Abstract
In inflammation, the post-transcriptional regulation of transiently expressed genes provides a potential therapeutic target. Tristetraprolin (TTP) is of the factors regulating decay of cytokine mRNAs. The aim of the present study was to identify cytokines whose expression is regulated by TTP. We established a TTP knock-down cell line by expressing shRNA against TTP (shTTP cell line). A cytokine antibody array was used to measure cytokine production in macrophages exposed to lipopolysaccharide (LPS). Cytokines IL-6, IL-12, TNF-α, and MIP-2 (a homologue to human IL-8) were expressed at higher levels whereas MIP-3α was produced at lower levels in LPS-treated shTTP cells than in control cells suggesting that the expression of these cytokines is regulated by TTP. The present data provide IL-12, MIP-2, and MIP-3α as novel inflammatory cytokine targets for TTP-mediated mRNA decay and stress the role of TTP in the regulation of the inflammatory process.
INTRODUCTION
In inflammation, the post-transcriptional regulation of transiently expressed genes provides a potential therapeutic target. The regulation of mRNA stability through AU-rich element (ARE)-containing areas in the 3′-untranslated region has been found an important means to regulate cytokine production. Tristetraprolin (TTP) is one of the factors known to regulate mRNA stability and expression of proinflammatory cytokines especially tumor necrosis factor (TNF)-α. TTP (synonyms: Nup475, TIS11, G0S24, and Zfp36) expression is induced by inflammatory and related stimuli including bacterial products, growth factors, 12-O-tetradecanoylphorbol-13-acetate ester and serum [1–6]. TTP is a member of a CCCH tandem zinc finger protein family that also contains Zfp36-like 1, Zfp36-like 2 and the recently found Zfp36-like 3 [7, 8]. TTP has been reported to bind the ARE of certain mRNAs, which leads to mRNA deadenylation and degradation [9, 10]. Recent articles suggest that TTP is a component of both stress granules and processing bodies [11] and that the zinc finger domain is needed to localize TTP into the stress granules, which are sites of stalled translational preinitiation complexes [12]. TTP also recruits and activates enzymes needed in ARE-containing mRNA decay [13], and TTP seems to have a multifunctional role in mRNA degradation.
Inflammatory tissues such as spleen, lymph nodes, and thymus express TTP mRNA [2, 3]. The significant role of TTP in inflammation was first discovered in TTP knock-out mice, which developed a set of severe inflammatory symptoms due to high levels of TNF-α [14]. In TTP deficient animals, the levels of TNF-α were elevated because of increased TNF-α mRNA stability [9, 15]. The mRNAs of granulocyte macrophage colony stimulating factor (GM-CSF), interleukin (IL)-2, IL-3, IL-6, cyclooxygenase-2 (COX-2), and plasminogen activator inhibitor type 2 have also been reported to be destabilized by TTP [16–21]. In contrast, TTP has been shown to inhibit human inducible nitric oxide synthase (iNOS) mRNA degradation. TTP did not bind to the iNOS mRNA but its effect was mediated through interaction with the KH-type splicing regulatory protein (KSRP) [22].
In the present study, we established a cell line expressing shRNA against TTP resulting in reduced TTP expression in response to inflammatory stimulus. In the further studies, we used a cytokine antibody array to measure the effects of TTP down-regulation on cytokine production in macrophages exposed to LPS.
MATERIALS AND METHODS
Cell culture
J774 murine macrophages (American Type Culture Collection, Rockville, Md, USA) were cultured at 37°C in humidified 5% carbon dioxide atmosphere in Dulbecco's modified Eagle medium with Ultraglutamine 1 (DMEM/U1, Cambrex Bioproducts Europe, Verviers, Belgium) supplemented with 10% heat-inactivated FBS (EuroClone, Wetherby, UK), penicillin (100 units/mL), streptomycin (100 μg/mL) and amphotericin B (250 ng/mL) (Gibco, Paisley, Scotland, UK).
Cell lines expressing short hairpin RNAs (shRNA) against TTP or a negative control sequence
The nucleotides (Table 1) (Metabion, Planegg-Martinsried, Germany) were annealed and ligated into the pSilencer neo vector (Ambion Inc, Austin, Tex, USA) with T4 DNA ligase (Fermentas Inc, Burlington, Ontario, Canada). One Shot TOP10 Competent Cells (Invitrogen, Paisley, UK) were chemically transformed according to the manufacturer's instructions. Plasmids were isolated with Plasmid Mini kit (QIAGEN Inc, Santa Clarita, Calif, USA) and transfected with FuGENE 6 Transfection Reagent (Roche Diagnostics Corporation, Indianapolis, Ind, USA) into J774 macrophages. G418 disulfide salt (Sigma Chemical Co, St Louis, Mo, USA) was used to select and maintain the J774 cell lines expressing shRNA against TTP (shTTP) and negative control shRNA (shNEG).
Table 1.
Target sequences and primers of shTTP and shNEG.
| shTTP target sequence 5′-AACAUAAACUCGGACUCCAUC-3′ |
| shTTP sense 5′-GATCCGCATAAACTCGGACTCCATCTTCAAGAGAGATGGAGTCCGAGTTTATGTTTTTTGGAAA-3′ |
| shTTP antisense 5′-AGCTTTTCCAAAAAACATAAACTCGGACTCCATCTCTCTTGAAGATGGAGTCCGAGTTTATGCG-3′ |
| shNEG target sequence 5′-AAACUACCGUUGUUAUAGGUG-3′ |
| shNEG sense 5′-GATCCACTACCGTTGTTATAGGTGTTCAAGAGACACCTATAACAACGGTAGTTTTTTTGGAAA-3′ |
| shNEG antisense 5′-AGCTTTTCCAAAAAAACTACCGTTGTTATAGGTGTCTCTTGAACACCTATAACAACGGTAGTG-3′ |
Stimulation of shTTP and shNEG cell lines
For the cytokine protein array, shTTP and shNEG cells were plated on 6 well plates 24 h prior to the experiment. Cells were first incubated in DMEM/U1 + FBS with or without LPS (100 ng/mL) (Sigma, St Louis, Mo, USA). After 1 h of incubation medium without FBS was changed to the wells and incubation was continued for 48 h. Thereafter, cell culture mediums were collected and stored at −20°C until assayed.
For Western blot shTTP and shNEG cells were plated on 6 well plates and grown to confluence. Cells were treated with or without LPS (100 ng/mL) for 6 h and proteins were extracted as described [23].
Western blotting
The protocol for Western blotting was described in [23]. The gels were loaded with 50 μg of protein. Actin antibody was purchased from Santa Cruz Biotechnology, Santa Cruz, Calif, USA and the mouse TTP antibody was a kind gift from Dr Perry Blackshear (NIEHS, Research Triangle Park, NC, USA). The bound antibodies were detected using Super Signal West Pico (for actin) or Dura (for TTP) chemiluminescent substrate for HRP detection (Pierce, Cheshire, UK) and FluorChem 8800 imaging system (Alpha Innotech, San Leandro, Calif, USA). The chemiluminescent signals were measured with FluorChem software v. 3.1.
TNF-α enzyme-linked immunosorbent assay (ELISA)
TNF-α concentrations in culture media were determined by mouse TNF-α DuoSet ELISA kit (R&D Systems, Inc, Minneapolis, Minn, USA) according to the manufacturer's instructions.
Cytokine antibody array
Cytokines were detected in cell culture media with Mouse Cytokine Antibody Array III (RayBiotech, Inc, Norcross, Ga, USA), which measures 62 cytokines and other inflammatory mediators. The array membranes were blocked with 2 mL of 1X blocking buffer for 30 min and then incubated with the sample (1 mL) for 2 h at room temperature. The membranes were washed three times with 2 mL of 1X wash buffer I and twice with 2 mL of 1X wash buffer II at room temperature. The membranes were then incubated in diluted primary antibodies over night at +4°C. The membranes were washed as described earlier and incubated with diluted HRP-conjugated streptavidin for 2 h at room temperature and washed. Detection buffer C and detection buffer D were combined and applied on the membranes for 2 min. Each membrane was exposed for 1 min and images were taken with FluorChem 8800 imaging system (Alpha Innotech Corp, San Leandro, Calif, USA) and chemiluminescent signals for each spot were measured with FluorChem software v. 3.1. The average chemiluminescence of each cytokine and control was calculated for all the treatments separately. The average of positive controls of each treatment was set to 100 and all cytokines of the same treatment were compared to that.
Statistics
Results are expressed as the mean ± standard error of mean (SEM). The significance of differences was calculated by analysis of variance supported by Dunnett's adjusted significance levels. A difference between treatment groups was considered significant when P < .05.
RESULTS
Down-regulation of TTP expression in macrophages transfected with shTTP
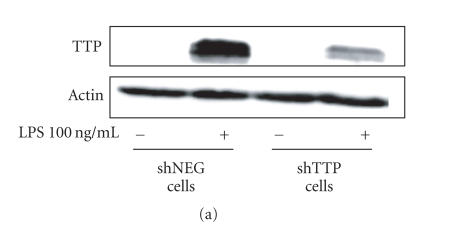
J774 macrophages were transfected with shTTP expression vector (shTTP cell line) or with shNEG negative control vector (shNEG cell line) and maintained under G418 disulfide salt selection. Neither of the cell lines expressed detectable amounts of TTP protein when cultured in the absence of lipopolysaccharide (LPS). When LPS (100 ng/mL) was added into the culture, TTP was clearly expressed in shNEG cell line whereas TTP protein expression was markedly lower in shTTP cells (Figure 1(a)).
Figure 1.
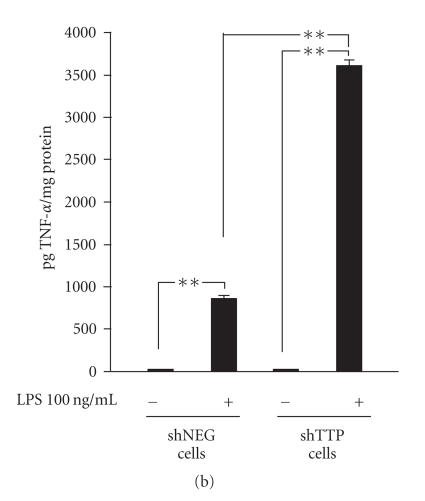
Down-regulation of TTP expression and enhancement of TNF-α production in shTTP cells. (a) shNEG and shTTP cells were stimulated with LPS (100 ng/mL) and proteins were extracted after 6 h of incubation. TTP and actin were detected by Western blot. The blot is a representative of three blots with similar results. (b) shNEG and shTTP cells were stimulated with LPS (100 ng/mL) for 1 h. Thereafter the medium was changed and the cells were incubated for another 48 h. TNF-α concentrations in the culture media were measured by ELISA. Values are mean ± SEM (n = 3). ** = P < .01.
TTP knock-out mice have been shown to have increased levels of circulating TNF-α due to increased TNF-α mRNA stability in the absence of TTP [15, 24]. LPS-induced TNF-α levels produced by shNEG and shTTP cell lines were determined by ELISA. The results show that the TNF-α levels were more than three fold higher in LPS-treated shTTP cells than in shNEG cells (Figure 1(b)) confirming the functional consequences of TTP knock-down in shTTP cells.
Cytokine production in shNEG and shTTP cell lines
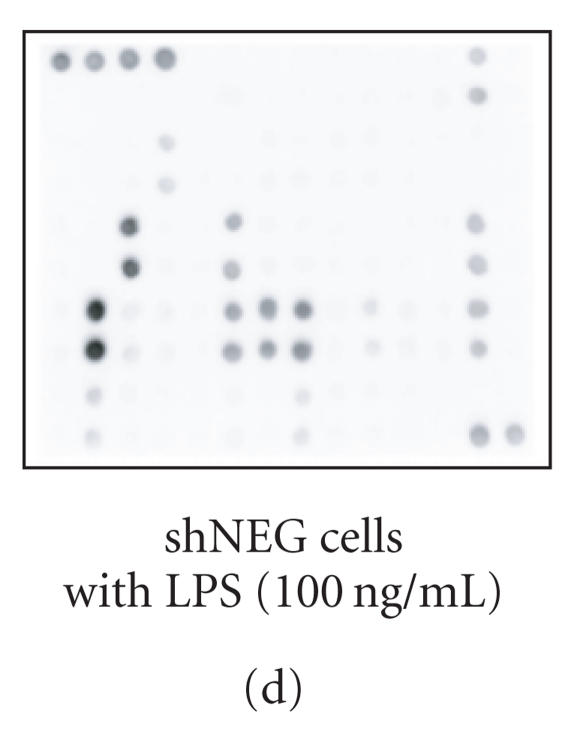
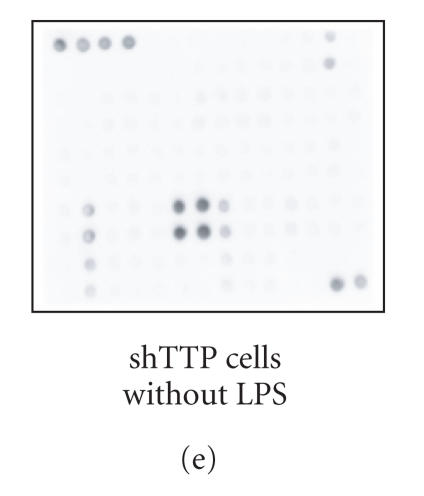
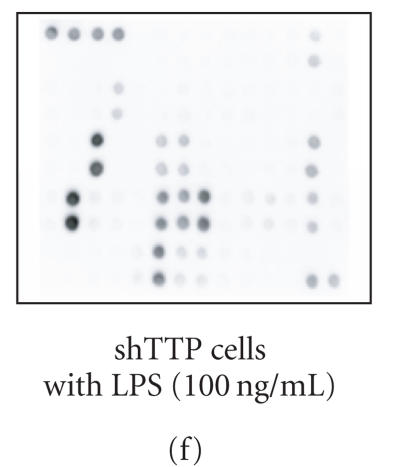
Cytokine production in shNEG and shTTP cell lines was measured by using an antibody array that detects 62 cytokines and other inflammatory mediators (Figure 2(a)). shNEG and shTTP cell lines were incubated with or without LPS (100 ng/mL) and cytokines produced into the culture medium were measured after 48 h incubation. An example membrane of each treatment is shown in Figures 2(b)–2(f).
Figure 2.
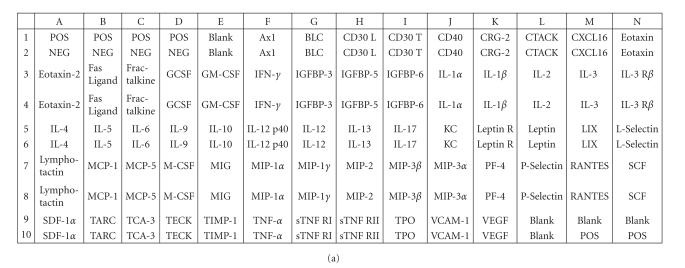
Cytokine antibody array. (a) A schematic diagram of the Mouse Cytokine Antibody Array III shows the locations of controls and the duplicate spots of cytokines. (b)–(f) Images of the membranes treated with cell culture media from the following experiments: (b) culture medium without cells, (c) shNEG cells, 49 h incubation, (d) shNEG cells, stimulated for 1 h with LPS (100 ng/mL) and incubated thereafter for 48 h, (e) shTTP cells, 49 h incubation, (f) shTTP cells, stimulated for 1 h with LPS (100 ng/mL) and incubated thereafter for 48 h. Representative membranes of three with similar results are shown. POS = positive control, NEG = negative control.
When analyzing the results of the cytokine antibody array the positive control was set as 100 and the cytokine results were related to the positive control. The average of negative controls and blanks obtain values < 2.5. The immunoreactivity of different cytokines in the cell culture medium was < 5 when compared to the positive controls.
In the absence of LPS, shNEG, and shTTP cell lines produced five out of the 62 measured cytokines into the culture medium, that is, cutaneous T-cell attracting chemokine (CTACK), CXCL16, MIP-1α, MIP-1γ and thymus and activation-regulated chemokine (TARC). Between the shNEG and shTTP cell lines, CXCL16 was expressed at higher levels in shNEG cell line whereas only minor differences between shNEG and shTTP cell lines were found in the other cytokines. The results are shown in Table 2.
Table 2.
Cytokines secreted spontaneously by shNEG and shTTP cell lines. Arbitrary units compared to positive controls (100) are presented. Mean ± SEM (n = 3).
| Cytokine | Medium | shNEG | shTTP |
| without cells | cell line | cell line | |
| CTACK | 1.9 | 5.8 | 6.3* |
| CXCL16 | 1.1 | 72.3** | 35.7* |
| MIP-1α | 2.9 | 82.3* | 117.5** |
| MIP-1γ | 1.8 | 118.4** | 128.8** |
| TARC | 2.5 | 41.3** | 28.1* |
** = P < .01, * = P < .05 as compared to the medium without cells.
LPS induced changes in cytokine production in shNEG and shTTP cell lines
In shNEG cell line the expression of 11 out of the 62 measured cytokines was changed following LPS treatment (Table 3). With LPS treatment the expression of 9 cytokines [GCSF, IL-6, LPS induced C-X-C chemokine (LIX), MCP-1, MCP-5, MIP-2, MIP-3α, RANTES, and sTNF RII] and the IL-12 p40 subunit increased. IL-12 p40 detects p40 subunit in the active IL-12 p70 as well as all otherwise engaged p40 subunits. On the contrary, the expression of CXCL16 in LPS-treated shNEG cells decreased as compared to untreated shNEG cells.
Table 3.
Cytokines secreted by untreated and LPS- (100 ng/mL) treated shNEG cells during 48 h incubation. Arbitrary units compared to positive controls (100) are presented. Mean±SEM (n = 3).
| Cytokine | Medium without cells | shNEG cells | |
| Untreated | LPS-treated | ||
| CXCL16 | 1.1 | 72.3 | 43.0* |
| GCSF | 2.3 | 3.3 | 21.4** |
| IL-6 | 2.4 | 3.8 | 172.3** |
| IL-12 p40 | 1.8 | 2.7 | 68.0** |
| LIX | 4.0 | 6.1 | 62.5** |
| MCP-1 | 4.4 | 81.2 | 293.6** |
| MCP-5 | 3.3 | 4.3 | 13.5** |
| MIP-2 | 4.2 | 51.0 | 128.1** |
| MIP-3α | 3.9 | 5.3 | 18.1** |
| RANTES | 3.0 | 4.3 | 53.8** |
| sTNF RII | 3.9 | 11.4 | 20.6* |
** = P < .01, * = P < .05 for the difference between untreated and LPS-treated shNEG cells.
In shTTP cell line, the production of 10 cytokines (GCSF, IL-6, IL-12, LIX, MCP-1, MCP-5, MIP-2, RANTES, sTNF RII, and TNF-α) and IL-12 p40 subunit were increased following LPS treatment (Table 4).
Differences in LPS-induced cytokine expression in shNEG and shTTP cell lines
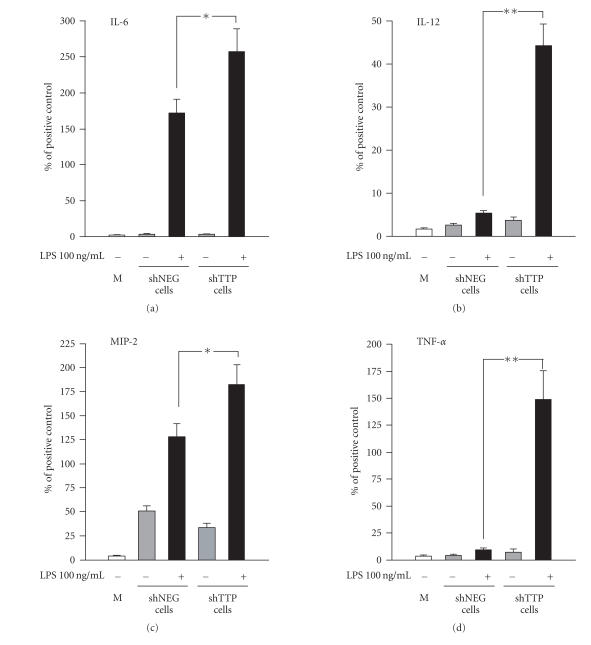
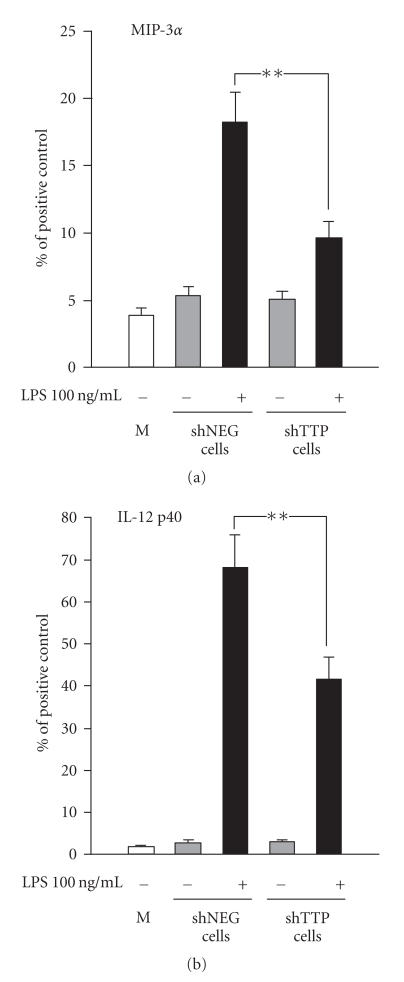
The LPS-induced production of six cytokines (IL-6, IL-12, IL-12 p40 subunit, MIP-2, MIP-3α, and TNF-α) was altered in shTTP cells as compared to shNEG cells suggesting that the expression of these six cytokines is regulated by TTP. Cytokines IL-6, MIP-2, IL-12, and TNF-α were expressed at higher levels in shTTP cell line than in shNEG cell line (Figure 3). In contrast, MIP-3α and IL-12 p40 subunit were expressed at lower levels in LPS-treated shTTP than in LPS-treated shNEG cell lines (Figure 4).
Figure 3.
TTP down-regulation (shTTP cell line) enhanced the production of IL-6, IL-12, MIP-2, and TNF-α in response to LPS. Cytokine antibody array membranes were incubated in medium control (M) or in media from cultures of TTP knock-down cells (shTTP cell line) or control cells (shNEG cell line) after treatment with or without LPS (100 ng/mL). Values are arbitrary units compared to positive controls (100) and are presented as mean ± SEM (n = 3). ** = P < .01, * = P < .05.
Figure 4.
TTP down-regulation (shTTP cell line) reduced the production of MIP-3α and IL-12 p40 subunit in response to LPS. Cytokine antibody array membranes were incubated in medium control (M) or in media from cell cultures of TTP knock-down cells (shTTP cell line) or control cells (shNEG cell line) after treatment with or without LPS (100 ng/mL). Values are arbitrary units compared to positive controls (100) and are presented as mean ± SEM (n = 3). ** = P < .01.
DISCUSSION
The role of mRNA turnover in the regulation of inflammatory gene expression has been recognized pathophysiologically and therapeutically important. TTP is one of the factors regulating mRNA decay. Here we report that TTP down-regulation resulted in increased expression of TNF-α, MIP-2, IL-6, and IL-12 in macrophages exposed to bacterial LPS. On the other hand, the levels of MIP-3α were reduced in LPS-treated TTP knock-down cells. The possible new targets for TTP discovered in the present study are IL-12, MIP-2 (a homologue to human IL-8) and MIP-3α.
TTP is a regulator of the stability of some transiently expressed ARE-containing cytokine mRNAs. TTP has been reported to down-regulate the expression of TNF-α, GM-CSF, IL-2, IL-3, and IL-6 by destabilizing their mRNAs [9, 15–19]. In the present study, we established a cell line where TTP was knocked down by expressing an shRNA against TTP, and evaluated the levels of 62 cytokines secreted by the cells into the culture medium by cytokine antibody array. Protein antibody array has proved to be a useful tool to measure the expression of multiple proteins in cells and tissues at the same time. The results obtained are more significant than when using cDNA microarrays as mRNAs are subjected to post-transcriptional and post-translational processes and the amount of mRNA may not correlate with protein expression [25]. The advantage of protein array as compared to ELISA is the simultaneous measurement of multiple proteins, but it is regarded as a semiquantitative method. In the present study, when TTP expression was induced by LPS, the TTP knock-down cell line showed clearly reduced TTP protein levels as compared to control cells. The reduction was functionally significant as it resulted in a clear increase in TNF-α production that is in line with the previous literature on the effects of TTP on TNF-α production [14, 24].
In experiments with TTP knock-out mice and cells derived from them, TTP has been shown to mediate the degradation of TNF-α and GM-CSF mRNAs [15, 16]. Both transcripts contain AREs [26] and TTP binds to the nonamer UUAUUUAUU in these structures with its zinc finger domains [9, 27–30]. ARE-binding proteins such as TTP and KSRP seem to accelerate the degradation of ARE-mRNAs via the exosome pathway by recruiting the exosome to the mRNA [31]. It has also been shown that TTP can promote mRNA deadenylation by stimulating poly(A) ribonuclease [32]. The N-terminal domain of TTP also associates with mRNA decay enzymes involved in decapping, deadenylation, and exonucleolytic decay [13]. In addition, TTP has been proposed to direct certain mRNAs through stress granules to processing bodies to be degraded there [11]. However, the detailed mechanisms involved in the TTP-mediated mRNA decay are not known.
IL-6 is another proinflammatory cytokine produced by macrophages that is known to be regulated by TTP. It was shown that IL-6 mRNA degradation was impaired in a HT1080-derived mutant cell line. When the cells were then stably transfected with TTP, the IL-6 mRNA decay restored to the wild-type levels [19]. Here, we show that down-regulation of TTP expression in activated macrophages resulted in increased IL-6 production. These data suggest that in wild-type cells IL-6 mRNA is destabilized by TTP.
In addition to TNF-α and IL-6 [9, 15, 19], TTP has been reported to down-regulate the expression of GM-CSF, IL-2, and IL-3 by destabilizing their mRNAs [16–18]. In the present experiments, we were not able to detect these cytokines in our macrophage cultures and could not draw any conclusions on the effects of TTP deficiency on those cytokines.
IL-12 is a proinflammatory cytokine involved, for example, in the pathogenesis of autoimmune diseases [33, 34]. IL-12 is a heterodimer comprised of p35 and p40 subunits and designated as IL-12 p70 [34]. The subunits are encoded by two distinct genes and they need to be expressed simultaneously by the same cell to generate the biologically active heterodimer p70. Neither of the subunits is active on its own. The p40 subunit can also form homodimers (IL-12 p80), which act as endogenous IL-12 antagonists by binding to the IL-12 receptor without inducing a cellular response. The p40 subunit is also a part of IL-23. Our results showed that IL-12 was produced at higher levels in TTP-knock-down cells than in control cells in response to LPS. The result suggests a similar pattern of regulation of one or both of the subunits of IL-12 as with TNF-α and IL-6 where TTP destabilizes the mRNA and promotes its rapid degradation. To our knowledge, this is the first time to report that the expression of IL-12 is regulated by TTP.
In addition, we found a decrease in the expression of IL-12 p40 subunit in LPS-treated shTTP cells as compared to shNEG cells. In macrophages, the p40 subunit is secreted in several-fold excess as compared to the p35–p40 heterodimer, and the formation of active IL-12 (p35 and p40 heterodimer) is regulated by the synthesis of p35 subunit [35]. The mRNA of p35 (GenBank accession number NM_008351) contains one copy of UUAUUUAUU nonamer which is the preferred TTP binding site [30], where as none of those are found in p40 mRNA (GenBank accession number NM_008352). Therefore we suggest that the target for TTP might be the p35 subunit, which could be expressed at lower levels in the LPS-treated shNEG cells than in shTTP cells. Further studies are needed to understand the mechanisms how TTP regulates IL-12 production.
A member of the CXC chemokine superfamily, MIP-2 (a homologue to human IL-8), is a potent chemoattractant for neutrophils [36]. In the present study, we found that MIP-2 production in response to LPS was higher in TTP knock-down cells than in control cells. We are not aware of earlier reports on the regulation of MIP-2 by TTP, and the results suggest a role for TTP as a destabilizer of MIP-2 mRNA. The sequence of MIP-2 mRNA (GenBank accession number NM_009140) contains three copies of the preferred nonamer binding site of TTP (UUAUUUAUU), which further supports MIP-2 mRNA as a target of the destabilizing effect of TTP.
MIP-3α (synonyms: CC chemokine ligand 20, CCL20; liver and activation-regulated chemokine, LARC; and Exodus-1) is a chemokine which has a potential role in rheumatoid arthritis [37]. In the present study, we found that MIP-3α production in response to LPS was lower in cells with reduced TTP expression than in control cells. That is interesting, because TTP has been recognized as a factor that down-regulates the expression of certain inflammatory genes by destabilizing their mRNAs. MIP-3α seems to be an exception to that rule. Our result is supported by the fact that MIP-3α mRNA (GenBank accession number NM_016960) contains no nonamer binding sites for TTP (UUAUUUAUU), and therefore it is unlikely a direct target of TTP. Recently, Fechir and coworkers reported that TTP was able to enhance human iNOS expression by stabilizing its mRNA in cytokine-treated bronchial epithelial cell line [22]. TTP did not bind to human iNOS mRNA but was shown to enhance the half-life of iNOS mRNA by an indirect mechanism. TTP was found to interact with KSRP and it was proposed to inhibit the degradation of iNOS mRNA and enhance iNOS expression by capturing KSRP [22]. In the present study, we found that down-regulation of TTP resulted in reduction of MIP-3α production. To our knowledge, this is the first report to show that TTP regulates the production of MIP-3α. It remains to be studied if TTP regulates MIP-3α expression in a similar mechanism as has been reported for human iNOS.
The results presented here provide IL-12, MIP-2, and MIP-3α as novel inflammatory cytokine targets for TTP-regulated mRNA decay. The data are implicated in the understanding of basic mechanisms of the inflammatory process and in the development of novel anti-inflammatory drugs.
Table 4.
Cytokines secreted by untreated and LPS- (100 ng/mL) treated shTTP cells during 48 h incubation. Arbitrary units compared to positive controls (100) are presented. Mean±SEM (n = 3).
| Cytokine | Medium without cells | shTTP cells | |
| Untreated | LPS-treated | ||
| GCSF | 2.3 | 3.6 | 32.0** |
| IL-6 | 2.4 | 3.5 | 258.0** |
| IL-12 p40 | 1.8 | 2.9 | 41.6** |
| IL-12 | 1.7 | 3.6 | 44.3** |
| LIX | 4.0 | 6.2 | 85.2** |
| MCP-1 | 4.4 | 46.3 | 349.1** |
| MCP-5 | 3.3 | 3.5 | 11.0** |
| MIP-2 | 4.2 | 33.6 | 182.3** |
| RANTES | 3.0 | 4.6 | 57.5** |
| sTNF RII | 3.9 | 12.1 | 27.3** |
| TNF-α | 3.7 | 7.2 | 149.0** |
** P < .01 for the difference between untreated and LPS-treated shTTP cells.
ACKNOWLEDGMENTS
We thank Dr Perry Blackshear (NIEHS, Research Triangle Park, NC, USA) for the tristetraprolin antibody, Mrs Niina Ikonen and Mrs Marja-Leena Lampén for excellent technical assistance, and Mrs Heli Määttä for skilled secretarial help. This study was supported by Tampere Graduate School in Biomedicine and Biotechnology, The Academy of Finland, and The Medical Research Fund of Tampere University Hospital.
References
- 1.Varnum BC, Lim RW, Sukhatme VP, Herschman HR. Nucleotide sequence of a cDNA encoding TIS11, a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4(1):119–120. [PubMed] [Google Scholar]
- 2.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. Journal of Biological Chemistry. 1990;265(27):16556–16563. [PubMed] [Google Scholar]
- 3.DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. Journal of Biological Chemistry. 1990;265(31):19185–19191. [PubMed] [Google Scholar]
- 4.Taylor GA, Lai WS, Oakey RJ, et al. The human TTP protein: sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Research. 1991;19(12):3454. doi: 10.1093/nar/19.12.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Q, Herschman HR. A corrected sequence for the predicted protein from the mitogen-inducible TIS11 primary response gene. Oncogene. 1991;6(7):1277–1278. [PubMed] [Google Scholar]
- 6.Heximer SP, Forsdyke DR. A human putative lymphocyte G0/G1 switch gene homologous to a rodent gene encoding a zinc-binding potential transcription factor. DNA and Cell Biology. 1993;12(1):73–88. doi: 10.1089/dna.1993.12.73. [DOI] [PubMed] [Google Scholar]
- 7.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochemical Society Transactions. 2002;30(6):945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 8.Blackshear PJ, Phillips RS, Ghosh S, Ramos SVB, Richfield EK, Lai WS. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the tristetraprolin family of CCCH tandem zinc finger proteins. Biology of Reproduction. 2005;73(2):297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 9.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Molecular and Cellular Biology. 1999;19(6):4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai WS, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. Journal of Biological Chemistry. 2001;276(25):23144–23154. doi: 10.1074/jbc.M100680200. [DOI] [PubMed] [Google Scholar]
- 11.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. Journal of Cell Biology. 2005;169(6):871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murata T, Morita N, Hikita K, Kiuchi K, Kiuchi K, Kaneda N. Recruitment of mRNA-destabilizing protein TIS11 to stress granules is mediated by its zinc finger domain. Experimental Cell Research. 2005;303(2):287–299. doi: 10.1016/j.yexcr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes and Development. 2005;19(3):351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor GA, Carballo E, Lee DM, et al. A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 15.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281(5379):1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 16.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95(6):1891–1899. [PubMed] [Google Scholar]
- 17.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. Journal of Immunology. 2005;174(2):953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 18.Stoecklin G, Ming X-F, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Molecular and Cellular Biology. 2000;20(11):3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoecklin G, Stoeckle P, Lu M, Muehlemann O, Moroni C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001;7(11):1578–1588. [PMC free article] [PubMed] [Google Scholar]
- 20.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3′-untranslated region of cyclooxygenase-2 mRNA: a polyadenylation variant in a cancer cell line lacks the binding site. Journal of Biological Chemistry. 2003;278(16):13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 21.Yu H, Stasinopoulos S, Leedman P, Medcalf RL. Inherent instability of plasminogen activator inhibitor type 2 mRNA is regulated by tristetraprolin. Journal of Biological Chemistry. 2003;278(16):13912–13918. doi: 10.1074/jbc.M213027200. [DOI] [PubMed] [Google Scholar]
- 22.Fechir M, Linker K, Pautz A, et al. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Molecular Pharmacology. 2005;67(6):2148–2161. doi: 10.1124/mol.104.008763. [DOI] [PubMed] [Google Scholar]
- 23.Jalonen U, Lahti A, Korhonen R, Kankaanranta H, Moilanen E. Inhibition of tristetraprolin expression by dexamethasone in activated macrophages. Biochemical Pharmacology. 2005;69(5):733–740. doi: 10.1016/j.bcp.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 24.Carballo E, Gilkeson GS, Blackshear PJ. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (-/-) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFα overproduction. Journal of Clinical Investigation. 1997;100(5):986–995. doi: 10.1172/JCI119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Templin MF, Stoll D, Schrenk M, Traub PC, Vöhringer CF, Joos TO. Protein microarray technology. Drug Discovery Today. 2002;7(15):815–822. doi: 10.1016/s1359-6446(00)01910-2. [DOI] [PubMed] [Google Scholar]
- 26.Chen C-YA, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends in Biochemical Sciences. 1995;20(11):465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 27.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. Journal of Biological Chemistry. 2002;277(50):48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 28.Blackshear PJ, Lai WS, Kennington EA, et al. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. Journal of Biological Chemistry. 2003;278(22):19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- 29.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of AU-rich mRNA-destabilizing motifs. Journal of Biological Chemistry. 2004;279(27):27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 30.Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. Journal of Biological Chemistry. 2005;280(40):34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 31.Chen C-Y, Gherzi R, Ong S-E, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107(4):451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 32.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Molecular and Cellular Biology. 2003;23(11):3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McInnes IB, Gracie JA. Targeting cytokines beyond tumor necrosis factor-α and interleukin-1 in rheumatoid arthritis. Current Rheumatology Reports. 2004;6(5):336–342. doi: 10.1007/s11926-004-0007-2. [DOI] [PubMed] [Google Scholar]
- 34.Becker C, Wirtz S, Neurath MF. Stepwise regulation of TH1 responses in autoimmunity: IL-12-related cytokines and their receptors. Inflammatory Bowel Diseases. 2005;11(8):755–764. doi: 10.1097/01.mib.0000172808.03877.4d. [DOI] [PubMed] [Google Scholar]
- 35.Snijders A, Hilkens CMU, van der Pouw Kraan TCTM, Engel M, Aarden LA, Kapsenberg ML. Regulation of bioactive IL-12 production in lipopolysaccharide-stimulated human monocytes is determined by the expression of the p35 subunit. Journal of Immunology. 1996;156(3):1207–1212. [PubMed] [Google Scholar]
- 36.Driscoll KE. TNFα and MIP-2: role in particle-induced inflammation and regulation by oxidative stress. Toxicology Letters. 2000;112-113:177–183. doi: 10.1016/s0378-4274(99)00282-9. [DOI] [PubMed] [Google Scholar]
- 37.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine and Growth Factor Reviews. 2003;14(5):409–426. doi: 10.1016/s1359-6101(03)00049-2. [DOI] [PubMed] [Google Scholar]