubMed] [Google Scholar]
- Cáceres, M., M. Puig and A. Ruiz, 2001. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 11: 1353–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals, F., M. Cáceres and A. Ruiz, 2003. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol. Biol. Evol. 20: 674–685. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., and G. M. Rubin, 2003. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 4: 89–117. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., P. Sniegowski and W. Stephan, 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Cirera, S., J. M. Martín-Campos, C. Segarra and M. Aguadé, 1995. Molecular characterization of the breakpoints of an inversion fixed between Drosophila melanogaster and D. subobscura. Genetics 139: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A., and K. H. Wolfe, 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A., E. Eichler, S. Oliver, A. Paterson and L. Stein, 2005. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet. 21: 673–682. [DOI] [PubMed] [Google Scholar]
- Coluzzi, M., A. Sabatini, A. della Torre, M. A. Di Deco and V. Petrarca, 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. [DOI] [PubMed] [Google Scholar]
- Durando, C. M., R. H. Baker, W. J. Etges, W. B. Heed, M. Wasserman et al., 2000. Phylogenetic analysis of the repleta species group of the genus Drosophila using multiple sources of characters. Mol. Phylogenet. Evol. 16: 296–307. [DOI] [PubMed] [Google Scholar]
- Eichler, E. E., and D. Sankoff, 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793–797. [DOI] [PubMed] [Google Scholar]
- Froenicke, L., M. Garcia, A. Graphodatsky, S. Müller, L. A. Lyons et al., 2006. Are molecular cytogeneticist and bioinformatics suggesting diverging models of ancestral mammalian genomes. Genome Res. 16: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, J., J. M. Ranz and A. Ruiz, 2002. Chromosomal elements evolve at different rates in the Drosophila genome. Genetics 161: 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, J., M. Nefedov, I. Bosdet, F. Casals, O. Calvete et al., 2005. A BAC-based physical map of the Drosophila buzzatii genome. Genome Res. 15: 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H., 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16: 461–468. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., and E. R. Lozovskaya, 1995. The Drosophila Genome Map: A Practical Guide. Springer, New York.
- Kazazian, Jr., H. H., 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Lagercrantz, U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lim, J. K., and M. J. Simmons, 1994. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays 16: 269–275. [DOI] [PubMed] [Google Scholar]
- MacIntyre, R. J., and G. E. Collier, 1986. Protein evolution in the genus Drosophila, pp. 39–146 in The Genetics and Biology of Drosophila, Vol. 3e, edited by M. Ashburner, L. Carson and J. N. Thompson, Jr. Academic Press, New York.
- Matzkin, L. M., Th. J. S. Merritt, C.-T. Zhu and W. Eanes, 2005. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics 170: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, E., B. Charlesworth and C. H. Langley, 1987. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet. Res. 49: 31–41. [DOI] [PubMed] [Google Scholar]
- Murphy, W. J., D. M. Larkin, A. Everts-van der Wind, G. Bourque, G. Tesler et al., 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617. [DOI] [PubMed] [Google Scholar]
- Nadeau, J. H., and B. A. Taylor, 1984. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. USA 81: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Grady, P. M., R. H. Baker, C. M. Durando, W. J. Etges and R. DeSalle, 2001. Polytene chromosomes as indicators of phylogeny in several species groups of Drosophila. BMC Evol. Biol. 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaceit, M., M. Aguadé and C. Segarra, 2006. Chromosomal evolution of elements B and C in the Sophophora subgenus of Drosophila: evolutionary rate and polymorphism. Evolution 60: 768–781. [PubMed] [Google Scholar]
- Pardue, M. L., K. Lowenhaupt, A. Rich and A. Nordheim, 1987. (dC-dA)n(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 6: 1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D., E. R. Lozovskaya and D. L. Hartl, 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Pevzner, P., and G. Tesler, 2003. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. USA 100: 7672–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J. R., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, New York.
- Ranz, J. M., C. Segarra and A. Ruiz, 1997. Chromosomal homology and molecular organization of Muller's elements D and E in the Drosophila repleta species group. Genetics 145: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., M. Cáceres and A. Ruiz, 1999. Comparative mapping of cosmids and gene clones from a 1.6 Mb chromosomal region of Drosophila melanogaster in three species of the distantly related subgenus Drosophila. Chromosoma 108: 32–43. [DOI] [PubMed] [Google Scholar]
- Ranz, J. M., F. Casals and A. Ruiz, 2001. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res. 11: 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., J. González, F. Casals and A. Ruiz, 2003. Low occurrence of gene transposition events during the evolution of the genus Drosophila. Evolution 57: 1325–1335. [DOI] [PubMed] [Google Scholar]
- Richards, S., Y. Liu, B. R. Bettencourt, P. Hradecky, S. Letovsky et al., 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, A., and M. Wasserman, 1993. Evolutionary cytogenetics of the Drosophila buzzatii species complex. Heredity 70: 582–596. [DOI] [PubMed] [Google Scholar]
- Ruiz, A., A. Fontdevila and M. Wasserman, 1982. The evolutionary history of Drosophila buzzatii. III. Cytogenetic relationships between two sibling species of the buzzatii cluster. Genetics 101: 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, C. A., N. Takezaki and M. Nei, 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12: 391–404. [DOI] [PubMed] [Google Scholar]
- Sankoff, D., 2003. Rearrangements and chromosomal evolution. Curr. Opin. Genet. Dev. 13: 583–587. [DOI] [PubMed] [Google Scholar]
- Sankoff, D., 2004. Conserved segment statistics and rearrangement inferences in comparative genomics, pp. 1–24 in Mathematics of Evolution and Phylogeny, edited by O. Gascuel. Clarendon Press, Oxford.
- Schulze, D. H., and C. S. Lee, 1986. DNA sequence comparison among closely related Drosophila species in the mulleri complex. Genetics 113: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, C., E. R. Lozovskaya, G. Ribo, M. Aguade and D. L. Hartl, 1995. P1 clones from Drosophila melanogaster as markers to study the chromosomal evolution of Muller's A element in two species of the obscura group of Drosophila. Chromosoma 104: 129–136. [DOI] [PubMed] [Google Scholar]
- Shaffer, L. G., and J. R. Lupski, 2000. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 34: 297–329. [DOI] [PubMed] [Google Scholar]
- Shapiro, J., 2005. A 21st century view of evolution: genome system architecture, repetitive DNA, and natural genetic engineering. Gene 345: 91–100. [DOI] [PubMed] [Google Scholar]
- Sharakhov, I. V., A. C. Serazin, O. G. Grushko, A. Dana, N. Lobo et al., 2002. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science 298: 182–185. [DOI] [PubMed] [Google Scholar]
- Sharakhov, I. V., B. J. White, M. V. Sharakhova, J. Kayondo, N. F. Lobo et al., 2006. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc. Natl. Acad. Sci. USA 103: 6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. D., and D. A. Petrov, 2004. Rapid sequence turnover at an intergenic locus in Drosophila. Mol. Biol. Evol. 21: 670–680. [DOI] [PubMed] [Google Scholar]
- Sperlich, D., and P. Pfriem, 1986. Chromosomal polymorphism in natural and experimental populations, pp. 257–309 in The Genetics and Biology of Drosophila, Vol. 3a, edited by M. Ashburner, L. Carson and J. N. Thompson, Jr. Academic Press, London.
- Spicer, G. S., 1988. Molecular evolution among some Drosophila species groups as indicated by two-dimensional electrophoresis. J. Mol. Evol. 27: 250–260. [DOI] [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kumar, 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Tesler, G., 2002. GRIMM: genome rearrangements web server. Bioinformatics 18: 492–493. [DOI] [PubMed] [Google Scholar]
- Tonzetich, J., T. W. Lyttle and H. L. Carson, 1988. Induced and natural break sites in the chromosomes of Hawaiian Drosophila. Proc. Natl. Acad. Sci. USA 85: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, J., C. P. Vieira, D. L. Hartl and E. R. Lozovskaya, 1997. Discordant rates of chromosome evolution in the Drosophila virilis species group. Genetics 147: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela, C. R., 1983. A revision of the Drosophila repleta species group (Diptera, Drosophilidae). Revta. Bras. Entomol. 27: 1–114. [Google Scholar]
- Wasserman, M., 1954. Cytological studies of the repleta group. Univ. Texas Publ. 5422: 130–152. [Google Scholar]
- Wasserman, M., 1962. Cytological studies of the repleta group of the genus Drosophila: V. The mulleri subgroup. Univ. Texas Publ. 6205: 85–117. [Google Scholar]
- Wasserman, M., 1963. Cytology and phylogeny of Drosophila. Am. Nat. 97: 333–352. [Google Scholar]
- Wasserman, M., 1982. Evolution of the repleta group, pp. 61–139 in The Genetics and Biology of Drosophila, Vol. 3b, edited by M. Ashburner, H. L. Carson and J. N. Thompson, Jr. Academic Press, New York.
- Wasserman, M., 1992. Cytological evolution of the Drosophila repleta species group, pp. 455–552 in Drosophila Inversion Polymorphism, edited by C. B. Krimbas and J. R. Powell. CRC Press, Boca Raton, FL.
- Wesley, C. S., and W. F. Eanes, 1994. Isolation and analysis of the breakpoint sequence of chromosomal inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton, L. T., 1942. Analysis of the repleta group of Drosophila. Univ. Texas Publ. 4228: 23–52. [Google Scholar]
- Wharton, L. T., 1944. Interspecific hybridization in the repleta group. Univ. Texas Publ. 4445: 175–193. [Google Scholar]
- White, M. J. D., 1973. Animal Cytology and Evolution. Cambridge University Press, Cambridge, UK.
- Yogeeswaran, K., A. Frary, T. L. York, A. Amenta, A. H. Lesser et al., 2005. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E. M., C. von Mering, I. Letunic, D. Torrents, M. Suyama et al., 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149–159. [DOI] [PubMed] [Google Scholar]
- Zody, M. C., M. Garber, D. J. Adams, T. Sharpe, J. Harrow et al., 2006. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature 440: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]