Abstract
Heroin use is postulated to act as a cofactor in the neuropathogenesis of human immunodeficiency virus (HIV-1) infection. Astrocytes, integral components of the CNS, are reported to be susceptible to HIV-1 infection. Upon activation, astrocytes release a number of immunoregulatory products or modulate the expression of a number of proteins that foster the immunopathogenesis of HIV-1 infection. However, the role of heroin on HIV-1 infectivity and the expression of the proteome of normal human astrocytes (NHA) have not been elucidated. We hypothesize that heroin modulates the expression of a number of proteins by NHA that foster the neuoropathogenesis of HIV-1 infection. We utilized LTR amplification and the p24 antigen assay to quantitate the effect of heroin on HIV-1 infectivity while difference gel electrophoresis (DIGE) combined with protein identification through high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) to analyze the effects of heroin on the proteomic profile of NHA. Results demonstrate that heroin potentiates HIV-1 replication in NHA. Furthermore, heroin significantly increased protein expression levels for protein kinase C (PKC), reticulocalbin 1 precursor, reticulocalbin 1, tyrosine 3-monooxgenase/tryptophan 5-monooxgenase activation protein, chloride intracellular channel 1, cathepsin D preproprotein, galectin 1 and myosin light chain alkali. Heroin also significantly decreased protein expression for proliferating cell nuclear antigen, proteasome beta 6 subunit, tropomyosin 3, laminin receptor 1, tubulin alpha 6, vimentin, EF hand domain family member D2, Tumor protein D54 (hD54), ATP synthase, H+ transporting, mitochondrial F1 complex and ribosomal protein S14. Identification of unique, heroin-induced proteins may help to develop novel markers for diagnostic, preventative and therapeutic targeting in heroin using subjects.
Keywords: Heroin, normal human astrocytes (NHA), difference gel Electrophoresis (DIGE), high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS)
INTRODUCTION
Heroin is one of the most widely abused and rapidly acting opiates[1]. According to the 2004 National Survey on Drug Use and Health, approximately 3.1 million Americans ages 12 and older reported trying heroin at least once during their lifetimes[2]. Parenteral drug abuse is a significant risk factor for contracting human immunodeficiency virus (HIV-1) infection[3,4]. HIV-1-infected individuals who are injecting drug users may undergo an accelerated rate of progression to acquired immunodeficiency syndrome (AIDS)[4–7].
Several AIDS patients develop severe neurological symptoms referred to as HIV-1 encephalopathy (HIVE)[6]. HIVE is characterized by multinucleated giant cells, microglial nodules and astrogliosis[6–9]. Macrophage and microglial cells are the primary sources of HIV-1 replication in central nervous system (CNS)[10–12]. Astrocytes are also reported to be susceptible to HIV-1 infection albeit at lower levels[13–17]. Astrocytes are integral components of the CNS; they maintain a homeostatic environment and actively participate in a bi-directional communication with neurons[13,18,19]. Upon activation, HIV-1 infected astrocytes may spread the infection or induce cellular damage to neighboring cells of the CNS through the release of viral and cellular products. Consequently, disruption of astrocyte function could lead to severe neuropathogenesis. Because a significant number of astrocytes can be infected with HIV-1 in the CNS and heroin may act as a cofactor in HIVE, we hypothesize that heroin-induced increases in HIV-1 susceptibility and progression to HIVE are mediated via dysregulation of specific proteins that foster the immunopathogenesis of HIV-1 infection.
In the present application, we studied the effect of heroin on HIV-1 susceptibility of normal human astrocytes (NHA) by using LTR amplification and the p24 antigen assay and heroin-induced differences in protein profiling of NHA cultures by the proteomic method of Difference Gel Electrophoresis (DIGE). The identification of unique, heroin specific responsive proteins by proteomic analyses may identify novel markers for diagnostic, preventive and therapeutic targeting in heroin-using HIV-1 seropositive populations.
MATERIALS AND METHODS
Primary NHA cultures isolated from human fetal brain cortex tissue (3 independent cultures) were obtained from Cell Systems (Kirkland, WA). NHA cells were initially grown in complete Astrocyte Basal Medium (Cell Systems) as recommended by the manufacturer. Astrocytes were plated in 6 well tissue culture plates at densities of 1x106cells mL−1 in DMEM + 10% FBS. By immunocytochemistry, astrocytes were > 95% GFAP positive and were > 98% viable by trypan blue exclusion criteria.
Drug treatment
A methanol solution of heroin-hydrochloride was purchased from Sigma-Aldrich (St. Louis, Mo) which was subsequently diluted in media to the required concentrations. For all experiments, cells treated with vehicle alone (media alone) was used as the untreated control. NHA were treated with and without heroin at 10−6 to 10−9M for 24 hr.
Treatment of human NHA with HIV-1 Isolate
NHA were treated with heroin for 24 hr washed and then infected with native HIV-1 Ba-L (NIH AIDS Research and Reference Reagent Program, Cat# 510) at a concentration of 10 3.5 TCID 50/ml cells, overnight and washed 3 times with Hank’s balanced salt solution (Invitrogen, Grand Island, NY) before being returned to culture. A post infection period of 48 hr was used in the study to amplify the LTR-R/U5 region which represents early stages of reverse transcription of HIV-1[20]. In separate experiments, NHA were treated with heroin for 24 hr, infected with HIV-1 Ba-L overnight and washed, cultured for 15 days. The culture supernatants were quantitated for p24 antigen using a p24 ELISA kit (ZeptoMetrix Corporation, Buffalo, NY) on day 15.
RNA extraction and Real Time, Quantitative PCR (Q-PCR)
Cytoplasmic RNA was extracted using an acid guanidinium-thiocyanate-phenol-chloroform method[21]. The final RNA pellet was dried and resuspended in diethyl pyrocarbonate (DEPC) water and the concentration of RNA was determined using a spectrophotometer at 260 nm. DNA contamination in the RNA preparation was removed by treating the RNA preparation with DNAse (1 IU μg −1 of RNA, Promega, Madison, WI) for 30 min at 37ºC, followed by proteinase K digestion at 37ºC for 15 min and subsequent extraction with phenol/chloroform and NH4OAc/ETOH precipitation. DNA contamination of the RNA preparation was checked by including a control in which reverse transcriptase enzyme was not added in the PCR amplification procedure. The isolated RNA was stored at −70°C until used. Gene expressions for protein kinase C substrate 80K-H, laminin receptor 1, chloride intracellular channel 1, vimentin, 18s RNA (Ambion, Austin, TX) and β-actin were quantitated using Q-PCR (Table 1). Relative abundance of each mRNA species was assessed using the SYBR green master mix from Stratagene (La Jolla, CA) to perform Q-PCR using the ABI Prism 5700 instrument that detects and plots the increase in fluorescence versus PCR cycle number to produce a continuous measure of PCR amplification. To provide precise quantification of initial target in each PCR reaction, the amplification plot is examined at a point during the early log phase of product accumulation. This is accomplished by assigning a fluorescence threshold above background and determining the time point at which each sample’s amplification plot reaches the threshold (defined as the threshold cycle number or CT). Differences in threshold cycle number are used to quantify the relative amount of PCR target contained within each tube[22]. Relative mRNA species expression was quantitated and expressed as transcript accumulation index (TAI= 2 − delta delta CT), calculated using the comparative CT method[23]. All data were controlled for quantity of RNA input by performing measurements on an endogenous reference gene, β-actin. In addition, results on RNA from treated samples were normalized to results obtained on RNA from the control, untreated sample. As a control, 18s RNA was also used in amplification procedure.
Table 1.
Primer sequences for Real Time Q-PCR
| Primer | Primer sequences |
|---|---|
| β-actin | 5’, 5’-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3’
3’, 5-AGT CAT AGT CCG CCTA GAA GCA TTT GCG GT-3’ |
| HIV-1 LTR RU/5 | 5’ 5′-TCT CTC TGG TTA GAC CAG ATC TG-3’
3’ ′-ACT GCT AGA GAT TTT CCA CAC TG-3’ |
| protein kinase C | 5’, 5’-TGA TTG TTG AGC ACC TCT CG-3’
3’ ′-GCC AGC AGA CAG AAA CTT CC-3’ |
| chloride intracellular channel 1 | 5’, 5’-ACC ATG GCT GAA GAA CAA CC-3’
3’ ′CCC TTG AGC CAC AGT ACC AT-3’ |
| laminin receptor 1 | 5’, 5’-GTT TGA TGT GGT GGA TGC TG-3’
3’ ′CGC TCC AGT CTT CAG TAG GG-3’ |
| vimentin | 5’, 5’-GAG AAC TTT GCC GTT GAA GC-3’
3’ ′TCC AGC AGC TTC CTG TAG GT-3’ |
DIGE
After stimulation, cells were washed 2 times with 1X PBS (Invitrogen, Grand Island, NY). Total protein was extracted using standard cell lysis buffer [30 mM TrisCl; 8 M Urea; 4% (w/v) CHAPS, adjusted to pH 8.5] for 10 min on ice. Cell lysate was centrifuged at 4ºC for 10 min at 12,000 g and lysate was further purified by precipitation with chloroform/methanol as described[24]. Samples were resuspended in standard cell lysis buffer. Final cell lysate protein concentrations were determined using Coomassie Protein Reagent (Bio-Rad, Hercules Ca) and used for protein determination by DIGE analysis. NHA cultured identically without heroin served as the control.
Ettan DIGE technique developed by Amersham Pharmacia Biotech[25] was used to detect differences in protein abundance between normal and experimental samples. The Ettan DIGE system uses three CyDye DIGE fluors (Cy2, Cy3, Cy5), each with a unique fluorescent wavelength, matched for mass and charge. CyDyes form a covalent bond with the free epsilon amino group on lysine residues of the sample proteins. CyDyes label approximately 2% of the lysine residues. This system allows for two experimental samples and an internal standard to be simultaneously separated on the same gel. The internal standard is comprised a pool of an equal amount of all the experimental samples. The use of an internal standard facilitates accurate inter-gel matching of spots and allows for data normalization between gels to minimize gel to gel experimental variability[25,26].
Cell lysates were labeled with CyDye as per the manufacturer. All reagents used were from GE Healthcare (Amersham Biosciences, Piscataway, NJ) Briefly, 50 μg of cell lysate was labeled with 400 pmol of either Cy3 or Cy5 or Cy2 (Cy2 was used to label the internal standard) on ice for 30 min and then quenched with a 50 fold molar excess of free lysine. Cy3, Cy5 and Cy2 labeled samples and unlabelled protein (500–800 μg) were then pooled. An equal volume of 2X sample buffer was added [8 M Urea; 2% (v/v) Pharmalytes 3–10; 2% (w/v) dithiothreitol (DTT); 4% (w/v) CHAPS] incubated on ice for 10 min. The total volume of sample was adjusted to 450 μl with rehydration buffer [4% (w/v) CHAPS; 8 M Urea; 1% (v/v) Pharmalytes 3–10 nonlinear (NL); 13 mM DTT]. Samples were applied to immobilized pH gradient (IPG) strips (24 cm, pH 3–10 NL) and absorbed by active rehydration at 30 V for 13 hr. Isoelectric focusing was carried out using an IPGphor IEF system with a three phase program; first phase at 500 V for 1 hr, second phase at 1000 V for 1 hr and third phase (linear gradient) 8000 V to 64000 V for 2hr (50 uA maximum per strip). Prior to separation in the second dimension, strips were equilibrated for 15 min in equilibration buffer I [50 mM Tris-HCl, 6 M Urea, 30% (v/v) glycerol, 2% (w/v) SDS, 0.5% (w/v) DTT]. The strips were again equilibrated for 15 min in the equilibration buffer II [50 mM Tris-HCl, 6 M Urea, 30% (v/v) glycerol, 2% (w/v) SDS, 4.5% (w/v) iodoacetamide] and the equilibrated IPG strips were then transferred onto 18 x 20 cm, 12.5% uniform polyacrylamide gels poured between low fluorescence glass plates. Gels were bonded to inner plates using Bind-Silane solution according to the manufacturer. Strips were overlaid with 0.9% agarose in 1X running buffer containing bromophenol blue and were run for 16 hr (1.8 W per gel, overnight) at 15ºC, in Ettan DALT electrophoresis system. After run was completed, the 2D gels were scanned three times with a Typhoon 9410 imager, each time at different excitation wavelengths [Cy3, 580 BP 30/green (532 nm); Cy5, 670 BP 30/red (633 nm); Cy2, 520 BP 40/blue (488 nm)]. Images were cropped in ImageQuant v5.2 and then imported into DeCyder Differential In-gel Analysis (DIA) software v5.0 from GE Healthcare for spot identification and normalization of spot intensities within each gel.
Gels were fixed in 30% (v/v) methanol, 7.5% (v/v) acetic acid for 3 hr and then stained with SYPRO-Ruby dye (Molecular Probes, Eugene, OR) overnight at room temperature. Gels were destained in water and then scanned using the Typhoon 9410 scanner. Spots of interest were excised from the gel using the Ettan Spot Picker.
The DeCyder software (GE Healthcare) has been specifically developed for use with Ettan DIGE. The DeCyder software allows for automatic detection of spots, background subtraction, quantitation, normalization, internal standardization and integral matching. DeCyder, DIA draws boundaries around spots in a composite gel image obtained from the intra-gel overlap of the Cy2, Cy3 and Cy5 scanned images and normalizes the data from each CyDye to account for differences in dye fluorescence intensity, scanner sensitivity. The abundance difference between samples run on the same gel is then analyzed. The biological variation analysis (BVA) component of DeCyder is then used to match all image comparisons from in-gel analysis for a cross-gel statistical analysis. DeCyder BVA initially calculates normalized intensities (“standard abundance”) for all spots by comparison to the internal standard and from this an average volume ratio and a Student’s paired t-test derived p value are calculated for each spot. A paired t-test derived p value of ≤0.05 was considered statistically significant[25,26].
HPLC-MS/MS
Excised spots were sent to the Proteomic Analysis Laboratory at the University of Arizona for analysis. In-gel digestion and HPLC-MS/MS were performed as described in Breci et al.[27]. Briefly, gel slices were destained[27–29] and digested with trypsin[30]. The tryptic peptides were extracted with 5% formic acid/50% CH3CN. HPLC was performed using a microbore system (Surveyor, ThermoFinnigan, San Jose, CA). The HPLC column eluate was directed into a ThermoFinnigan LCQ Deca XP Plus ion trap mass spectrometer. Automated peak recognition, dynamic exclusion and daughter ion scanning of the two most intense ions were performed using the Xcalibur software[31,32]. Spectra were scanned over the range 400–1400 mass units. MS/MS data are analyzed using SEQUEST software and searched against the latest version of the National Center for Biotechnology’s public non-redundant protein database[27]. Statistical significance was determined using Student’s t-test (Sigmastat, SPSS Inc.). p <0.05 was considered significant.
RESULTS
Heroin enhances HIV-1 replication in NHA
Data presented in Fig. 1 show the stimulatory effects of heroin on HIV-1 replication in NHA. NHA were treated with and without heroin for 24 hr, infected with HIV-1 Ba-L washed and returned to culture for 48 hr at which RNA was subsequently extracted. The HIV-LTR-R/U5 region was amplified by real time, Q-PCR using primers specific for a 180 bp fragment of the region as described[20]. This method is designed to detect early stages of reverse transcription of HIV-1. Data show that heroin at 10−9 (TAI = 2.55, p = 0.001) 10−7 (TAI = 3.07, p = 0.001) and 10−5 M (TAI = 3.20, p ≤ 0.001) significantly upregulated HIV-LTR-R/U5 region compared to the untreated HIV-1-infected control culture (TAI = 1.0).
Fig 1.
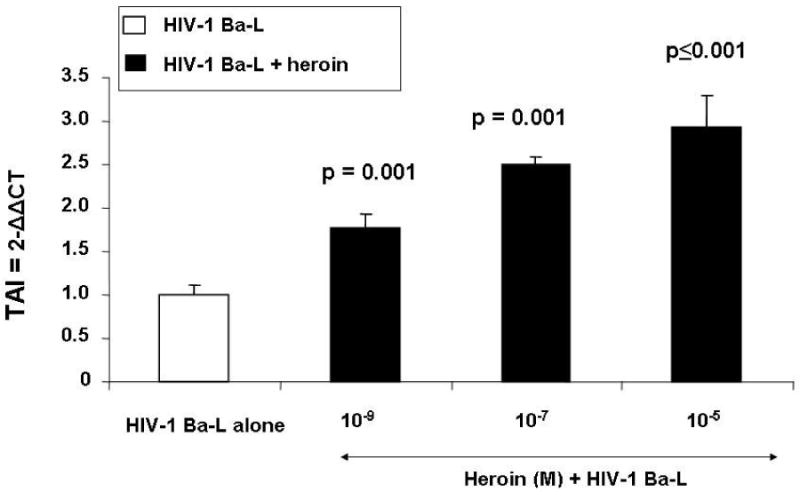
Effect of Heroin on LTR gene expression. NHA were treated with or without heroin (10−5-10−9 M, 24 hr), infected with HIV-1 and then cultured for 48 hr. RNA was extracted and reverse transcribed followed by quantitative, real time Q-PCR using primers specific for the LTR region of the HIV-1 genome. Data are presented as the mean ± SD of 3 independent experiments. Statistical significance was calculated by Students’ ‘ t’ - test
In subsequent experiments, NHA were treated with and without heroin for 24 hr, infected with HIV-1 Ba-L washed and returned to culture for 15 days at which levels of p24 antigen were measured. Data presented in Fig. 2 show the levels of p24 Antigen in the culture supernatants of NHA infected with HIV-1 in the presence or absence of heroin (10−7 M). NHA infected with HIV-1 in the absence of heroin produced 0.22 ng mL−1 of p24 Antigen whereas NHA infected with HIV-1 in the presence of heroin significantly upregulated the production of p24 Antigen (0.91 ng mL−1, p = 0.002). Although NHA showed only low level of infection with HIV-1 which is consistent with other studies[13,14], the production of p24 Antigen was significantly upregulated by heroin and thus supports the LTR amplification as presented in Fig. 1.
Fig. 2.
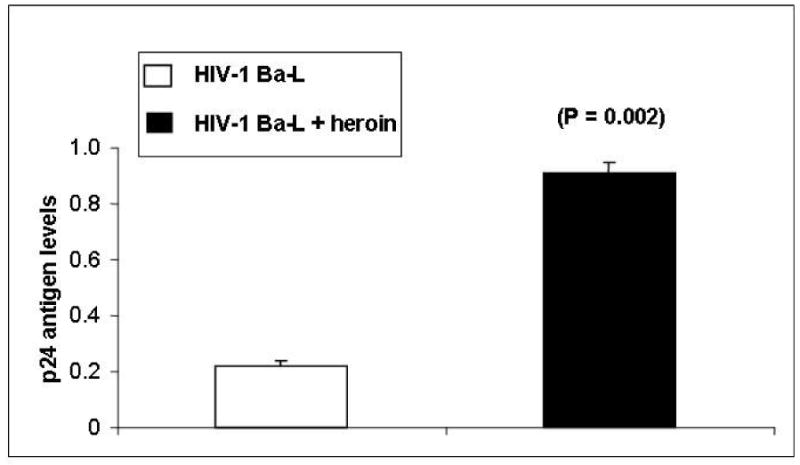
NHA were treated with or without heroin (10−7 M, 24 hr), infected with HIV-1 and then cultured for 15 days. The culture supernatants were quantitated for p24 antigen (ng mL−1) using Retrotek, HIV-1 p24 ELISA from ZeptoMetrix (Cat # 0801111, Buffalo, NY) with a minimum detection range of 1 pg p24 antigen mL−1. Data are presented as the mean ± SD of 3 independent experiments. Statistical significance was calculated by Students’ ‘t’ – test
Heroin differentially induces the expression of several proteins in NHA as measured by proteomic studies
Data presented in Fig. 3 show differences in protein expression between heroin treated and untreated NHA cultures. Several protein spots were identified that were differentially expressed. The protein spots were excised from the gel and identified by HPLC-MS/MS. Table 2 lists proteins that were identified by HPLC-MS/MS. The protein spots that were identified by HPLC-MS/MS that had significantly increased expression levels (Table 3) in NHA treated with heroin compared to control were: protein kinase C, reticulocalbin 1 precursor, reticulocalbin 1, cathepsin D preproprotein, chloride intracellular channel 1, tyrosine 3-monooxgenase/tryptophan 5-monooxgenase activation protein, myosin light chain alkali smooth-muscle isoform and galectin 1. The protein spots that were identified by HPLC-MS/MS that had significantly decreased expression levels (Table 3) in NHA treated with heroin compared to control were: proliferating cell nuclear antigen, tropomyosin 3, proteasome beta 6 subunit, laminin receptor 1, tubulin alpha 6, vimentin, EF hand domain family member D2, Tumor protein D54 (hD54), ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit and ribosomal protein S14.
Fig. 3.

Two-dimensional analyses of heroin-induced differentially expressed proteins in NHA cells. Cells (1 x 106 cells mL−1) were treated with 10−7M heroin for 24 hr. Total protein was isolated and subjected to DIGE analyses as described in methods, (n = 3). Data are shown as a representative 2D SYPRO-Ruby stained gel image of heroin treated NHA cells. Arrows represent differentially expressed proteins. The pH increases from left to right and the molecular mass decreases from the top to the bottom of the gels
Table 2.
NHA were cultured with and without heroin (10−7 M) for 24 hr (n = 3 independent experiments using 3 different NHA cultures). Protein was extracted and subjected to DIGE as described in methods section. Data represent statistically significant differentially expressed proteins (Student’s t-test) that were identified using HPLC-MS/MS. Data are represented as protein name, gene accession number (Gi No.), theoretical isoelectric point (pI), theoretical mass and function.
| Spot # | Protein Name | Accession Number | Theoretical pI | Theoretical mass | Biological function |
|---|---|---|---|---|---|
| 1456 | protein kinase C | gi|4506067| | 4.59 | 731215.07 | kinase |
| 2434 | Myosin light chain alkali, smooth-muscle isoform | gi|16924329| | 4.56 | 16930.05 | Regulatory light chain of myosin |
| 2235 | proliferating cell nuclear antigen | gi|4505641| | 4.57 | 28768.78 | Cell cycle control protein |
| 2331 | Tyrosine 3-monooxgenase/tryptophan 5-monooxgenase activation protein | gi|4507953| | 4.73 | 27745.10 | signal transduction |
| 2212 | tropomyosin 3 | gi|24119203| | 4.75 | 29032.66 | Cytoskeletal protein |
| 2057 | Laminin receptor 1 | gi|9845502| | 4.79 | 32854.08 | Ribosomal subunit |
| 2398 | proteasome beta 6 subunit | gi|23110925| | 4.80 | 25357.72 | Ubiquitin proteasome system protein |
| 1942 | reticulocalbin 1 precursor | gi|4506455| | 4.86 | 38890.00 | Calcium binding protein |
| 1932 | reticulocalbin 1 | gi|4506455| | 4.86 | 38890.00 | Calcium binding protein |
| 1804 | tubulin alpha 6 | gi|14389309| | 4.96 | 49895.33 | Cytoskeletal protein |
| 2231 | chloride intracellular channel 1 | gi|14251209| | 5.09 | 26922.73 | Intracellular ligand gated channel |
| 1160 | Vimentin | gi|2119204| | 5.06 | 53651.68 | Cytoskeletal protein |
| 2417 | EF hand domain family, member D2 | gi|20149675| | 5.15 | 26697.28 | unknown |
| 2275 | Tumor protein D54 (hD54) | gi|20141658| | 5.26 | 22237.73 | regulator of cell proliferation |
| 2502 | Galectin 1 | gi|4504981| | 5.33 | 14715.70 | beta-galactoside-binding protein |
| 2432 | ATP synthase, H+transporting, mitochondrial F1 complex, delta subunit | gi|13325214| | 5.38 | 17489.96 | Transport protein |
| 2223 | cathepsin D preproprotein | gi|4503143| | 6.1 | 44552.22 | aspartic protease |
| 2374 | ribosomal protein S14 | gi|5032051| | 10.07 | 16272.71 | Ribosomal subunit |
Table 3.
NHA were cultured with and without heroin (10−7 M) for 24 hr. Protein was extracted and subjected to DIGE as described in Materials and Methods. Data represent statistically significant up and down regulated spots that were identified using HPLC-MS/MS (n = 3 independent experiments using 3 different NHA cultures, Student’s t-test).
| Spot Number | Protein Name | Volume Ratio | P value |
|---|---|---|---|
| Up Regulated Proteins | |||
| 1456 | protein kinase C | 2.14 | 0.001 |
| 1942 | reticulocalbin 1 precursor | 1.96 | 0.05 |
| 1932 | reticulocalbin 1 | 1.52 | 0.05 |
| 2231 | chloride intracellular channel 1 | 2.58 | 0.001 |
| 2223 | cathepsin D preproprotein | 1.89 | 0.001 |
| 2331 | tyrosine 3-monooxgenase/tryptophan 5-monooxgenasea ctivation protein | 3.85 | 0.0097 |
| 2434 | Myosin light chain alkali, smooth-muscle isoform | 2.62 | 0.024 |
| 2502 | galectin 1 | 1.49 | 0.022 |
| Down Regulated proteins | |||
| 2235 | proliferating cell nuclear antigen | −1.82 | 0.033 |
| 2212 | tropomyosin 3 | −1.49 | 0.0063 |
| 2398 | proteasome beta 6 subunit | −1.45 | 0.01 |
| 2057 | laminin receptor 1 | −1.68 | 0.0029 |
| 1804 | tubulin alpha 6 | −1.58 | 0.0035 |
| 1160 | vimentin | −1.74 | 0.05 |
| 2417 | EF hand domain family member D2 | −2.36 | 0.0056 |
| 2275 | Tumor protein D54 (hD54) | −1.44 | 0.035 |
| 2432 | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | −2.83 | 0.0069 |
| 2374 | ribosomal protein S14 | −1.54 | 0.0051 |
Heroin produces differential modulation of gene expression by NHA
The effect of heroin on mRNA levels of selected proteins was investigated for mRNA analyses by Q-PCR. NHA were cultured for 24 hr with 10−7 M heroin, RNA was extracted, reverse transcribed and cDNA was amplified by Q-PCR using the primers shown in Table 1. Data presented in Fig. 4 show that heroin treatment had no effect on 18s control RNA gene expression which is used as an internal control. However, gene expression for protein kinase C (TAI = 1.89, p = 0.017) and chloride intracellular channel 1 (TAI = 2.52, p = 0.005) were significantly upregulated following treatment of NHA with heroin compared to the untreated control. Further, gene expression for laminin receptor 1 (TAI = 0.48, p ≤ 0.001) and vimentin (TAI = 0.40, p ≤ 0.001) were significantly down regulated in heroin treated NHA compared to the untreated control, confirming our proteomic analyses.
Fig. 4.
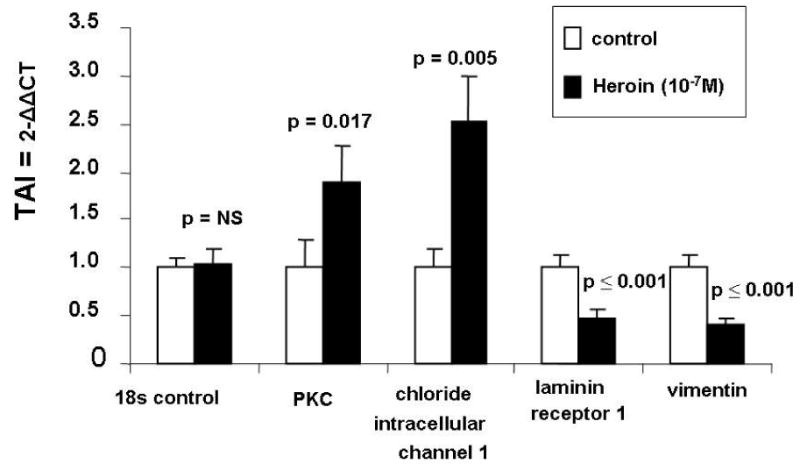
Heroin-induced changes in gene expression. Cells (1 x 106 cells mL−1) were treated with 10−7 M heroin for 24 hr. Total RNA was isolated and subjected to Q-PCR as described in methods. Data are presented as the mean ± SD of 3 independent experiments. Statistical significance was calculated by Students’ ‘ t’ - test
DISCUSSION
Heroin use is postulated to act as a cofactor in the neuropathogenesis of HIV-1 infection. Astrocytes are reported to be susceptible to HIV-1 infection[13–15]. Upon activation, astrocytes release a number of immunoregulatory products or modulate the expression of a number of proteins that foster the immunopathogenesis of HIV-1 infection. However, the role of heroin on HIV-1 replication in NHA has not been elucidated. The current study demonstrates for the first time that pretreatment of NHA with heroin prior to HIV-1 infection potentiates viral replication as demonstrated by a significant increase in LTR–R/U5 gene expression as well as increased p24 antigen levels compared to the non-heroin treated HIV-1 infection control. These studies demonstrate that heroin potentiates HIV-1 replication in NHA and possibly acts as a co-factor in HIV-1 neuropathogenesis. However, the mechanisms whereby heroin potentiates HIV-1 replication in NHA have not been elucidated. We report that heroin modulates the expression of a number of proteins by NHA that foster the neuropathogenesis of HIV-1 infection.
Protein kinase C (PKC) is a family of serine and threonine protein kinases that phosphorylate a wide variety of protein targets and are involved in diverse cellular signaling pathways[33]. The current study demonstrates that heroin upregulates PKC protein expression in NHA. Previous studies have shown that PKC phosphorylates the HIV-1 proteins Gag, Tat and Rev[34–36]. Studies have also demonstrated that Tat-mediated transactivation of the viral LTR promoter requires PKC suggesting that PKC may play a role in the transition of HIV-1 from latency to productive growth[37]. Therefore, increased production of PKC induced by heroin may induce latent HIV-1 infected NHA to become active viral producers.
Chloride channels are proteins that regulate stabilization of cell membrane potential, transepithelial transport, maintenance of intracellular pH and regulation of cell volume[38]. Chloride intracellular channel 1 is localized to the cell nucleus and has both nuclear and plasma membrane chloride ion channel activity[39]. The present study demonstrates that heroin enhances protein expression for chloride intracellular channel 1 in NHA. Previous studies have shown that Gp120 from HIV-1 viral strains JRFL (R5) and IIIB (X4) activate chloride channels which are mediated by CCR5 and CXCR4[40]. The antiviral drugs AZT and acyclovir block the swelling-dependent chloride current and chloride flux in fibroblasts[41]. Therefore, enhanced infection of NHA by HIV-1 may be induced by increased chloride channel activity, however future studies are necessary to determine this effect.
Cathepsin D is a lysosomal aspartyl protease released by several immune cells. Cathepsin D is located in the acid endosomal/lysosomal compartment of immune cells and plays a pivotal role in antigen processing[42,43]. Previous studies showed that polymorphonuclear cells (PMNs) isolated from HIV-1+ patients have significantly higher cathepsin D activity than normal controls and these cells are suggested to play a role in spreading HIV-1 infection to neighboring cells[44]. Further studies have also shown that vaginal secretions contain cathepsin D and exposure of lymphocyte cultures to these secretions increased HIV-1 replication in these cells[45]. Furthermore, human breast milk also enhanced HIV-1 infection of lymphocyte cultures which was inhibited by anti-cathepsin D antibodies or pepstatin A, an inhibitor of cathespin D suggesting that cathepsin D may interact with gp120 to modify the affinity of HIV-1 co-receptors thereby enhancing HIV-1 infectivity[45,46]. The current study demonstrates for the first time that heroin enhances the expression of the preprotein form of cathepsin D in NHA. The preprotein form of cathepsin D is cleaved to produce the active form of cathepsin D. This increase in expression of the preprotein may ultimately lead to an increase in the production of the active form of cathepsin D upon cell activation by HIV-1 in NHA. An increase in cathepsin D activity may therefore increase the susceptibility of NHA to HIV-1.
Galectin-1 is a member of a family of lectins that are defined by a shared amino acid sequence in their carbohydrate recognizing domain and β-galactosidase affinity. Galectin-1 is widely expressed in mammals including the lung, brain, heart, spleen and lymph nodes[47]. Galictin-1 is located both extracellularly and intracellularly. Galectin-1 modulates cell proliferation, apoptosis, cell cycle arrest, cell-matrix adhesion and cell to cell adhesion[47–49]. Previous studies demonstrate that galectin-1 acts as a soluble HIV-1 binding protein that can stabilize virus-cell interactions and promote virus replication in PBMC and CD4+ T cells[50]. The current study demonstrates that heroin upregulates the expression of galectin-1 in NHA perhaps enhancing the susceptibility of these cells to HIV-1 infection by providing a mechanism of virus to cell interactions. Future studies are necessary to determine the role of galectin-1 in NHA infectivity.
Vimentin is an intermediate filament that plays a role in mechanical and biological functions such as cell contractility, migration, stiffening and proliferation[51]. HIV-1 utilizes the host cytoskeletal system for infection and replication. HIV-1 and HIV-2 proteases have been shown to cleave vimentin[52–55]. Furthermore, cleavage of vimentin by HIV-1 proteases is necessary for changes in chromatin organization and distribution induced by HIV-1[56]. In HeLa cells, the HIV-1 viral infectivity factor (Vif) colocalizes with vimentin in perinuclear aggregates[57,58]. Additionally, vimentin expression was shown to be decreased in SVGA-Tat expressing cells[59]. The current study demonstrates that heroin decreases the expression of vimentin in NHA suggesting that heroin can modulate the expression of host proteins associated with viral infection.
In summary, we were able to identify several proteins in NHA that were differentially regulated by heroin which directly or indirectly play a supportive role in the neuropathogenesis of HIV-1 infection. Further studies are needed to investigate the relationships of these proteins in the neuropathogenesis of HIVE.
Acknowledgments
This work was supported in part by NIDA grants 1RO1-DA14218, 1RO1-DA10632, 1RO1-DA12366, 1RO1-DA15628, 1RO1DA02153701, 1F32DA02153501 and the Margaret Duffy and Robert Cameron Troup Memorial Fund of Kaleida Health, Buffalo, NY.
References
- 1.Office of National Drug Control Policy (ONDCP) www.whitehousedrugpolicy.gov/drugfact/heroin/index.html.
- 2.Anonymous. Substance Abuse and Mental Health Services Administration, Results from the 2004 National Survey on Drug Use and Health: National Findings. 2005. [Google Scholar]
- 3.Des JDC, Friedman SR. HIV infection among persons who inject illicit drugs: problems and prospects. J Acquir Immune Defic Syndr. 1998;1:267–273. [PubMed] [Google Scholar]
- 4.Nath A, Avison MJ, Maragos WF, Schmitt FA, Berger JR. Acceleration of HIV dementia with methamphetamine and cocaine. J Neurovirol. 2001;7:66–71. doi: 10.1080/135502801300069737. [DOI] [PubMed] [Google Scholar]
- 5.Nath A, Hauser KF, Wojna V, Booze RM, Maragos W, Prendergast M, Cass W, Turchan JT. Molecular basis for interactions of HIV and drugs of abuse. J Acquir Immune Defic Syndr. 2002;31S2:S62–69. doi: 10.1097/00126334-200210012-00006. [DOI] [PubMed] [Google Scholar]
- 6.Nath A. Human immunodeficiency virus (HIV) proteins in neuropathogenesis of HIV dementia. Infect Dis. 2002;186;(S2):S193–198. doi: 10.1086/344528. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz SA, Nair MP. Current concepts in human immunodeficiency virus infection and AIDS. Clin Diagn Lab Immunol. 1999;6:295–305. doi: 10.1128/cdli.6.3.295-305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Mattson MP, Galey D, Nath A. Molecular and cellular mechanisms of neuronal cell death in HIV dementia. Neurotox Res. 2005;8:119–134. doi: 10.1007/BF03033824. [DOI] [PubMed] [Google Scholar]
- 9.Gendelman HE, Tardieu M, Lipton SA, Bukrinsky MI, Nottet HS. The neuropathogenesis of HIV-1 infection. J Leukoc Biol. 1994;56:389–398. doi: 10.1002/jlb.56.3.389. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nature Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 11.Minagar A, Fujimura R, Shapshak P, Heyes M, Ownby R, Eisdorfer C. The role of macrophage/microglia and astrocytes in the pathogenesis of three neurologic disorders: HIV-associated dementia, Alzheimer disease and multiple sclerosis. J Neurol Sci. 2002;202:13–23. doi: 10.1016/s0022-510x(02)00207-1. [DOI] [PubMed] [Google Scholar]
- 12.Kramer-Hammerle S, Wolff H, Rothenaigner I, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111:194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Brack-Werner R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS. 1999;13:1–22. doi: 10.1097/00002030-199901140-00003. [DOI] [PubMed] [Google Scholar]
- 14.Canki M, Chao W, Thai JN, Potash MJ, Ghorpade A, Volsky DJ. Highly productive infection with pseudotyped human immunodeficiency virus type 1 (HIV-1) indicates no intracellular restrictions to HIV-1 replication in primary human astrocytes. J Virol. 2001;75:7925–7933. doi: 10.1128/JVI.75.17.7925-7933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conant K, Atwood W, Tornatore C, Traub R, Meyers K, Major OE. In vivo and in vitro infection of the astrocyte by HIV-1. Adv Neuroimmunol. 1994;4:287–289. doi: 10.1016/s0960-5428(06)80269-x. [DOI] [PubMed] [Google Scholar]
- 16.Speth C, Dierich MP, Sopper S. HIV-infection of the central nervous system: the tightrope walk of innate immunity. Mol Immunol. 2005;42:213–228. doi: 10.1016/j.molimm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Clarke JN, Lake J, Burrell CJ, Wesselingh SL, Gorry PR, Li P. Novel pathway of human immunodeficiency virus type 1 uptake and release in astrocytes. Virology. 2006 doi: 10.1016/j.virol.2005.12.004. Epub. [DOI] [PubMed] [Google Scholar]
- 18.Dong Y, Benveniste EN. Immune function of astrocytes. GLIA. 2001;36:180–190. doi: 10.1002/glia.1107. [DOI] [PubMed] [Google Scholar]
- 19.Hansson E, Ronnback L. Glial neuronal signaling in the central nervous system. FASEB J. 2003;17:351–348. doi: 10.1096/fj.02-0429rev. [DOI] [PubMed] [Google Scholar]
- 20.Secchiero P, Curreli S, Zella D, Capitani S, Mirandola P, Gallo RC, Zauli G. Engagement of CD28 modulates CXC chemokine receptor 4 surface expression in both resting and CD3-stimulated CD4+ T cells. J Immunol. 2000;164:4018–4024. doi: 10.4049/jimmunol.164.8.4018. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Shively L, LeBon JM, Chang L, Riggs AD, Liu Q, Singer-Sam J. Real-time PCR assay for quantitative mismatch detection. Biotechniques. 2003;34:498–502. doi: 10.2144/03343st01. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan S, Schwartz S, Nair MP. Immunological assays for chemokine detection in in vitro culture of CNS cells. Biol Proced Online. 2003;5:90–102. doi: 10.1251/bpo50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wessel D, Flugge UI. Method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 25. http://www1.amershambiosciences.com/aptrix/upp00919.nsf/Content/Proteomics+DIGE+Protocols.
- 26.Tonge R, Middleton B, Shaw J, Rayner S, Rowlinson R, Pognan F, Young J, Currie I, Hawkins E, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 27.Breci L, Keeler M, Hattrup E, Johnson R, Letarte J, Haynes PA. Comprehensive proteomics in yeast using chromatographic fractionation, gas phase fractionation, protein gel electrophoresis and isoelectric focusing. Proteomics. 2005;5:2018–2028. doi: 10.1002/pmic.200401103. [DOI] [PubMed] [Google Scholar]
- 28.Gharahdaghi F, Weinberg CR, Imai BS, Meagher DA, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Cooper B, Andon NL, Eckert D, Yates JR, Haynes PA. Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J Am Soc Mass Spectrom. 2003;14:736–741. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 30.Wilm M, Houthaeve T, Shevchenko A, Schweigerer L, Breit S, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 31.Haynes PA, Figeys D, Gygi SP, Aebersold R. Proteome analysis: biological assay or data archive? Electrophoresis. 1998;19:1862–1871. doi: 10.1002/elps.1150191104. [DOI] [PubMed] [Google Scholar]
- 32.Andon NL, Koller A, Hollingworth S, Yates JR, Greenland AJ, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.O’Brian CA, Bornmann WG, Chu F, Maxwell DS. Protein kinase Calpha and epsilon small-molecule targeted therapeutics: a new roadmap to two Holy Grails in drug discovery? Expert Rev Anticancer Ther. 2006;6:175–86. doi: 10.1586/14737140.6.2.175. [DOI] [PubMed] [Google Scholar]
- 34.Burnette B, Yu G, RL Felsted. Phosphorylation of HIV-1 gag proteins by protein kinase C. J. Biol. Chem. 1993;268:8698–703. [PubMed] [Google Scholar]
- 35.Fouts DE, Cengel KA, True HL, Celander DW. Site-specific phosphorylation of the human immunodeficiency virus type-1 Rev protein accelerates formation of an efficient RNA-binding conformation. Biochemistry. 1997;36:13256–61322. doi: 10.1021/bi971551d. [DOI] [PubMed] [Google Scholar]
- 36.Holmes AM. In vitro phosphorylation of human immunodeficiency virus type 1 Tat protein by protein kinase C: evidence for the phosphorylation of amino acid residue serine-46. Arch Biochem Biophys. 1996;335:8–12. doi: 10.1006/abbi.1996.0476. [DOI] [PubMed] [Google Scholar]
- 37.Jakobovits A, Rosenthal A, Capon DJ. Trans-activation of HIV-1 LTR-directed gene expression by tat requires protein kinase C. EMBO J. 1990;9:3413. doi: 10.1002/j.1460-2075.1990.tb08223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh KS, Yuspa SH. Intracellular chloride channels: critical mediators of cell viability and potential targets for cancer therapy. Curr Pharm Des. 2005;11:2753–2764. doi: 10.2174/1381612054546806. [DOI] [PubMed] [Google Scholar]
- 39.Jentsch TJ, Neagoe I, Scheel O. CLC chloride channels and transporters. Curr Opin Neurobiol. 2005;15:319–325. doi: 10.1016/j.conb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Liu QH, McManus C, Williams DA, Doms RW, Baribaud F, De Clercq E, Schols D, Collman RG, Kotlikoff MI, Freedman BD. HIV-1 gp120 and chemokines activate ion channels in primary macrophages through CCR5 and CXCR4 stimulation. Proc Natl Acad Sci USA. 2000;97:4832–1837. doi: 10.1073/pnas.090521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gschwentner M, Woll E, Susanna A, Nagl UO, Ritter M, Laich A, Schmarda A, Ellemunter H, Pinggera GM, Huemer H, et al. Antiviral drugs from the nucleoside analog family block volume-activated chloride channels. Mol Med. 1995;1:407–417. [PMC free article] [PubMed] [Google Scholar]
- 42.Riese RJ, Chapman HA. Cathepsins and compartmentalization in antigen presentation. Curr Opin Immunol. 2000;12:107–113. doi: 10.1016/s0952-7915(99)00058-8. [DOI] [PubMed] [Google Scholar]
- 43.Fonteneau JF, Lirvall M, Kavanagh DG, Cover TL, Sanders C, Bhardwaj N, Larsson M. Characterization of the MHC class I cross-presentation pathway for cell-associated antigens by human dendritic cells. Blood. 2003;02:4448–4455. doi: 10.1182/blood-2003-06-1801. [DOI] [PubMed] [Google Scholar]
- 44.PriFouts DE, Cengel KA, True HL, n-Mathieu C, Celander DW, Faure G, Bary V, Kolopp-Sarda MN, Schumacher H, Canton P, May T, Bene MC. Assessment by flow cytometry of peripheral blood leukocyte enzymatic activities in HIV patients. J Immunol Methods. 2001;252:139–146. doi: 10.1016/s0022-1759(01)00348-9. [DOI] [PubMed] [Google Scholar]
- 45.El Messaoudi K, Van Tieghem N, Thiry L, Englert Y, Liesnard C, Moguilevsky N, Bollen A. HIV-1 infectivity and host range modification by cathepsin D present in human vaginal secretions. AIDS. 1999;13:333–339. doi: 10.1097/00002030-199902250-00005. [DOI] [PubMed] [Google Scholar]
- 46.El Messaoudi K, Liesnard C, Thiry LF, Bollen A, Van Tieghem N, Moguilevsky N. A human milk factor susceptible to cathepsin D inhibitors enhances human immunodeficiency virus type 1 infectivity and allows virus entry into a mammary epithelial cell line. J Virol. 2000;74:1004–1007. doi: 10.1128/jvi.74.2.1004-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elola MT, Alberti AF, Chiesa ME, Mordoh J, Fink NE. Galectin-1 receptors in different cell types. J Biomed Sci. 2005;12:13–29. doi: 10.1007/s11373-004-8169-5. [DOI] [PubMed] [Google Scholar]
- 48.Almkvist J, Karlsson A. Galectins as inflammatory mediators. Glycoconj J. 2004;19:575–581. doi: 10.1023/B:GLYC.0000014088.21242.e0. [DOI] [PubMed] [Google Scholar]
- 49.Scott K, Weinberg C. Galectin-1: a bifunctional regulator of cellular proliferation. Glycoconj J. 2004;19:467–477. doi: 10.1023/B:GLYC.0000014076.43288.89. [DOI] [PubMed] [Google Scholar]
- 50.Ouellet M, Pelletier I, Mercier S, Roy J, Bounou S, Sato S, Hirabayashi J, Tremblay MJ. Galectin-1 acts as a soluble host factor that promotes HIV-1 infectivity through stabilization of virus attachment to host cells. J Immunol. 2005;174:4120–4126. doi: 10.4049/jimmunol.174.7.4120. [DOI] [PubMed] [Google Scholar]
- 51.Wang N, Stamenovic D. Mechanics of vimentin intermediate filaments. J Muscle Res Cell Motil. 2002;23:535–540. doi: 10.1023/a:1023470709071. [DOI] [PubMed] [Google Scholar]
- 52.Snasel J, Horejsi M, Shoeman R, Sedlacek J, Hruskova-Heidingsfeldova O, Ruml T, Pichova I. Cleavage of vimentin by different retroviral proteases. Arch Biochem Biophys. 2000;377:241–245. doi: 10.1006/abbi.2000.1776. [DOI] [PubMed] [Google Scholar]
- 53.Honer B, Shoeman RL, Traub P. Human immunodeficiency virus type 1 protease microinjected into cultured human skin fibroblasts cleaves vimentin and affects cytoskeletal and nuclear architecture. J Cell Sci. 1991;00 (Pt 4):799–807. doi: 10.1242/jcs.100.4.799. [DOI] [PubMed] [Google Scholar]
- 54.Honer B, Shoeman RL, Traub P. Degradation of cytoskeletal proteins by the human immunodeficiency virus type 1 protease. Cell Biol Int Rep. 1992;16:603–612. doi: 10.1016/s0309-1651(06)80002-0. [DOI] [PubMed] [Google Scholar]
- 55.Shoeman RL, Kesselmeier C, Mothes E, Traub P. Intermediate filament assembly and stability in vitro: effect and implications of the removal of head and tail domains of vimentin by the human immunodeficiency virus type 1 protease. Cell Biol Int Rep. 1990;14:583–594. doi: 10.1016/0309-1651(90)90038-z. [DOI] [PubMed] [Google Scholar]
- 56.Shoeman RL, Hartig R, Huttermann C, P Traub. Amino-terminal polypeptides of vimentin are responsible for the changes in nuclear architecture associated with human immunodeficiency virus type 1 protease activity in tissue culture cells. Mol Biol Cell. 2001;12:143–154. doi: 10.1091/mbc.12.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karczewski MK, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henzler T, Herrmann H, Harmache A, SuzanM M, Spring H, Panek T, Audoly G, Bosch V. Fully functional, naturally occurring and C-terminally truncated variant human immunodeficiency virus (HIV) Vif does not bind to HIV Gag but influences intermediate filament structure. J Gen Virol. 2001;82(Pt 3):561–573. doi: 10.1099/0022-1317-82-3-561. [DOI] [PubMed] [Google Scholar]
- 59.Pocernich CB, Poon HF, Boyd-Kimball D, Lynn BC, Thongboonkerd V, Calebrese V, Klein JB, Nath A, Butterfield DA. Proteomics analysis of human astrocytes expressing the HIV protein Tat. Brain Res Mol Brain Res. 2005;133:307–316. doi: 10.1016/j.molbrainres.2004.10.023. [DOI] [PubMed] [Google Scholar]


