Abstract
The mimp1 element previously identified in the ascomycete fungus Fusarium oxysporum has hallmarks of miniature inverted-repeat transposable elements (MITEs): short size, terminal inverted repeats (TIRs), structural homogeneity, and a stable secondary structure. Since mimp1 has no coding capacity, its mobilization requires a transposase-encoding element. On the basis of the similarity of TIRs and target-site preference with the autonomous Tc1-like element impala, together with a correlated distribution of both elements among the Fusarium genus, we investigated the ability of mimp1 to jump upon expression of the impala transposase provided in trans. Under these conditions, we present evidence that mimp1 transposes by a cut-and-paste mechanism into TA dinucleotides, which are duplicated upon insertion. Our results also show that mimp1 reinserts very frequently in genic regions for at least one-third of the cases. We also show that the mimp1/impala double-component system is fully functional in the heterologous species F. graminearum, allowing the development of a highly efficient tool for gene tagging in filamentous fungi.
MINIATURE inverted-repeat transposable elements (MITEs) are a particular group of transposable elements (TEs), first described in plants (Bureau and Wessler 1992, 1994b; Casacuberta et al. 1998; Zhang et al. 2000, 2001; Santiago et al. 2002), but later found in a wide range of organisms including Caenorhabditis elegans (Oosumi et al. 1996), mosquitoes (Tu 1997, 2001; Feschotte and Mouches 2000), zebrafish (Plasterk et al. 1999), humans (Smit and Riggs 1996), and fungi (Yeadon and Catcheside 1995). MITEs share common structural features: the presence of terminal inverted repeats (TIRs), a small size (usually <600 bp), and no coding region. While they resemble previously characterized nonautonomous DNA elements, such as Ds (reviewed in Kunze and Weil 2002), the first MITEs identified in plants, Tourist and Stowaway, displayed other specific features: a stable secondary structure, AT richness, particular target-site duplications (TSD), a high copy number, and a great uniformity of size and sequence. However, as new miniature elements were identified, some exceptions were observed: GC-rich elements (Tu 2001), lack of secondary structure (Bureau et al. 1996), or moderate copy number (Holyoake and Kidwell 2003), suggesting that only some characteristics are shared by all MITEs.
More recently, a link between MITE families and well-characterized DNA transposons, belonging mainly to the Tc1-mariner superfamily (Feschotte et al. 2002), has been established on the basis of TIRs and TSD sequence similarities, supporting the idea that MITEs are a particular type of nonautonomous class II transposons. However, the origin of these short elements is still unclear: some exhibiting extended regions of similarity with full-length elements may have originated by internal deletions; others with homology limited to the TIRs or to terminal regions may originate de novo by capture of DNA sequences between the TIRs.
The classification of MITEs within class II TEs also implies that they are mobilized by a transposase from autonomous members of their own (or a related) family by a cut-and-paste mechanism. The recent studies on a rice MITE (Jiang et al. 2003; Kikuchi et al. 2003) confirmed the cut-and-paste mechanism and indicated that amplification could be influenced by specific (environmental or genomic) conditions (Jiang et al. 2003; Shan et al. 2005). However, although putative autonomous partners have been proposed for several MITE families, a clear demonstration that they are indeed necessary for MITE transposition is still lacking.
In Fusarium oxysporum, numerous DNA transposon families have been identified, mainly by trapping into a target gene or sequencing of some genomic regions (Daboussi and Langin 1994; Hua-Van et al. 2000; Daboussi and Capy 2003). Autonomous members of the Tc1-mariner superfamily have been identified: Fot1 is a pogo-like element that may be present in a relatively high copy number (∼100 copies), essentially full-length copies (Daboussi et al. 1992; Migheli et al. 1999); impala is a representative of the Tc1 family (Robertson 2002) present in low copy number and composed of at least five different subfamilies (E, D, F, K, and P) (Hua-Van et al. 2001a). Two of these (E and D) contain autonomous members presenting up to 20% divergence at the DNA level (Hua-Van et al. 1998, 2001b). By sequencing genomic regions surrounding copies of the D subfamily, five elements of ∼200 bp were identified. These elements, called mimp, have no coding capacity but display TIRs very similar to those of impala (Hua-Van et al. 2000). We wondered whether these short elements are mobile and, if so, whether they could be transactivated by an impala transposase. Here we show that the mimp1 family is present in a number of related species and that its distribution follows the distribution of impala. Using a phenotypic excision assay in two different species, we demonstrate that they are trans-mobilized by the impala transposase and that transposition occurs through a cut-and-paste mechanism. The functionality of the double-component system in the heterologous species F. graminearum, the genome sequence of which is available, provided the opportunity to analyze the insertion site preference of newly transposed copies. Preliminary results suggest that this system is an interesting alternative tool for gene tagging in filamentous fungi.
MATERIALS AND METHODS
Fungal strains:
F. oxysporum strains FOM150 nia9 and FO5 nia13 are nitrate-reductase-deficient mutants usually used as recipient strains in transformation experiments (Migheli et al. 1999; Hua-Van et al. 2001b). The F. graminearum strain Fg820 was kindly provided by G. H. J. Kema. Four nitrate-reductase mutants, named Fg820 nia1, Fg820 nia5, Fg820 nia6, and Fg820 nia14, were obtained as previously described and also used as recipient strains in transformation experiments. Strains belonging to the F. oxysporum complex (FO complex) and used in this study (FOM24, FOM150, FOM7, FOM466A, FOMP2, FO5, FO47, FOR4, FORL28, FOA1, FOLn3, FOVR1, FOVR3, FOD11, FOL15, and FOMK419) have been described in Hua-Van et al. (2001a). Strains from related species, i.e., F. foetens (NRRL31852), F. redolens (NRRL25600 and 28381), and F. hostae (NRRL29642), were obtained from K. O'Donnell (National Center for Agricultural Utilization Research, U. S. Department of Agriculture, Peoria, IL). More distant species from the Discolor (F. culmorum) and Martiella (F. caucasicum and Neocosmospora sp.) sections have also been used (Daboussi et al. 2002).
Plasmids:
pHEO62 contains the ORF encoding the impalaE transposase (Hua-Van et al. 1998), cloned between the gpdA promoter and the trpC terminator of Aspergillus nidulans. The entire cassette was extracted from plasmid pEO62 (Li Destri Nicosia et al. 2001) as an EcoRI/HindIII fragment and cloned into plasmid pBC1004 (Carroll et al. 1994), which permits hygromycin selection. pNm1H18 was constructed by introduction of a mimp1 element into the first intron of the niaD gene (Figure 3). This mimp1 copy corresponds to the reamplification of the mimp1.1 element (AF076624) using the HindImp primer (5′-GCCCTAAGCTTACAGTGGGGTGCAATAAGTTTG-3′). This primer anneals to the TIRs of mimp1 and impala and contains a HindIII site (underlined) at the 5′-end. PCR conditions were 1 min denaturation at 95°, followed by 30 cycles of 1 min at 94°, 1 min at 59°, and 30 sec at 72°, and then an elongation step of 10 min at 72°. The PCR product was then digested with HindIII and inserted into the HindIII site of the first intron of the niaD gene in plasmid pAN301 (Malardier et al. 1989), deleted from a dispensable NdeI fragment (in which a NdeI fragment located downstream of the niaD gene has been removed), resulting in the niaD∷mimp1 construct.
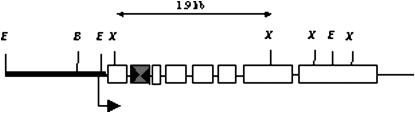
Figure 3.—
Structure of plasmid pNm1H18 used for the phenotypic assay for excision. The 1.3-kb -niaD promoter is indicated as a thick line. niaD exons are shown as open rectangles. mimp1 is the shaded rectangle in which large arrowheads represent TIRs. Restriction enzyme sites are indicated as E, EcoRI; B, BamHI; X, XbaI.
Transformation experiments:
The transformation procedure was as described (Hua-Van et al. 2001b). Plasmid pNm1H18 was introduced into strain FOM150 nia9, which contains different sources of transposases, including impalaE and D (Hua-Van et al. 2001b), together with plasmid pAN7-1 (Punt et al. 1987) carrying the selectable marker hph conferring resistance to hygromycin B. Plasmid pNm1H18 was also introduced in the FO5 nia13, Fg820 nia1, Fg820 nia5, Fg820 nia6, and Fg820 nia14 strains, devoid of all types of TEs identified within the FO complex (Migheli et al. 1999; Hua-Van et al. 2001b; Chalvet et al. 2003), together with plasmid pAN7-1 or plasmid pHEO62, which carries the source of impalaE transposase together with the selectable marker hph. In all three genetic contexts, cotransformants containing plasmid pNm1H18 were identified among hygromycin-resistant transformants through colony hybridization, using a niaD probe from plasmid pAN301 (Malardier et al. 1989).
Revertant selection:
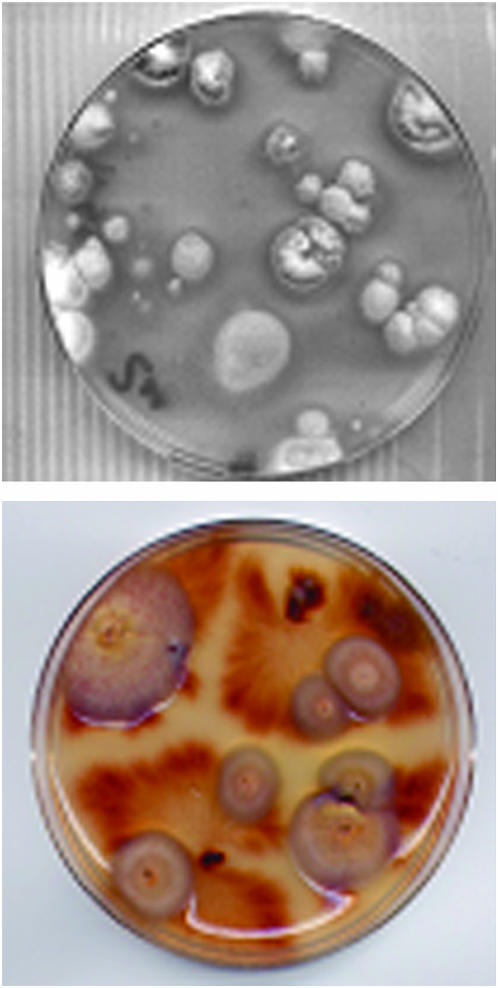
Spores of cotransformants were recovered by washing and filtration from a single spore culture grown on solid medium and spread at different dilutions on nitrate minimal agar medium supplemented with triton X100 (0.06%) as previously described (Hua-Van et al. 2001b). Alternatively, plugs of each cotransformant were picked on plates of nitrate minimal agar medium. Revertants are easily detected as patches of aerial mycelium with a wild-type phenotype on a background of sparse mycelium corresponding to a niaD mutant (Figure 4).
Figure 4.—
Selection of excision events. The high number of niaD+ colonies that can be selected from an FO5 (top) or a Fg820 (bottom) transformant carrying the niaD∷mimp construct and an exogenous source of transposase. Plates were inoculated with 103 conidia or with four mycelia plugs and incubated for 1 month at 26°.
DNA preparation and Southern blot analysis:
DNA extractions from F. oxysporum strains were conducted as previously described (Langin et al. 1990). Extraction of genomic DNA from F. graminearum strains was performed using another procedure. Approximately 106 spores were spread on a cellophane disk onto a potato dextrose agar plate. After overnight growth at 26°, germlings were scraped and frozen in liquid nitrogen. After adding 600 μl of lysis solution (100 mm Tris pH 9.0, 10 mm EDTA, 1% sarkosyl) and ∼100 μl of glass beads (200- to 500-μm diameter, Fisher Scientific, Illirch, France), the mycelium was ground using the Fastprep101 machine (twice for 30 sec at 4000 rpm). DNA was purified by successive extractions using phenol, phenol/chloroform (1/1), and chloroform and then precipitated by adding 1/20 vol 3 m sodium acetate and 0.6 vol isopropanol. After a washing step in 70% ethanol and drying, genomic DNA was resuspended in sterile distilled water plus RNAse A at a final concentration of 1 mg/ml and incubated 1 hr at 37° for RNA digestion. Ten micrograms of genomic DNA was digested with EcoRI or XbaI, separated electrophoretically on 0.7% agarose gels, and transferred on nylon membranes, using a vacuum blotter. DNA templates were 32P-labeled using the redi primeII kit (Amersham Biosciences). Hybridizations were conducted under standard conditions (Sambrook et al. 1989).
Polymerase chain reaction and primer sequences:
Hybridization probes were obtained by PCR. Primers SpeE3 and SpeE5 (Hua-Van et al. 2001a) were used to generate a PCR product corresponding to impala elements of the E subfamily, with 50 ng of genomic DNA from the FOM24 strain as a template. PCR conditions were as described in Hua-Van et al. (2001a). The HindImp primer was used, as described above, to generate either PCR products from different genomic DNAs (see Figure 1D) or the mimp1 PCR product used as a probe. The 419-bp niaD probe was generated with primers niaCG1 (5′-CACTAGTATGTGCAGGCAAC-3′) and niaCG2 (5′-TTCAGCCACTTGACACTG-3′), using the pAN301 plasmid as a template. PCR conditions were as follows: 2 min at 94°, then 30 cycles of 30 sec at 94°, 30 sec at 59°, and 2 min at 72°.
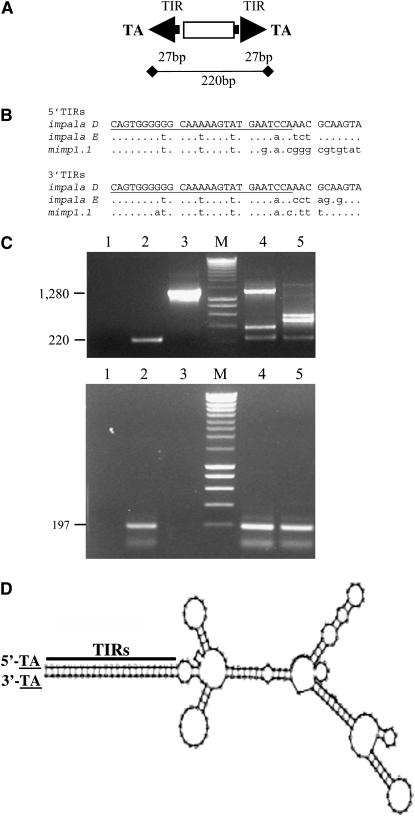
Figure 1.—
(A) mimp1 structure. (B) Sequence comparison of TIRs between impala elements and the mimp1.1 copy. Dots indicate sequences that are identical to the impala D TIRs sequence (37 bp). Lowercase letters indicate sequence variation. mimp1 and impala TIRs align over the first 27 nucleotides (underlined in the impala D sequences). (C) Agarose gel electrophoresis of PCR products obtained from two different F. oxysporum strains. Genomic DNAs of FOM24 (lane 4) and FOL15 (lane 5) were amplified using either the HindImp primer specific for TIRs (top) or for the mimp1-specific primers (bottom; see materials and methods). Lanes 1–3 correspond to controls: 1, no DNA control; 2, pNm1H18 plasmid carrying the mimp1.1 copy; 3, pNI160 plasmid carrying impala160 (Hua-Van et al. 2001b). Lanes labeled as M were loaded with a molecular weight marker (SmartLadder, Eurogentec). (D) Predicted secondary structure of the mimp1 element, using the RNAFOLD program (http://www.infobiogen.fr).
The size homogeneity of mimp1 elements in the different Fusarium strains was determined using mimp1-specific primers: SPEmimp1-5′ (5′-CAATAAGTTTGAATACCGGGCGTG-3′) and SPEmimp1-3′ (5′-GTTTGAATACCTTTTGATTTG-3′), both with overlapping TIRs (italics) and with internal sequences (underlined). In reference to the full-length mimp1.1 element previously identified (Hua-Van et al. 2000), the expected size of the PCR product is 197 bp. PCR conditions enabling the amplification of larger DNA molecules were used: a denaturation step of 1 min at 95° followed by 35 cycles of 1 min at 94°, 1 min at 53°, 2 min 30 sec at 72° and a final elongation step of 10 min at 72°.
mimp1 amplification in different strains was obtained using primer mi1 (5′-TACAGTGGGATGCAATAAGTTTGAATAC-3′) located within TIRs, alone or in combination with primers SacR (5′-CTGAGGAGGGAGCTCGATCTAGCC-3′) or SacF (5′-GGCTAGATCGAGCTCCCTCCTCAG-3′). A denaturation step of 3 min at 95° followed by 35 cycles of 30 sec at 95°, 30 sec at 59°, 1 min at 72°, and a final extension of 5 min at 72° was used.
Characterization of the FO5 endogenous mimp1 copy was performed using the HindImp primer alone or in combination with SacR or SacF. PCR conditions were 5 min at 95° and then 45 cycles of 1 min at 94°, 1 min at 59°, and 1 min at 72° and a final elongation step of 10 min at 72°.
Excision events were controlled by PCR on 50 ng of genomic DNA of putative revertants and of the corresponding transformants, using primers niaD144 (5′-GTTCATGCCGTGGTCGCTGC-3′) and niaD754r (5′-AGTTGGGAATGTCCTCGTCG-3′) under the following conditions: 4 min at 94° and then 30 cycles of 1 min at 94°, 1 min at 59°, and 2 min at 72°. The sizes of the expected PCR products are 717 bp for a transformant and 485 bp for a revertant.
Amplification of mimp1 flanking sequences:
Two approaches were used to recover mimp1 flanking sequences. For nonsequenced Fusarium strains, inverse PCR was performed. A 100-ng aliquot of genomic DNA of a given strain was digested with either one or two restriction enzymes at 37° during 30 min. The enzymes were then inactivated by incubation at 65° for 10 min. A total of 30 ng of the digested DNA was self-circularized during 30 min at room temperature using T4 DNA ligase (Biolabs) in a final volume of 10 μl. The ligase was then inactivated by incubation at 65° for 10 min. Approximately 7.5 ng of ligated DNA was used in PCR experiments with the primers Div149 (5′-GCAGGCTAAACTCCAAATAGGC-3′) and Div53 (5′-GTAGCGTGGCTCAAAGAGGC-3′), which correspond to internal regions of the canonical mimp1 element and are directed toward the TIRs. The amplification program was as follows: a denaturation step of 1 min at 95° and then 1 min at 94°, 45 sec at 58°, and 4 min at 72° for 35 cycles, followed by an elongation step of 10 min at 72°.
For revertants from the F. graminearum genetic background, mimp1 flanking sequences were recovered using a modified thermal asymetric interlaced (TAIL)–PCR approach.(Liu and Whittier 1995). Two rounds of PCR were performed on 50 ng of genomic DNA of the revertants. In the primary PCR, the arbitrary degenerate (AD) primer AD2, 5′-AG(A/T)GNAG(A/T)ANCA(A/T)AGA-3′, and the specific primer m1Div53F, 5′-GCCTCTTTGAGCCACGCTAC-3′, were used with reduced-stringency and high-stringency annealing temperatures of 44° and 66°, respectively. The secondary PCR was set up on 2 μl of a 1/100 dilution of the primary PCR, using the same AD primer and Div149 (see above) as the second specific primer. The annealing temperatures were the same as for the primary PCR. PCR programs were as described in Liu and Whittier (1995).
Cloning of PCR products and DNA sequencing:
PCR products were directly cloned into the pGEM-T Easy vector (Promega, Madison, WI) using 3 μl of either rough or purified PCR products, following the manufacturer's instructions. Sequencing of PCR products cloned into the pGEM-T Easy vector was performed by Genome Express (Meylan, France) using an ABI Big Dye terminator kit (Perkin-Elmer, Norwalk, CT) and the universal M13/pUC sequencing primer (−20) (forward) and pR primer (5′-GGAAACAGCTATGACCATG-3′; reverse).
Sequence analysis:
Multiple alignments were performed using Clustal W (Thompson et al. 1994) and slightly modified at both 5′- and 3′-ends for improvement. The parsimony analysis was realized on 179nt using PAUP4.0b10 (Swofford 2002) with default parameters. The bootstrap analysis comprised 100 replicates. The potential of the sequences to form stable secondary structures was analyzed with the RNAFOLD program (http:www.infobiogen.fr) using standard parameters. Searches for matches of nucleotide sequences in the current database (nonredundant GenBank) or in the F. graminearum Genome Database available at the Munich Information Center for Protein Sequences (MIPS) (http://mips.gsf.de/genre/proj/fusarium/) used the BlastN or BlastX algorithms (Altschul et al. 1997).
RESULTS
The mimp1 MITE family:
We previously described five copies of a putative MITE family, called mimp, discovered in the genome of F. oxysporum (strain FOM24) by sequencing genomic regions nested with TEs (Hua-Van et al. 2000). They are characterized by a small size (∼220 bp), the lack of coding capacity, and the presence of short terminal inverted repeats of 27 bp (Figure 1A). They have been grouped into two subfamilies (mimp1 and mimp2) on the basis of an absence of internal sequence homology. The mimp TIRs present a strong similarity with the first 27 nt of impala ends (Figure 1B), a F. oxysporum Tc1-like family organized in different subfamilies (Hua-Van et al. 1998, 2001b). Furthermore, mimp are flanked by TA dinucleotides, as is impala, which presumably correspond to the target duplication generated upon integration. Apart from the TIRs, no obvious sequence similarities were found between mimp and impala, or between mimp and any other known repetitive element.
To gain insight into the structure and the evolution of the mimp1 family, we used a PCR approach to investigate sequence and size variability of mimp1 elements in different genetic contexts belonging to the Fusarium genus. Primers located in the TIRs were first used and allowed amplification of fragments of different sizes in several strains. It was then determined that the larger ones (>220 bp) corresponded to full-length (1300 bp) or internally deleted (400–600 bp) impala (Figure 1C, top, lanes 4 and 5) while the 220-bp product was mimp1 amplicon. In contrast, the use of mimp1-specific primers (see materials and methods) under the same conditions each time revealed only the expected 197-bp PCR product (Figure 1C, bottom).
In addition to the uniformity in size of the different amplified full-length copies, the mimp1 family also contains some truncated elements at either the 5′- or the 3′-end. Indeed, three truncated copies were previously identified (Hua-Van et al. 2000). In this analysis, we obtained one other truncated copy among 18 independent inverse-PCR products recovered from different species (data not shown).
A total of 37 new mimp1 sequences were obtained either by sequencing of FOM24 genomic clones (6) or by PCR (31) for other strains, including 12 from various strains of F. oxysporum and 19 from other closely related species. The latter ones were obtained using a combination of specific internal and TIR primers. These sequences corresponded to 23 different nucleotide sequences that were aligned with the four known mimp1 sequences and a sequence retrieved from the GenBank database (AJ608703). The nucleotide differences calculated on 179 internal nucleotides (excluding the primer sequences) ranged from 0 to 20% within species as well as between species. This relatively high sequence identity has allowed us to perform a conventional phylogenetic analysis. A tree deduced from this alignment permits us to define clearly only one well-supported clade, containing sequences closely related mainly from FOM24. Others were more scattered and did not show clustering according to the species (data not shown).
Finally, the mimp1 nucleotide sequence showed a strong potential to form a stable secondary structure (see Figure 1D). Although some exceptions exist (Bureau et al. 1996), a number of MITE elements display this property. The mimp1 predicted a ΔG0 value of −87.7 kcal/mol that corresponds to the lowest values reported for other families (Bureau and Wessler 1994a; Casacuberta et al. 1998).
In conclusion, the mimp1 elements showed an inverted terminal repeated structure that was able to fold into a stable secondary structure. The family appears homogeneous in size, in contrast with most nonautonomous elements such as internally deleted impala. The sequence of the TIRs (similar to those of impala) and of the TSD indicated a relationship with the Tc1-mariner superfamily of class II transposons, a group known to be associated with numerous MITE families (Feschotte et al. 2002).
Copy number and genomic representation in the Fusarium genus:
We first examined mimp1 distribution in a wide collection of strains of the Fusarium oxysporum complex (FOC), previously studied for the presence of different TEs (Hua-Van et al. 2001a; Daboussi et al. 2002; Chalvet et al. 2003). This analysis was extended to different Fusarium species either phylogenetically closely related to FOC (Baayen et al. 2001; Skovgaard et al. 2003) or more distantly related.
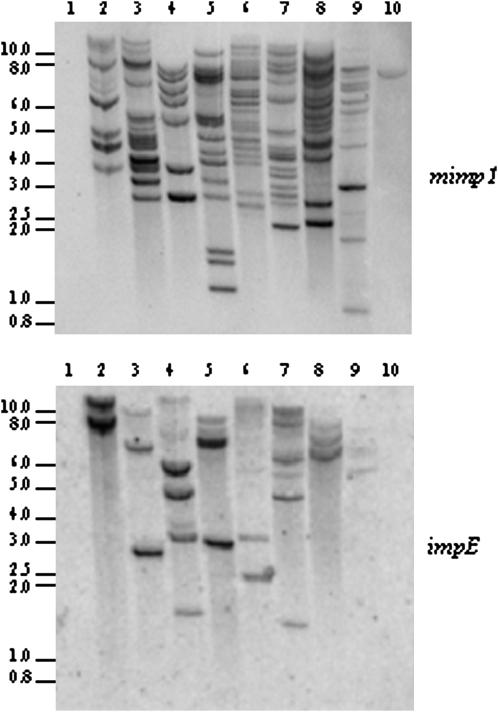
Southern blot experiments performed on 16 FOC strains, representative of seven formae speciales and one nonpathogenic strain, revealed that all contained mimp1 elements (Figure 2; data not shown). mimp1 was also detected in the three strains from FOC-related species (Figure 2, lanes 2–4), but not in more distant species (Figure 2, lane 1; data not shown).
Figure 2.—
Distribution of mimp1 and impala elements in the genome of Fusarium strains. Genomic DNAs were digested by EcoRI and probed with the mimp1 element or the impala E amplification product under high-stringency conditions. 1, F. culmorum strain 1602; 2, F. foetens strain 31852; 3, F. redolens strain 29642; 4, F. hostae strain 28381; 5, FOr4; 6, FOa1; 7, FOLn3; 8, FOL15; 9, FOM K419; 10, FO5.
When detected, mimp1 appears to be repetitive. The copy number varied between 7 and 20, according to the number of hybridizing bands observed in the Southern blot (Figure 2). It is noteworthy that some hybridizing bands present a signal of stronger intensity, which could result either from the presence of several mimp copies on the same fragment or from a difference in the size of the mimp1 copy—full-length vs. truncated. Alternatively, weaker signals could result from nucleotide divergence preventing strong hybridization, as previously observed for the different impala subfamilies (Hua-Van et al. 1998, 2001b).
The TIR similarity of mimp1 and impala elements suggests that mimp1 could use the impala transposase to move and multiply. To clarify the functional relationship between mimp1 and impala, the presence of impala, the putative autonomous partner of mimp1, was also investigated in these genomes. The membranes used to detect mimp1 were thus hybridized with an impala probe allowing the detection of the impala E subfamily (see materials and methods). As observed for mimp1, the distant species appeared to be devoid of impala. In all the strains containing mimp1 elements, impala signals were observed except in FO5 (Figure 2, lane 10). Beyond the absence of impala signal, this context was already shown to lack any impala activity (Hua-Van et al. 2001b). We attempted to explain the unique faint mimp1 hybridization signal by conducting PCR experiments. No amplification was obtained using specific TIR primers, even with extended elongation time. Using a combination of specific internal and TIR primers, only the 3′ part could be amplified (data not shown), indicating that the mimp1-hybridizing band in FO5 likely corresponds to a truncated copy. Moreover, the corresponding sequence showed only one nucleotide difference with the mimp1 consensus sequence (>59 positions). The weakness of the hybridization signal in FO5 could thus reflect the truncation of this copy rather than a high sequence divergence.
Hence, mimp1 and impala seem to exhibit a similar distribution within the Fusarium genus except in the FO5 strain.
Mobilization of mimp1 by an autonomous impala element:
Given TIR and target-site similarities between mimp1 and impala, as well as the presence of mimp1 in strains containing impala elements, it was tempting to speculate that the nonautonomous mimp1 elements could be mobilized by the transposase encoded by an autonomous impala element (Migheli et al. 1999; Hua-Van et al. 2001b; Chalvet et al. 2003). To test this hypothesis, we performed a phenotypic assay for excision, previously used to demonstrate transposition of different TEs (Migheli et al. 1999; Hua-Van et al. 2001b; Chalvet et al. 2003). To this end, we constructed a plasmid, pNm1H18, in which a mimp1 element was inserted within the first intron of the niaD gene, leading to a nonfunctional gene (see materials and methods and Figure 3). This construct was introduced in three genetic backgrounds, which contained or lacked an impala source of transposase. The strain FOM150 nia9 contains different autonomous impala copies, and this context has been already used to verify the mobility of defective impala copies (Hua-Van et al. 2001b). Three [nia−] transformants carrying pNm1H18 yielded Nia+ revertant colonies (1–5/plate). Southern blot and PCR analyses performed on a sample of Nia+ colonies revealed that mimp1 is mobile and has been excised from niaD (data not shown). The excision events were confirmed by the presence of excision footprints at the empty sites (Table 1). The FO5 nia13 context is devoid of all types of TEs previously identified within the FO complex (Migheli et al. 1999; Hua-Van et al. 2001b; Chalvet et al. 2003). From seven transformants in which pNm1H18 was introduced alone, very few Nia+ colonies were observed (<1 colony/plate). Of the three recovered, only one showed an empty site, but sequencing revealed a 5-nt deletion instead of a typical footprint of two to five additional nucleotides (Table 1); the two others still showed the mimp1 copy into the niaD intron. When plasmid pNm1H18 was introduced into the same genetic background, together with the plasmid carrying the impalaE transposase under the control of a strong promoter, we obtained 6 cotransformants, 5 of them giving numerous Nia+ colonies (5–50/plate, see Figure 4). The remaining cotransformant gave very few Nia+ colonies, none of them resulting from mimp1 excision but more likely from reversion of the endogenous nia gene, as previously observed (Migheli et al. 1999; Hua-Van et al. 2001b; Chalvet et al. 2003). In the F. graminearum genetic background, similar results were observed. A total of 11 transformants carrying the pNm1H18 alone were obtained. None of them gave rise to Nia+ colonies even after 6 weeks of growth on nitrate minimal agar medium. On the contrary, among 41 cotransformants carrying the pNm1H18 plasmid, together with the pHEO62 plasmid carrying the exogenous source of impala transposase, 33 gave rise to Nia+ colonies, the number varying from 4 to 20/petri dish (data not shown).
TABLE 1.
Results of molecular analyses performed on a sample of NiaD+ colonies
| Genetic context | Source of transposase | No. of NiaD+ colories analyzeda | Excision event | Footprint |
|---|---|---|---|---|
| FOM150 nia9 | Endogenous | 5 (3) | Yes | cagTA (4) |
| ctgTA (1) | ||||
| FO5 nia13 | None | 3 (2) | Nob | — |
| Exogenous | 3 (1) | Noc | — | |
| 22 (5) | Yes | cagTA (11) | ||
| ctgTA (9) | ||||
| caTA (1) | ||||
| TA (1) | ||||
| Fg820nia | None | 0 | — | — |
| Exogenous | 12 (12) | Yes | cagTA (7) | |
| ctgTA (3) | ||||
| caTA (2) |
mimp1 excision events were detected by PCR analysis using primers (niaD144 and niaD754r), allowing amplification of a 485-bp fragment corresponding to an empty site (excision event, yes) in the niaD+ revertants instead of a 717-bp fragment in the transformant corresponding to the insertion site occupied by the mimp1 element. Footprints are composed of nucleotides from the ends of mimp1 (lowercase) and the TA duplicated target site.
The numbers of transformants from which the NiaD+ colonies were obtained are indicated in parentheses.
One revertant carries an empty site; however, sequencing revealed a 5-nt deletion instead of a typical footprint.
The NiaD+ colonies recovered from this transformant, which do not show an empty site, likely result from a reversion of the mutation in the recipient strain since we demonstrated that the exogenous source of transposase had been disrupted.
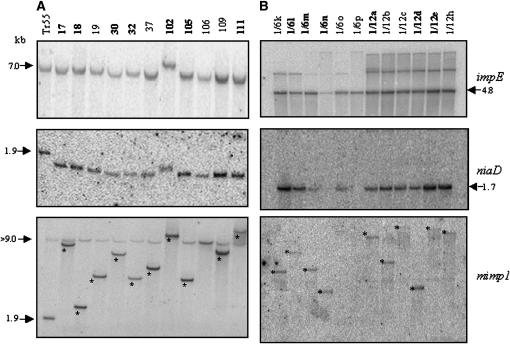
A molecular analysis of one FO5 nia13 transformant (TR55) and a sample of its revertants indicated that this transformant contained a single intact copy of the niaD∷mimp1 construct on the basis of the shift of the niaD-hybridizing band from 1.9 kb (occupied site) in TR55 to 1.7 kb (empty site) in all the revertants (Figure 5A, middle). We also observed at least one copy of the plasmid carrying the transposase gene (Figure 5A, top). The excision of mimp1 was confirmed using a rapid PCR screen, which revealed, in each case, the presence of an excision footprint (Table 1). These footprints were very similar to those obtained in the impala active context (FOM150 nia9) and were also similar to those left by impala excision. Generally, five additional nucleotides were left, two corresponding to the duplicated TA target site and three from one of the ends of the element (Hua-Van et al. 2001b).
Figure 5.—
Southern blot analysis of transformant TR55 and a sample of corresponding revertants in the FO5 genetic background (A) and of a sample of revertants obtained from two independent transformants in the Fg820 background (B). Genomic DNA was digested with XbaI and membranes were successively hybridized with an impalaE probe (top), a niaD probe (middle), and a mimp1 probe (bottom). The stars indicate the reinsertion of excised copies. Numbers in boldface type correspond to revertants submitted to IPCR (FO5) or TAIL–PCR (Fg820).
The reinsertion of mimp1 was assessed using a probe corresponding to the entire element (Figure 5A, bottom). A new hybridizing band, corresponding to a reinserted element, was observed in ∼91% (10/11) of the revertants analyzed.
Similarly, the molecular analysis of 12 revertants derived from two independent cotransformants from the Fg820nia background was performed. Hybridization with the impE probe showed that each strain contains at least one intact copy of the construct carrying the source of transposase as shown by the presence of a 4.8-kb hybridizing XbaI fragment (Figure 5B, top). Additional bands were observed, corresponding most likely to a disrupted copy of the same plasmid. The presence of a single 1.7-kb hybridization band using the niaD probe demonstrated that all correspond to an excision event (Figure 5B, middle). Another set of 12 revertants, each derived from a different transformant, was used to examine the presence of excision footprints at the empty site. Typical excision footprints were found in all cases (Table 1). Using the mimp1 element as a probe, reinsertion was observed in ∼83% (10/12) of the strains analyzed. These results tend to confirm that the reinsertion frequency appears higher than that observed for impala, which is in the range of 50–75% (Migheli et al. 2000; Hua-Van et al. 2001b).
Molecular analysis of mimp1 insertion sites:
Previous studies indicate that some MITE families insert preferentially into genic regions (Mao et al. 2000; Zhang et al. 2000) and/or have a propensity to insert into each other, giving nested structures (Jiang and Wessler 2001). In addition, for a given family, the distribution of ancient vs. recently inserted elements with respect to predicted genes can vary (Santiago et al. 2002), suggesting that selection can also shape the genomic distribution of MITEs.
To obtain data on the target-site preference of mimp1 elements, the regions flanking different mimp1 elements were cloned by inverse polymerase chain reaction (IPCR) or TAIL–PCR, sequenced, and used to seek homologies in databases. For those giving a match, we calculated the distance to putative initiator ATGs or stop codons of the closest predicted genes. Two types of mimp1 insertions were considered: (i) elements originally present in the genome of formae speciales of F. oxysporum or different Fusarium species closely related to the FOC and (ii) newly inserted elements either in the FO5 or in the Fg820 backgrounds, resulting from the mobilization of the mimp1 copy by the impala E transposase, totaling 43 mimp1-insertion sites.
The four copies of mimp1 previously identified in FOM24 (mimp1.1-mimp1-4) had been found in transposon nests (Hua-Van et al. 2000). From 18 other ancient insertions analyzed, 10 clones yielded no significant hits in the databases, 7 showed high homology with known transposable elements (Table 2), and 1 corresponded to the insertion of mimp1 in the 5′-untranslated region of a fungal gene, very close to the putative start codon (Table 2). It is interesting to note that in three cases mimp1 was found close to F. oxysporum solo-LTR Han or Han-like elements, which have a strong potential to form stable secondary structures (ΔG0 values of −70 and −55 kcal/mol, respectively).
TABLE 2.
Insertion sites of resident or newly reinserted mimp1 elements
| Clone | IPCR product size (sequenced) | Major BLAST hits | e-value | Distance to mimp1 (bp) |
|---|---|---|---|---|
| 21(M24)a | 695 | Fo solo LTR Han (AF076629) | 3.10−94 | 8 |
| 144(M24)a | 331 | Fo solo LTR Han (AF076629) | 2.10−75 | 3 |
| 104(MK)a | 1403 | Fo solo LTR Han (AF076629) | 3.10−81 | 578 |
| 109(Fh)a | 762 | Nh PiggyBac-like partial transposase [initially referred to as pathogenicity protein PEP1 (AAK18805)] | 6.10−27 | 277 (5′) |
| Fg hypothetical protein FG05946 | 7.10−20 | |||
| 119(Fh)a | 1023 | Fo mimp1 (AF76624) | 8.10−13 | 38 |
| 168(Fh)a | 480 | Fo mimp1 (AF76624) | 2.10−80 | 2 |
| Fo impalaD (AF363425) | 3.10−95 | 79 | ||
| Mimp1-9(L15)a | 682 | Fo mimp1 (AF76624) | 0.004 | 340 |
| Fo clone ACQ2_T7 transposon impala (AF285757) | 0.0015 | 244 | ||
| 26(R4)a | 1131 | Fg hypothetical protein FG03406 (EAA71224) | 6.10−26 | 135 (5′) |
| An pectin methylesterase (BAA75474) | 3.10−11 | 135 (5′) | ||
| 18(Fo5)b | 1800 (1185) | Fg hypothetical protein FG05987 (EAA75632) | 2.10−31 | 569 (5′) |
| At phosphoglycerate/biphosphoglycerate mutase protein (NP_187168) | 2.10−8 | 569 (5′) | ||
| Pc WRKY transcription factor (AAD55974) | 0.001 | ORF | ||
| 25(Fo5)b | 1502 | Fg hypothetical protein FG00406 (EAA67497) | 2.10−45 | 294 (5′) |
| 100(Fo5)b | 3500 (1486 + 127) | Fg hypothetical protein FG01165 (EAA68631); St DNA mismatch repair protein (AAK58517) | 7.10−42 0.023 | 277 (3′) >2000 (3′) |
| 111H(Fo5)b | 2800 (1080) | Fg hypothetical protein FG06828 (EAA76760) | 7.10−20 | >1000 (5′) |
| Sp MutS protein homolog 1 (O13921) | 4.10−4 | >1000 (5′) | ||
| 112(Fo5)b | 1861 | Fg hypothetical protein FG06184 (EAA74540) | 3.10−60 | >1000 (5′) |
| Ac Δ9-fatty acid desaturase (CAA59939) | 7.10−29 | >1000 (5′) | ||
| 120(Fo5)b | 1500 (890) | Fg hypothetical protein FG10674 (EAA70296) | 2.10−31 | 125 (5′) |
| Nc transcriptional factor PRO1 (XP_327678) | 1.10−3 | 125 (5′) |
If sequence information was not obtained for the whole IPCR product, the size of the sequenced region is indicated in parentheses. BlastN or BlastX algorithms (Altschul et al. 1997) were used to search for significant matches in the databases. When a putative ORF was detected, the relative mimp1 location was indicated as follows: 5′, 3′, and ORF indicate positions (nt) within the 5′ region, 3′ region, or the coding sequence of the putative gene, respectively. In such a case, two matches are listed for each alignment: the one giving the highest e-value and, when available, the first one giving information about gene function. Fo, F. oxysporum; Fg, F. graminearum; Fh, F. hostae; An, Aspergillus niger; Nh, Nectria haematococca; At, Arabidopsis thaliana; Pc, Petroselinum crispum; St, Streptococcus thermophilus; Sp, Schizosaccharomyces pombe; Ac, Ajellomyces capsulatus; Nc, N. crassa.
Resident mimp1 copies.
Newly reinserted mimp1 copies.
Sequence analysis of genomic regions flanking newly inserted mimp1 in the FO5 background revealed a rather different situation. Of 13 reinsertion events, 7 did not show any significant matches, even when BLASTed against fungal-specific databases. The remaining 6 flanking regions all showed significant matches to known coding sequences (Table 2). In four revertants (18, 25, 100, and 120), insertion events occurred in close vicinity of open reading frames (ORFs) (Table 2). In revertant 120, for example, mimp1 reinserted 125 bp upstream from an ORF encoding a putative Zn(2)Cys6 fungal transcription factor. Analysis of mimp1 reinsertion sites in the F. graminearum genomic background was greatly facilitated by the availability of the genome sequence. First, flanking sequences were recovered by a modified TAIL–PCR procedure, easier to set up than IPCR, and second, the reinsertion sites could all be located at the nucleotide level by alignment with the genome sequence at MIPS (http://mips.gsf.de/genre/proj/fusarium/). The data obtained from the analysis of 12 reinsertion sites are of particular interest. Indeed, as shown in Table 3, in seven strains (58%), mimp1 has reinserted <500 bp from a gene, a very promising result for use of mimp1 as a tool for insertional mutagenesis.
TABLE 3.
Insertion sites of newly reinserted mimp1 elements in the F. graminearum genetic context
| Clone | F. graminearum contig DNA giving BLASTN hita | Closest geneb | Distance to mimp1 (bp) |
|---|---|---|---|
| 1/6-1 | 1.450 (82,393) | FG10800 | 1025 (3′) |
| 1/6l | 1.415 (76,938) | FG09992 | 19 (5′) |
| 1/6m | 1.107 (33,781) | FG01974 | 115 (3′) |
| 1/6n | 1.193 (156,744) | FG04610 | 341 (5′) |
| 1/12-1 | 1.298 (37,311) | FG07090 | 91 (5′) |
| 1/12a | 1.367 (81,420) | fgd367-380 | 675 (5′) |
| 1/12d | 1.40 (5,392) | FG00905 | ORF |
| 1/12e | 1.74 (44,890) | FG01453 | 606 (3′) |
| 1/24-1 | 1.293 (61,535) | fgd293-250 | 56 (5′) |
| 1/24j | 1.207 (94,644) | fgd207-380 | 214 (5′) |
| 1/24k | 1.294 (33,844) | FG07062 | 865 (5′) |
| 5/19-1 | 1.254 (76,853) | FG06328 | 592 (5′) |
The BlastN algorithm (Altschul et al. 1997) was used to search for significant matches in the F. graminearum genome database at MIPS (http://mips.gsf.de/genre/proj/fusarium/).
Contig DNA from the F. graminearum genome sequence giving a hit with mimp1 flanking sequences. The location of mimp1 insertion in each contig DNA is indicated in parentheses. To determine the distance of mimp1 reinsertion sites to genes, the annotated version of the F. graminearum genome sequence available at MIPS (http://mips.gsf.de/genre/proj/fusarium/) was used.
The gene closest to the mimp1 insertion site. The relative mimp1 location was indicated as follows: 5′, 3′, and ORF indicate positions (nt) within the 5′ region, 3′ region, or the coding sequence of the gene, respectively.
DISCUSSION
Similarities between impala and mimp1 TIRs and TSD, which are key components for the transposition reactions of many class II elements, led us to hypothesize a functional relationship between these two elements.
By analyzing the distribution of impala and mimp1 elements across the Fusarium genus, we observed that both are widely distributed within the FOC complex, being present in closely related species and absent in more distantly related ones. The structural analysis of the mimp1 family revealed a size homogeneity but a nucleotide divergence ranging from 0 to 20%, suggesting an old, but nevertheless recently active, element.
Overall, mimp1 presented several features of MITEs except that its copy number appears unusually low. Indeed, most MITEs families in plant and animal species are associated with a high copy number (Tu 1997; Santiago et al. 2002), although rare examples of MITE families with moderate copy number have also been described (Holyoake and Kidwell 2003; Saito et al. 2005). The low copy number of mimp1 could simply reflect the low activity of impala. This element itself is usually present in low copy number. Moreover, it is the representative of a basal branch of Tc1-like elements containing few members (Plasterk et al. 1999). On the other hand, as for all fungal genomes, the genome of F. oxysporum, estimated to be in the range of 40–50 Mb (Davière et al. 2001), is very small compared to genomes in which MITEs were found in large copy numbers. Considering that the larger the genome, the higher the number of transposable elements (Kidwell and Lisch 2002), huge amounts of TEs are not expected. This is supported by data from recently sequenced fungal genomes and other studies that reported a moderate TE content ∼5–10% (Dean et al. 2005; Loftus et al. 2005) with a maximum of 20% for Neurospora crassa (Galagan et al. 2003). Finally, although a great diversity of TEs can be found (for review see Daboussi and Capy 2003), few MITE families have been identified in fungi up to now, suggesting that this kind of element is not very successful in such organisms.
Although a clear relationship between mimp1 and impala was found, the origin of mimp1 remains enigmatic. The simplest hypothesis is that mimp1 was derived from impala by internal deletion. This is the case for guest, identified as a severely deleted relic of a full-length element in N. crassa (Ramussen et al. 2004). Several deletion derivatives of impala were identified (Hua-Van et al. 2001a; this work) and could always be unambiguously related to a particular impala subfamily due to the few nucleotide differences observed. The situation for mimp1 is clearly different since no homology is detectable between mimp1 and any of the impala subfamilies, except for the TIRs, implying a high selective pressure on these sequences. A de novo formation is also possible. This model supposes that new transposons can be created following the fortuitous association of TIRs bordering a segment of genomic DNA. This scenario was proposed for the origin of Ds1 (MacRae and Clegg 1992) and the creation a new P element in Drosophila (Tsubota and Huong 1991).
Other mimp families with elements of the same size, exhibiting impala-like TIRs but with unrelated central regions, have been identified. Their analysis may help to clarify the origin of these elements.
In this study, we provide direct evidence that impala is the autonomous partner of mimp1 since transposition of mimp1 occurs only when a source of impala transposase is provided. The use of another genetic background free of active impala elements (H. C. Kistler, personal communication), F. graminearum, confirmed the functional relationship between mimp1 and impala. Although several MITE families could be linked to elements encoding a transposase on the basis of TIRs similarities (Jiang et al. 2003; Saito et al. 2005) and comobilization or in vitro physical interactions between the two suspected partners (Feschotte et al. 2005; Shan et al. 2005), there are very few cases of direct evidence of the mobilization of a MITE by an autonomous candidate (Rezsohazy et al. 1997; Jiang et al. 2003; Saito et al. 2005). The results presented here clearly connect a fungal MITE and the transposase of a Tc1-like element. This is the first functional system reported in fungi and a new example of a Tc1/mariner-related MITE.
We also show that mimp1 transposes by a cut-and-paste mechanism. The transposition mechanism of MITEs has long remained mysterious because excision was rarely observed (Wessler et al. 1995; Wessler 1998). Direct evidence of excision was obtained recently with the rice element mPing (Kikuchi et al. 2003), allowing the classification of MITEs as class II elements. Our results reinforce the fact that MITEs are capable of both excision and reinsertion.
The characteristics of impala transposition, excision/reinsertion, excision footprints, and target-site duplication are retrieved for mimp1. It is noteworthy that interesting transposition specificities appeared associated to mimp1. First, the frequency of mimp1 reinsertion, >80%, appears higher than that observed for impala. Second, one-third of the newly reinserted mimp1 elements in F. oxysporum lie <500 nt from an ORF. Even more promising results were obtained in the F. graminearum strain as more than half of the reinsertion sites were shown to be located <500 nt from an ORF. The discrepancy between the two results might be due to differences between the two genomes but may also be attributed to the poor sequence availability in F. oxysporum.
Previous studies have demonstrated that MITEs are frequently found flanking many plant genes. Tourist and Stowaway have been identified in association with >100 plant genes (Bureau and Wessler 1992, 1994a,b). A computer-based systematic survey reveals a predominance of MITEs in wild-type rice genes (Bureau et al. 1996). However, the association of MITEs with genes is not uniform among the different MITE families, i.e., only 10.5% for explorer, but nearly 98% for Stowaway (Mao et al. 2000). Some MITEs were found to insert into other MITEs or adjacent to other TEs (Jiang and Wessler 2001). This is what we observed in backgrounds containing TEs where mimp1 insertions were frequently found within or adjacent to solo-LTRs, mimp, or impala elements but not to DNA transposons such as Fot1 and Hop (Daboussi et al. 1992; Chalvet et al. 2003), although these elements constitute a much larger fraction of the genome. One explanation could be that mimp1 amplification preceded the amplification of the TEs now found in a high copy number. Other hypotheses include preference for self-insertions or targeting through secondary structure, as proposed by Jiang and Wessler (2001). The fact that Han, Han-like, and mimp elements all can be folded into a stable secondary structure favors this hypothesis. The difference observed between resident and newly inserted copies may result from the differential richness of the TEs or may reflect the effect of selection on ancient insertions.
In conclusion, the identification of this mobilizing MITE system offers great prospects to tag genes in filamentous fungi. First, as for the impala system, the double component mimp1/impala is expected to function in a large range of fungal species. Second, if the higher ability to reinsert in the vicinity of genes is confirmed on a larger set of revertants, this system should represent a powerful molecular tool for the functional analysis of fungal genomes.
Acknowledgments
We thank Thomas Ruby, Gilles Pinto, and Corentin Laulier for their contributions to some experiments. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS) and Groupement de Recherche CNRS no. 2157 “Evolution des Eléments Transposables: du Gène aux Populations.”
References
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baayen, R. P., K. O'Donnell, S. Breeuwsma, D. M. Geise and C. Waalwijk, 2001. Molecular relationships of fungi within the Fusarium redolens-F. hostae clade. Phytopathology 91: 1037–1044. [DOI] [PubMed] [Google Scholar]
- Bureau, T. E., and S. R. Wessler, 1992. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T. E., and S. R. Wessler, 1994. a Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91: 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T. E., and S. R. Wessler, 1994. b Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6: 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T. E., P. C. Ronald and S. R. Wessler, 1996. A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc. Natl. Acad. Sci. USA 93: 8524–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, A. M. C., J. A. Sweigard and B. Valent, 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41: 22. [Google Scholar]
- Casacuberta, E., J. M. Casacuberta, P. Puigdomenech and A. Monfort, 1998. Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterisation of the Emigrant family of elements. Plant J. 16: 79–85. [DOI] [PubMed] [Google Scholar]
- Chalvet, F., C. Grimaldi, F. Kaper, T. Langin and M. J. Daboussi, 2003. Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20: 1362–1375. [DOI] [PubMed] [Google Scholar]
- Daboussi, M., and P. Capy, 2003. Transposable elements in filamentous fungi. Annu. Rev. Microbiol. 57: 275–299. [DOI] [PubMed] [Google Scholar]
- Daboussi, M. J., and T. Langin, 1994. Transposable elements in the fungal plant pathogen Fusarium oxysporum. Genetica 93: 49–59. [DOI] [PubMed] [Google Scholar]
- Daboussi, M. J., T. Langin and Y. Brygoo, 1992. Fot1, a new family of fungal transposable elements. Mol. Gen. Genet. 232: 12–16. [DOI] [PubMed] [Google Scholar]
- Daboussi, M. J., J. M. Davière, S. Graziani and T. Langin, 2002. Evolution of the Fot1 transposons in the genus Fusarium: discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol. 19: 510–520. [DOI] [PubMed] [Google Scholar]
- Davière, J. M., T. Langin and M. J. Daboussi, 2001. Potential role of transposable elements in the rapid reorganization of the Fusarium oxysporum genome. Fungal Genet. Biol. 34: 177–192. [DOI] [PubMed] [Google Scholar]
- Dean, R. A., N. J. Talbot, D. J. Ebbole, M. L. Farman, T. K. Mitchell et al., 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., and C. Mouches, 2000. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol. Biol. Evol. 17: 730–737. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., X. Zhang and S. R. Wessler, 2002. Miniature inverted-repeat transposable elements and their relationship to established DNA transposons, pp. 1147–1158 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society of Microbiology, Washington, DC.
- Feschotte, C., M. T. Osterlund, R. Peeler and S. R. Wessler, 2005. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 33: 2153–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Holyoake, A. J., and M. G. Kidwell, 2003. Vege and Mar: two novel hAT MITE families from Drosophila willistoni. Mol. Biol. Evol. 20: 163–167. [DOI] [PubMed] [Google Scholar]
- Hua-Van, A., F. Héricourt, P. Capy, M. J. Daboussi and T. Langin, 1998. Three highly divergent subfamilies of the impala transposable element coexist in the genome of the fungus Fusarium oxysporum. Mol. Gen. Genet. 259: 354–362. [DOI] [PubMed] [Google Scholar]
- Hua-Van, A., J. M. Davière, T. Langin and M. J. Daboussi, 2000. Genome organization in Fusarium oxysporum: clusters of class II transposons. Curr. Genet. 37: 339–347. [DOI] [PubMed] [Google Scholar]
- Hua-Van, A., T. Langin and M. J. Daboussi, 2001. a Evolutionary history of the impala transposon in Fusarium oxysporum. Mol. Biol. Evol. 18: 1959–1969. [DOI] [PubMed] [Google Scholar]
- Hua-Van, A., J. A. Pamphile, T. Langin and M. J. Daboussi, 2001. b Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. 264: 724–731. [DOI] [PubMed] [Google Scholar]
- Jiang, N., and S. R. Wessler, 2001. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13: 2553–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N., Z. Bao, X. Zhang, H. Hirochika, S. R. Eddy et al., 2003. An active DNA transposon family in rice. Nature 421: 163–167. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., and D. R. Lisch, 2002. Transposable elements as sources of genomic variation, pp. 59–90 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society of Microbiology, Washington, DC.
- Kikuchi, K., K. Terauchi, M. Wada and H. Y. Hirano, 2003. The plant MITE mPing is mobilized in anther culture. Nature 421: 167–170. [DOI] [PubMed] [Google Scholar]
- Kunze, R., and C. F. Weil, 2002. The hAT and CACTA superfamilies of plant transposons, pp. 565–610 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society of Microbiology, Washington, DC.
- Langin, T., M. J. Daboussi, C. Gerlinger and Y. Brygoo, 1990. Influence of biological parameters and gene transfer technique on transformation of Fusarium oxysporum. Curr. Genet. 17: 313–319. [Google Scholar]
- Li Destri Nicosia, M. G., C. Brocard-Masson, S. Demais, A. Hua Van, M. J. Daboussi et al., 2001. Heterologous transposition in Aspergillus nidulans. Mol. Microbiol. 39: 1330–1344. [PubMed] [Google Scholar]
- Liu, Y. G., and R. F. Whittier, 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681. [DOI] [PubMed] [Google Scholar]
- Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo et al., 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae, A. F., and M. T. Clegg, 1992. Evolution of Ac and Dsl elements in select grasses (Poaceae). Genetica 86: 55–66. [DOI] [PubMed] [Google Scholar]
- Malardier, L., M. J. Daboussi, J. Julien, F. Roussel, C. Scazzocchio et al., 1989. Cloning of the nitrate reductase gene (niaD) of Aspergillus nidulans and its use for transformation of Fusarium oxysporum. Gene 78: 147–156. [DOI] [PubMed] [Google Scholar]
- Mao, L., T. C. Wood, Y. Yu, M. A. Budiman, J. Tomkins et al., 2000. Rice transposable elements: a survey of 73,000 sequence-tagged-connectors. Genome Res. 10: 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migheli, Q., R. Lauge, J. M. Daviere, C. Gerlinger, F. Kaper et al., 1999. Transposition of the autonomous Fot1 element in the filamentous fungus Fusarium oxysporum. Genetics 151: 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migheli, Q., C. Steinberg, J. M. Davière, C. Olivain, C. Gerlinger et al., 2000. Recovery of mutants impaired in pathogenicity after transposition of Impala in Fusarium oxysporum f. sp. melonis. Phytopathology 90: 1279–1284. [DOI] [PubMed] [Google Scholar]
- Oosumi, T., B. Garlick and W. R. Belknap, 1996. Identification of putative nonautonomous transposable elements associated with several transposon families in Caenorhabditis elegans. J. Mol. Evol. 43: 11–18. [DOI] [PubMed] [Google Scholar]
- Plasterk, R. H., Z. Izsvak and Z. Ivics, 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15: 326–332. [DOI] [PubMed] [Google Scholar]
- Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels and C. A. van den Hondel, 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56: 117–124. [DOI] [PubMed] [Google Scholar]
- Ramussen, J. P., A. H. Taylor, L. J. Ma, S. Purcell, F. Kempken et al., 2004. Guest, a transposable element belonging to the Tc1/mariner superfamily is an ancient invader of Neurospora genomes. Fungal Genet. Biol. 41: 52–61. [DOI] [PubMed] [Google Scholar]
- Rezsohazy, R., H. G. van Luenen, R. M. Durbin and R. H. Plasterk, 1997. Tc7, a Tc1-hitch hiking transposon in Caenorhabditis elegans. Nucleic Acids Res. 25: 4048–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., 2002. Evolution of DNA transposons in eukaryotes, pp. 1093–1110 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society of Microbiology Press, Washington, DC.
- Saito, M., J. Yonemaru, G. Ishikawa and T. Nakamura, 2005. A candidate autonomous version of the wheat MITE Hikkoshi is present in the rice genome. Mol. Genet. Genomics 273: 404–414. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Santiago, N., C. Herraiz, J. R. Goni, X. Messeguer and J. M. Casacuberta, 2002. Genome-wide analysis of the Emigrant family of MITEs of Arabidopsis thaliana. Mol. Biol. Evol. 19: 2285–2293. [DOI] [PubMed] [Google Scholar]
- Shan, X., Z. Liu, Z. Dong, Y. Wang, Y. Chen et al., 2005. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol. Biol. Evol. 22: 976–990. [DOI] [PubMed] [Google Scholar]
- Skovgaard, K., S. Rosendahl, K. O'Donnell and H. I. Nirenberg, 2003. Fusarium commune is a new species identified by morphological and molecular phylogenetic data. Mycologia 95: 630–636. [PubMed] [Google Scholar]
- Smit, A. F., and A. D. Riggs, 1996. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA 93: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota, S. I., and D. V. Huong, 1991. Capture of flanking DNA by a P element in Drosophila melanogaster: creation of a transposable element. Proc. Natl. Acad. Sci. USA 88: 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z., 1997. Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 94: 7475–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z., 2001. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. USA 98: 1699–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S., 1998. Transposable elements associated with normal plant genes. Physiol. Plant 103: 581–583. [Google Scholar]
- Wessler, S. R., T. E. Bureau and S. E. White, 1995. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5: 814–821. [DOI] [PubMed] [Google Scholar]
- Yeadon, P. J., and D. E. Catcheside, 1995. Guest: a 98 bp inverted repeat transposable element in Neurospora crassa. Mol. Gen. Genet. 247: 105–109. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., J. Arbuckle and S. R. Wessler, 2000. Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc. Natl. Acad. Sci. USA 97: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., C. Feschotte, Q. Zhang, N. Jiang, W. B. Eggleston et al., 2001. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc. Natl. Acad. Sci. USA 98: 12572–12577. [DOI] [PMC free article] [PubMed] [Google Scholar]