Abstract
The H19 gene is subject to genomic imprinting because it is methylated and repressed after paternal inheritance and is unmethylated and expressed after maternal inheritance. We recently identified a 1.1-kb control element in the upstream region of the H19 gene that functions as a cis-acting silencer element in Drosophila. Here we investigate the function of this element in mice. We demonstrate that both H19–lacZ and H19–PLAP reporter transgenes can undergo imprinting with repression and hypermethylation after paternal transmission at many integration sites. However, transgenes that were deleted for the 1.1-kb silencer element showed loss of paternal repression, but they did not show marked changes in the paternal methylation of the remaining upstream region. This study demonstrates that the 1.1-kb control element identified in Drosophila is required to silence paternally transmitted H19 minitransgenes in mice.
Genomic imprinting confers different functions on the two parental genomes during development by silencing one allele of each imprinted gene in a parent-of-origin-dependent manner (1–4). This transcriptional repression is brought about by epigenetic modifications that are thought to be erased and re-established during germ cell development. Methylation of the dinucleotide CpG has been extensively studied as a candidate for the imprinting mark, because it is both heritable and associated with transcriptional repression. In support of this model, the inactive paternal H19 allele is hypermethylated with a compacted chromatin structure over an 8-kb region encompassing the body of the gene and −4 kb of upstream sequence (5–9). By contrast, the active maternal allele is unmethylated with an open chromatin configuration. Methylation of the paternal allele is initiated early in the germ line and fully established in sperm (6, 7, 10). This is most pronounced in a region between −2 kb and −4 kb (the differentially methylated domain, henceforth called the DMD), which is resistant to the genome-wide demethylation that occurs during pre-implantation development (11–13). Futhermore, in embryos with the null mutation for the DNA methyltransferase gene (Dnmt1), there was a loss of DNA methylation at the locus and consequent biallelic expression of H19 (14).
A number of studies have used mouse transgenes as a stringent test for the presence of control elements. H19 transgenes with −3.8 kb of upstream sequence (and therefore containing the DMD) were subject to imprinting when present as multicopy loci at random integration sites. This was evidenced by transcriptional repression and hypermethylation after inheritance through the male germ line and demethylation and expression after maternal transmission (7, 15, 16). A larger 130-kb yeast artificial chromosome (YAC) transgene with both Igf2 and H19 genes reliably underwent imprinting as one- to two-copy loci (17). Although imprinting of the smaller H19 transgenes was variable, it nevertheless indicated the presence of a cis-control element within the −3.8-kb region, particularly because transgenes with only −1.8 kb of upstream flank (and no DMD) did not show repression (16). However, separate transgene modifications, by replacement of the H19 gene with the luciferase (luc) reporter gene or the deletion of 701 bp in the 5′ most portion of the first exon, also abrogated repression despite the presence of the −3.8-kb region (15). It was therefore inferred that the H19 transcript or genomic sequence were required for H19 imprinting. However, “knock-in” targeting of endogenous H19 with neomycin (neo) or luc resulted in appropriate imprinting of the replacement prokaryotic gene (18, 19). The reasons for these contradictory results are unclear.
We recently introduced an H19–lacZ reporter transgene into Drosophila to test for conserved function of the −3.8-kb upstream region in an organism that lacks DNA methylation or genomic imprinting. Using a genetic screen, we identified a 1.1-kb region lying between −2.9 and −1.8 kb from the transcriptional start site that acted as a silencer element (20). The location of this silencer region shows a striking overlap with the DMD described above. A similar silencer element was also detected in the Prader–Willi/Angelman syndrome locus (21). These data suggested the intriguing possibility of a mechanistic link between gene silencing in Drosophila and imprinting in mice.
As a first step toward elucidating this relationship, we examined the imprinting of H19 reporter constructs in mice together with the effects of deleting the 1.1-kb control element. This study demonstrates that the 1.1-kb cis element is required for silencing the paternally inherited H19 reporter transgenes but is not required for intiating or maintaining parent-of-origin-specific methylation in the remaining upstream region.
MATERIALS AND METHODS
Transgenes.
Genomic fragments were obtained from a λ2001 mouse 129/Sv library (a gift of A. Smith, Univ. of Edinburgh) and from the cosmid cAH (8). Subclones encompassing −0.25, −3.8, and −10.5 kb of upstream sequence were cloned into pBluescript KS+ (Stratagene) together with the H19 promoter (22). The plasmid pZ (a gift of P. Kastner, College de France), which contains a β-galactosidase (lacZ) reporter with a nuclear localization signal was further modified by the insertion of flanking loxP sites. The resulting reporter fragment was then inserted at a SmaI site just beyond the TATA box in the H19 promoter. HGF20 had a polymorphic H19 genomic fragment (+2041, AccI → SacI) inserted distal to the 3′ loxP site. A 2.2-kb SpeI–BglII fragment containing the downstream enhancers (23) was then inserted immediately 3′ to the poly(A) sequence of the reporter or the H19 sequence.
The H19–PLAP transgene was built in a manner identical to the corresponding −10.5-kb H19–lacZ construct, except that a full-length genomic clone of human placental alkaline phosphatase (PLAP, also known as ALPP) was used as a reporter. This contained introns but not loxP sites and was excised as a 5-kb HindIII–EcoRI fragment from pSV2Apap (24). This was blunt ligated into the DraIII site at +5 bp in exon 1 of H19 and therefore did not disrupt the promoter sequence beyond the TATA box. In addition a 4.2-kb SphI–EcoRV fragment from cAH was inserted into the −10.5-kb flank because the lacZ transgene did not include a 136-bp EcoRI–EcoRI fragment that lies at −4 kb (11).
The silencer deletion for the DEL–H19 transgene was created by BspEI–BamHI digestion and blunt ligation of a plasmid containing the SphI–EcoRV fragment, which was then inserted into H19–PLAP to make DEL–H19. This procedure resulted in a 1177-bp deletion with a 5′ boundary at −2999 bp that was only 81 bp larger than that identified in Drosophila. The BamHI site at the 3′ boundary was preserved.
Generation of Transgenic Mice.
DNA for pronuclear injection was purified away from vector sequences by NotI or NotI/XhoI digestion and agarose gel electrophoresis by using either elution from DE81 paper or dialysis and concentration with Elutip-D columns (Schleicher & Schüll). Fragments were filtered and dialyzed against injection buffer (25) with spin columns or drop dialysis (Millipore). DNA concentration was estimated by agarose gel electrophoresis after serial dilution and comparison to λ HindIII standards (New England Biolabs). Injections were carried out at 1 to 2 ng/ml. Pronuclear injection was performed as described into B6/CBA F2 eggs (25). For the HGF20 lines, similarly purified DNA was co-electroporated with the Pgkneo fragment p770 (a gift of A. Smith) into R1 ES cells (26). G418 selection was started at 24 hr post-electroporation and clones were selected at day 6 and 7. Two low copy number clones were chosen for blastocyst injection (27).
Mouse Strains.
Lines from founders derived by pronuclear injection were maintained on a mixed F1 (C57BL/6 × CBA) background. The ES cell-derived lines were maintained on an inbred 129/Sv background.
Embryo Collection and Reporter Gene Staining.
Embryos were collected at days 11.5 and 13.5 (counting the morning of the vaginal plug as day 0.5) from the uterus and placed in PBS. Whole mount staining with 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-gal) was carried out for day 11.5 embryos as described (17). Day 13.5 embryos were fixed for 1 hr and then cut sagitally on a −20°C cold stage. Fixation was continued for one further hour followed by staining as before. H19–PLAP embryos were fixed for 15–30 min in 4% paraformaldehyde and washed twice in PBS. Endogenous placental alkaline phosphatase (PLAP) was inactivated by incubating the embryos at 72°C for 1 hr. The embryos were allowed to cool, agitated in BM purple alkaline phosphatase substrate (Boehringer Mannheim) at 4°C overnight and post-fixed in 4% paraformaldehyde for 24 hours. All embryos were then washed in PBS and stored in 70% ethanol.
Dehydration, clearing, and embedding were carried out as described (28). Clearing with oil of wintergreen or rapid chloroform extraction was used to prevent decolorization of H19–lacZ embryos. Sections were cut at 8 μm and mounted on polylysine-coated slides. PLAP staining was performed directly on the slide after mounting with BM purple alkaline phosphatase substrate. Sections were counterstained with nuclear fast red or eosin. Embryos and sections were photographed with Ektachrome 160T or Fujichrome 64T film.
Genomic DNA Analysis.
DNA extraction, Southern blotting, and transgene-copy-number characterization were carried out as described (17). Genotyping of offspring and embryos was carried out by PCR (29) after lines had been characterized by Southern analysis. The primers HGFPRO1 (5′-GGCCATGTACTGATTGGTTGACA-3′), HGFPRO2 (5′-GAATTCCGGAAAACTTTATCCAT-3′), H19PLAP1 (5′-TTGGTTGACAGAGTAGGGGC-3′), and H19PLAP2 (5′-GAGCAAAGATCAGGTCAGCC-3′) were used to amplify products spanning the H19 promoter and the 5′ portion of the lacZ (HGFPRO1 + HGFPRO2) or PLAP (H19PLAP1 + H19PLAP2) reporters. Methylation analysis was performed on tissue from hemisected or decapitated day 11.5 and day 13.5 embryos as previously described (17).
RESULTS
Transgenes containing reporters allow comprehensive analysis of the extent of expression within individual tissues and cells in a large number of embryos. We used the lacZ reporter because it has proved an excellent marker of transgene expression. However, its high CpG content could influence epigenetic modifications on the transgenes (30). Therefore, we also used the mammalian gene PLAP (24, 31). H19–lacZ transgenes with −10.5 kb, −3.8 kb and −2.5 kb of flanking sequence were constructed to test the function of the upstream region in imprinting (Fig. 1). The transgene HGF22 with −3.8 kb, of upstream sequence had been previously introduced into Drosophila (20). Twenty-one transgenic lines were obtained, but 9 of the 16 H19–lacZ lines were excluded from the analysis (3 lines failed to transmit the transgene and the remaining 6 lines showed no expression) (Table 1). In addition, four DEL–H19 lines were generated that had a 1.2-kb deletion of the silencer region identified in Drosophila (Fig. 1 and Table 1).
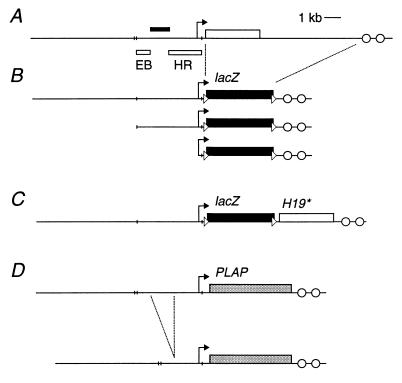
Figure 1.
Structure of the H19 genomic locus and reporter transgenes. (A) The H19 gene is depicted by the open rectangle with the arrow indicating the promoter (internal exon/intron boundaries not shown). The downstream enhancers are represented by open circles. The filled rectangle above the horizontal line indicates the position of the silencer region identified in Drosophila (20). The open rectangles below the line show the probes EB and HE that were used for methylation analysis. The short vertical lines indicate EcoRI sites. (B) H19–lacZ transgenes with −10.5 kb, −3.8 kb, and −0.25 kb of upstream region (HGF23, HGF22, and HGF21, respectively). The filled rectangle represents the bacterial lacZ gene and the dashed lines show the region of the wild-type locus that has been replaced by the reporter. The loxP sites are depicted by triangles. (C) HGF20 transgenes were identical to HGF23 except that a 4-kb H19 genomic fragment containing an artificial polymorphism was inserted downstream of the lacZ poly(A) and 3′ loxP site. (D) H19–PLAP and DEL–H19 transgenes with and without the silencer region. The gray rectangles depict the human PLAP genomic reporter (internal exon/intron boundaries not shown). The dashed lines show the 1.2-kb BspEI–BamHI deletion and the resulting DEL–H19 transgene has −9.3 kb of remaining upstream region.
Table 1.
Summary of H19–lacZ, H19–PLAP, and DEL–H19 transgenic lines
| Upstream region, kb | Transgene | Line | Copy no. | Transgenic embryos, total no. | Imprinting | Differential methylation |
|---|---|---|---|---|---|---|
| −10.5 | H19–PLAP | FL3 | 1 | 26 | No | No |
| FL5 | 7 | 56 | Yes | Yes | ||
| FL8 | 9 | 32 | Yes* | ND | ||
| FL15 | 1 | 43 | No | Yes | ||
| FL16 | 14 | 47 | Yes | Yes | ||
| HGF20 | MLK16.1 | 4 | 22 | Yes | Yes | |
| HGF23 | 3 | 2 | 34 | Yes | ND | |
| 16b | 1 | 41 | Yes | ND | ||
| −3.7 | HGF22 | 3.7RF3y | 6 | 14 | Yes | Yes |
| −0.2 | HGF21 | SCB01 | 5 | 3 | No | NA |
| SCB02 | >50 | 10 | No | NA | ||
| SCB10 | 10 | 5 | No | NA | ||
| −9.3 | DEL–H19 | DEL7 | 1 | 35 | No* | Yes |
| DEL16 | 10 | 23 | No* | Yes | ||
| DEL21 | 5 | 11 | No | Yes | ||
| DEL24 | 4 | 21 | No | Yes |
The copy number, total number of transgenic embryos examined, imprinting as judged by repression on paternal transmission, and differential methylation of the DMD region are indicated for each line. Nine H19–lacZ lines were excluded from the analysis because of failure of transmission or expression. Transgenes from line HGF20-23 each contain the lacZ reporter, but differ in the amount of upstream region. Asterisks in “Imprinting” column mark lines that showed some anomalous repression as described in the text. ND, not done. NA, not applicable.
Expression Pattern of the H19 Reporter Transgenes.
The H19–lacZ and H19–PLAP reporter transgenes contained two previously characterized downstream enhancers (22, 32). Their expression was first examined after maternal transmission in day 11.5 and 13.5 embryos. The overall pattern of expression between lines was highly consistent and was unaffected by the size of the upstream flank or by the choice of the reporter. At day 11.5, strong expression was seen in the liver and endothelium of the gut and blood vessels (Fig. 2). Expression in mesodermal and ectodermal tissues was also demonstrated by staining throughout the sclerotome and in the eye and periventricular tissues of the forebrain. The expression in brain parenchyma was unexpected (33), but in situ hybridization analysis on wild-type day 11.5 and day 13.5 embryos confirmed low-level expression of the endogenous gene in the lens epithelium and forebrain (data not shown). Low levels of expression were detected in the meninges but not in the choroid plexus. At day 13.5, endodermal expression was present throughout the gut, liver, pancreas, kidney, and gonad. Cartilage, which is derived from migrated sclerotome, was strongly stained. No expression was seen in skeletal muscle and only low-level expression was seen in the heart and base of the tongue. lacZ reporter transgenes containing the Igf2 P3 promoter and the identical downstream enhancer fragment showed similar patterns of expression (W. M. Rideout III and J.D.B., unpublished observations). These data confirmed that the two downstream enhancers are not solely endoderm specific, but have additional expression in the mesodermal tissues. Expression analysis from the Igf2–H19 YAC transgene suggests that those for skeletal muscle must lie within its 130-kb domain (17).
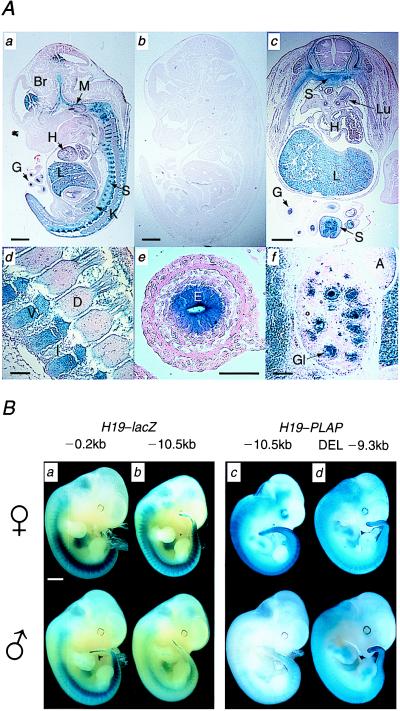
Figure 2.
(A) PLAP reporter gene expression in FL16 day 13.5 embryo after maternal and paternal transmission. (a) Midsagittal section after maternal transmission shows strong staining in liver, sclerotome, brain, meninges, and gut endoderm. Low levels of expression are seen in the heart. (b) Midsagittal section after paternal transmission shows complete repression of PLAP activity. (c) Transverse section through lower thoracic region after maternal transmission. The PLAP staining clearly shows the developing sclerotome, which is migrating dorsally to form the vertebral bodies and ventrally to form the cartilage and bones of the thoracic ribs. There is no expression in the spinal cord or dermatomyotome. Extracoloemic gut shows strong endothelial staining and some expression is also seen in the heart and bronchial epithelium of the lung. (d) Parasagittal view of upper thoracic vertebral column showing the formation of the vertebral bodies and intervertebral discs. (e) Sagittal section through extracoloemic gut showing high-level expression in the endoderm-derived endothelium. (f) Sagittal section through kidney showing PLAP expression in the developing glomeruli, but not in the tubules. Scale bar indicates 1 mm in a, b, and c and 0.1 mm in d, e, and f. A, adrenal gland; Br, brain; D, dorsal root ganglion; E, gut endothelium; G, gut; Gl, glomerulus; H, heart; I, intervertebral disk; L, liver; Lu, lung; K, kidney; M, meninges; S, sclerotome; V, vertebral body. (B) Repression of H19–lacZ and H19–PLAP transgenes requires the silencer region. Representative day-11.5 transgenic embryos stained as whole mount specimens for lacZ or PLAP activity after maternal (Upper) or paternal (Lower) transmission. The size of upstream region in the transgenic line is shown above the panel. (Scale bar indicates 1 mm.) The level of expression of the reporter gene was quantitated byusing Scion Image software (Scion, Frederick, MD). The mean density values for the embryos depicted were normalized against an equivalent embryo with no staining. (a) Line SCB01 shows very slight repression after paternal expression. Maternal expression = 64.41. Paternal expression = 62.04. (b) Paternal transmission of line MLK16.1 shows marked repression of lacZ, most notable in the sclerotome. Maternal expression = 55.16. Paternal expression = 34.64. (c) Line FL8 shows strong repression of PLAP after paternal transmission. Some activity can be seen in the caudal sclerotome. Maternal expression = 98.90. Paternal expression = 22.19. (d) Line DEL24, which does not contain the 1.2-kb silencer, shows minor repression on paternal transmission. Maternal expression = 109.05. Paternal expression = 102.65.
H19 Reporter Transgenes Are Repressed on Paternal Transmission.
To ask whether the transgenes showed evidence of imprinting, we compared the expression of H19–lacZ or H19–PLAP in transgenic embryos after male and female transmission. Embryos from eight lines with −10.5 kb and one line with −3.8 kb of upstream flank were examined at day 11.5 after whole mount staining. Seven of these transgenic lines showed marked repression of the reporters after paternal inheritance (Fig. 2B and Table 1). Indeed, one line, FL16, showed imprinting similar to the endogenous gene with complete repression after paternal inheritance and high-level expression after maternal inheritance (Fig. 2A). However, two lines (FL8 and FL15) also showed repression on maternal inheritance.
The degree of repression upon paternal inheritance was, however, variable between embryo littermates, with a minority of embryos apparently escaping transgene repression altogether. This variability was not unexpected because it has been reported previously by others who used H19 minitransgenes (7, 15, 16). Five lines (MLK16.1, FL5, FL8, FL15, and FL16) were studied in some detail for expression after sequential paternal and maternal transmission. The paternal repression was relieved after transmission through the female germ line, which is consistent with the effect of erasure on the imprinting signal in the paternal germ line. In the two lines where repression was still seen on maternal transmission, FL8 showed repression in 3 of 7 embryos, compared with 21 of 25 embryos after maternal inheritance, whereas in line FL15, 13 of 14 maternally transmitted embryos and 27 of 29 paternal embryos were repressed.
In contrast, no repression was seen in three lines containing only −0.25 kb of upstream flank (thus lacking the DMD and the 1.1-kb region). In the small number of embryos examined, high-level expression after paternal and maternal transmission was seen. Taken together, these data further confirmed that sequences between −3.8 and −0.25 kb contain a key control element and that transgenes containing this region are prone to repression after paternal inheritance but not after maternal inheritance.
DEL–H19 Reporter Transgenes Are Not Repressed on Paternal Transmission.
Having established that the H19 reporter transgenes showed evidence of imprinted expression, we went on to test the function of the 1.1-kb upstream silencer element previously identified in Drosophila. We deleted a 1.2-kb BspEI–BamHI fragment from the −10.5-kb flank of the H19–PLAP reporter transgene, resulting in a partial deletion of the DMD with 840 bp remaining in −9.3 kb of upstream flanking sequence.
Embryos from the four transgenic lines were analyzed on day 11.5 after maternal and paternal transmission. The most striking difference we found was the absence of silencing of the paternally inherited transgene in the DEL–H19 lines compared with those that contained the intact flanking region (Fig. 2B). Only 4 of 90 embryos showed any repression of the reporter. In line DEL7, repression was detected in 2 of 22 embryos after paternal inheritance; repression was also detected in line DEL16 for 1 of 14 paternally transmitted and 1 of 9 maternally transmitted transgenes.
DNA Methylation Analysis of Transgenes.
We then examined methylation of the DMD (−1.8 to −3.8 kb) of the H19 transgenes to ask whether the changes in expression were accompanied by appropriate parent-of-origin methylation changes (Fig. 3). This region shows differential methylation within the H19 locus and has been proposed to confer the parent-of-origin-dependent mark that is required for imprinting (11, 12). DNA was obtained from day-13.5 embryos after maternal and paternal transmission. The methylation status of CfoI sites within the DMD of the H19–PLAP lines was examined by using the probe EB and digestion with SphI, which gave a transgene-specific band of 4477 bp (Fig. 3A). Because the H19–lacZ lines did not have a suitable polymorphism within the DMD, the intensity of the −1.8- to −3.8-kb region was compared after maternal and paternal transmission by using a full-length probe and digestion with EcoRI, BamHI, and CfoI. Six lines (MLK16.1, FL3, FL5, FL8, FL15, and FL16) with −10.5 kb of upstream flank were examined. The single-copy line FL3, which did not exhibit paternal repression, showed no methylation for either maternal and paternal transmission (Fig. 3B). The remaining lines showed relative hypermethylation of transgenes after paternal inheritance when compared with the hypomethylation seen after maternal inheritance (Fig. 3B and data not shown). Although this represented a switch in the epigenetic state of the transgene, it was not as complete as that demonstrated at the locus or on the Igf2–H19 YAC transgene. Incomplete differential methylation has been observed in other H19 mini transgenes (7, 15, 16) and appears to become more complete with successive generations.
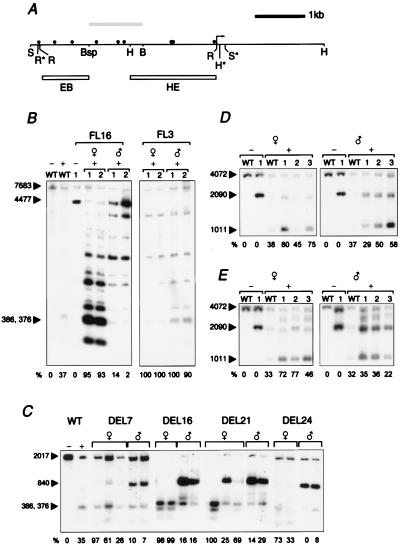
Figure 3.
Methylation of transgenes after maternal and paternal transmission. The relative degree of demethylation is quantiatated for all methylation analyses by measuring the intensity of the fully methylated transgene-specific and fully unmethylated bands with Scion Image (except in wild-type control lanes, where the wild-type bands are compared). Values are corrected for copy number of the transgene and expressed as the percentage unmethylated DNA for each lane. (A) Methylation-sensitive CfoI sites in the 4.0-kb region upstream of the H19 gene. The arrow indicates the position of the H19 promoter (gene body not shown). The lollipops above the line represent CfoI sites. Polymorphic restriction sites within the transgenes are marked with an asterisk. The grey box above the line indicates the silencer region that is deleted in the DEL lines. The boxes below the line indicate the HindIII–EcoRI probe HE and the EcoRI–BspEI probe EB. [Restriction enzyme sites are labeled Bsp (BspEI), B (BamHI), H (HindIII), R (EcoRI), and S (SphI).] (B) Distal methylation in the region of the DMD for lines FL16 and FL3. WT indicates wild-type littermate; transgenic littermates are numbered. The plus sign (+) indicates the addition of CfoI. Maternal and paternal transmission of the transgene is indicated by symbols above the lanes. Ten micrograms of DNA from day-13.5 embryos was digested sequentially with SphI and CfoI. The transgene-specific band of 4477 bp is indicated. Imprinted FL16 transgenes show low levels of methylation on maternal transmission and high levels of methylation on paternal transmission. The bi-allelic-expressing FL3 transgenes show the absence of methylation on maternal and paternal transmission. (C) Deletion of the silencer region does not alter parent-specific methylation. DNA from day-13.5 embryos from all four DEL lines was sequentially digested with BamHI/EcoRI and CfoI, and was then probed with EB. The transgene-specific band of 840 bp is indicated; it contains the 5′-most portion of the DMD. The transgenes were predominantly unmethylated on maternal transmission and methylated on paternal transmission. (D) Proximal methylation for line 3.7RF3y. DNA from day-13.5 embryos was digested sequentially with HindIII and the methylation-sensitive enzyme CfoI, and was then probed with HE. The arrows indicate the wild-type HindIII fragment of 4072 bp and the transgene-specific band of 2090 bp. The transgene band is unmethylated on maternal transmission and partially methylated on paternal transmission. (E) Proximal methylation for line MLK16.1 containing −10.5 kb of upstream region. The digest was carried out as described before. The transgene-specific band shows predominate methylation on paternal transmission and less methylation after maternal transmission.
We next asked whether the apparent lack of repression of the transgene in DEL–H19 lines after paternal inheritance was accompanied by changes in the degree of methylation of the upstream region. The DEL–H19 transgenes contained a polymorphic 840-bp fragment within the DMD that was not removed by the silencer deletion. Using probe EB, we examined the methylation of the two remaining CfoI sites (Fig. 3A). Strikingly, this partial DMD sequence showed appropriate paternal methylation and was predominantly unmethylated on maternal transmission (Fig. 3C). Partial maternal methylation was seen in lines DEL7 and DEL21 (Fig. 3C). This apparent gain of methylation was unexpected and it is unclear whether it was an effect of the silencer deletion or an aberrant methylation imprint as has been shown by other nondeleted H19 minitransgenes (7).
Methylation analysis was also carried out for the promoter-proximal region of the transgenes. It is important to note that this region shows considerable variability in the degree of methylation even at the wild-type locus, and it is therefore less informative than the distal region (11, 12). We first examined methylation proximal to the transgene H19 promoter by using day-13.5 embryonic material from the same six lines with −10.5 kb and one line with −3.8 kb of upstream flank. The H19–lacZ lines contained a 2090-bp HindIII transgene-specific fragment that was probed with HE. For the H19–PLAP lines, the analysis was based on the comparison of intensity differences within the −1.8-kb region after EcoRI, BamHI, and CfoI digestion. As shown in Fig. 3 D and E (and data not shown), digestion with the methylation-sensitive enzyme CfoI showed relative hypermethylation of transgenes after paternal inheritance compared with that shown after maternal inheritance, although there was variation among individuals in the degree of methylation.
The four DEL–H19 lines were then analyzed. Methylation analysis of the proximal H19 region was carried out as done previously for the H19–PLAP lines. Differential methylation was observed in only one line (DEL16) with the remainder of embryos examined showing partial methylation irrespective of parental origin (data not shown).
Taken together, these data showed that both H19–lacZ and H19–PLAP transgenes were predominantly paternally methylated and maternally unmethylated, confirming that reporter minitransgenes can undergo parental-specific methylation. Importantly, removal of the silencer element from the H19–PLAP transgenes abrogated imprinting as judged by transgene expression, but did not have a marked effect on the paternal-specific DNA methylation of the remaining upstream region.
DISCUSSION
Our results demonstrate that H19 transgenes with either the lacZ or PLAP reporter genes can undergo appropriate imprinting at many integration sites. The H19 gene was not required for imprinting to occur; these data differ from previous studies on H19–luc reporter transgenes, which did not show repression or parent-of-origin methylation changes (15). The reasons for this discrepancy are unclear but it is important to emphasize that our analysis allowed us to determine the degree of repression within all expressing tissues in developing embryos. Previous studies have restricted their analysis to expression in neonatal liver (7, 15, 16), which may be a disadvantage because expression from the downstream enhancers shows profound down-regulation shortly after birth (23). We also tested the function in mice of a 1.1-kb cis silencing element that had been independently identified in the DMD region with a genetic screen in Drosophila (20). Deletion of this element abrogated gene silencing previously seen after paternal inheritance of the H19 transgenes. This finding is similar to previously published data that showed a lack of imprinting for H19 transgenes truncated at −1.8 kb (and therefore not containing the 2-kb DMD) (16). Most suprisingly, however, our deletion did not alter the paternal hypermethylation for the remaining DMD, suggesting that the silencer element is not essential for the initiation of DNA methylation in this region.
The imprinting of our H19 minitransgenes was variable, although comparable to results reported previously (7, 15, 16). Our results may be explained in part by the mixed genetic background used in most of the matings; genetic background has been shown to be an important determinant of the basal level of H19 expression (34). However, line MLK16.1 was maintained on the inbred 129/Sv strain and still showed variation. It is also possible that the substitution of the H19 sequence with the reporter genes may have altered the persistence of the repressive signal, because targeted replacement of the H19 gene with luc showed varying paternal repression (19). This imperfect imprinting differs greatly from that of the endogenous locus and the Igf2–H19 transgene (17), which implies that the minitransgenes lack other cis-elements present within the 130-kb YAC that protect imprinting signals from position effects at the integration locus. However, this insulating effect may be simply a function of the larger transgene. It is interesting that the imprinting of minitransgenes seems to be shown best by multiple-copy loci and that one- to two-copy lines are either silenced or do not imprint. The YAC transgenes showed the opposite effect; they were imperfectly imprinted only when integrated in high copy numbers (although this result may have been related to the limiting effects of trans factors). One explanation for the improved imprinting of high-copy number minitransgenes may be augmentation of the repressive signal by the silencing effects of transgene arrays (35, 36).
How could the silencing of paternally inherited H19 transgenes in mice and silencing in Drosophila be linked mechanistically? There is evidence for conservation of silencing mechanisms in eukaryotes, as exemplified by the conserved functions of the homologues of the Polycomb group proteins (37). However, there is no evidence yet to suggest that these proteins are responsible for the silencing/imprinting function in Drosophila and mice. The Drosophila and mouse data together suggest a model in which the initial repression in imprinting is mediated by protein–DNA interaction at the silencer element, which then acts as a template for subsequent methylation that permanently stabilizes silencing (38). However, this model is not supported by our important finding that deletion of the 1.1-kb silencer did not abrogate methylation after paternal inheritance of the transgene. This finding implies that the two processes may function associatively rather than sequentially. To investigate this further and to exclude an artefactual methylation effect from the multicopy transgene arrays, we have deleted the silencer at the endogenous locus (R.A.D., J.D.B., J.F.-X.A., S.C.B., K.J.H., L. Dandolo, and M.A.S., unpublished work). From preliminary studies it is apparent that this mutation acts in a manner similar to the deletion in the transgenic DEL lines. Further studies are needed to identify the trans-acting factors in both Drosophila and mice so that the precise mechanism by which this cis control element functions can be explored.
It is possible that our experiments have identified only part of the control element needed for imprinting, albeit that portion with the most evolutionarily conserved epigenetic function. A partial deletion may explain why DNA methylation is unaffected and also why some repression was observed despite deletion of the silencer. It is very likely that other control elements may reside within the 8-kb locus or immediately upstream of the silencer region (9). Thorvaldsen et al. (39) have recently deleted the major portion of the DMD in mice and showed that the paternal copy of H19 is no longer repressed. However, they also noted the loss of parent-specific methylation of the remaining DMD and the promoter proximal region. In contrast, our targeted deletion of the endogenous 1.1-kb region is in agreement with the methylation data reported in this reporter transgene study. The role of the 5′ portion of the DMD in imprint initiation and maintenance requires further investigation.
We also noted a small but detectable, higher expression level after maternal inheritance of the DEL–H19 and −0.25-kb H19–lacZ lines (which did not contain the DMD or the silencer), when compared with paternal inheritance. This effect has previously been described by using quantitative analysis of Igf2–luc transgenes in mice, which showed higher levels of expression on maternal inheritance (23). The analysis suggests the possibility of an important interaction between the H19 silencer element and the downstream enhancers. The enhancers (or some other regulatory element within the downstream-3′ region) may be required to overcome the activity of the silencer element after passage through the female germ line and could be a crucial step in the imprint switching of H19. This possibility may be tested by using other reporter transgenes that are entirely devoid of the 3′ region.
These experiments demonstrate that a conserved silencing/imprinting function acts at the H19 locus and also suggests that this may be independent of DNA methylation. Further approaches are required to show which cis-elements are sufficient for faithful imprinting and to characterize the trans effectors of the silencing mechanisms. Methods that use the targeted integration of single-copy transgenes into defined genomic loci (40) and modifications of large YAC or bacterial artificial chromosome inserts should improve our understanding of this complex locus.
Acknowledgments
J.D.B. received a Cancer Research Campaign Research Fellowship for a Clinician [CRC]; R.A.D. was funded by a Wellcome Prize Studentship; and M.A.S. was supported by a grant from the Wellcome Trust.
ABBREVIATIONS
- DMD
differentially methylated domain
- PLAP
placental alkaline phosphatase
- YAC
yeast artificial chromosome
References
- 1.Surani M A. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 2.Reik W, Walter J. Curr Opin Genet Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- 3.Ohlsson R, Tycko B, Sapienza C. Trends Genet. 1998;14:435–438. doi: 10.1016/s0168-9525(98)01583-2. [DOI] [PubMed] [Google Scholar]
- 4.Tilghman S M. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 5.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Nature (London) 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 7.Bartolomei M S, Webber A L, Brunkow M E, Tilghman S M. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 8.Koide T, Ainscough J, Wijgerde M, Surani M A. Genomics. 1994;24:1–8. doi: 10.1006/geno.1994.1574. [DOI] [PubMed] [Google Scholar]
- 9.Hark A T, Tilghman S M. Hum Mol Genet. 1998;7:1979–1985. doi: 10.1093/hmg/7.12.1979. [DOI] [PubMed] [Google Scholar]
- 10.Tada T, Tada M, Hilton K, Barton S C, Sado T, Takagi N, Surani M A. Dev Genes Evol. 1998;207:551–561. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- 11.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 12.Tremblay K D, Duran K L, Bartolomei M S. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olek A, Walter J. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 14.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 15.Pfeifer K, Leighton P A, Tilghman S M. Proc Natl Acad Sci USA. 1996;93:13876–13883. doi: 10.1073/pnas.93.24.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elson D A, Bartolomei M S. Mol Cell Biol. 1997;17:309–317. doi: 10.1128/mcb.17.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ainscough J F, Koide T, Tada M, Barton S, Surani M A. Development (Cambridge, UK) 1997;124:3621–3632. doi: 10.1242/dev.124.18.3621. [DOI] [PubMed] [Google Scholar]
- 18.Ripoche M A, Kress C, Poirier F, Dandolo L. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 19.Jones B K, Levorse J M, Tilghman S M. Genes Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyko F, Brenton J D, Surani M A, Paro R. Nat Genet. 1997;16:171–173. doi: 10.1038/ng0697-171. [DOI] [PubMed] [Google Scholar]
- 21.Lyko F, Buiting K, Horsthemke B, Paro R. Proc Natl Acad Sci USA. 1998;95:1698–1702. doi: 10.1073/pnas.95.4.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo-Warren H, Pachnis V, Ingram R S, Tilghman S M. Mol Cell Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward A, Fisher R, Richardson L, Pooler J A, Squire S, Bates P, Shaposhnikov R, Hayward N, Thurston M, Graham C F. Genes Funct. 1997;1:25–36. doi: 10.1046/j.1365-4624.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 24.Henthorn P, Zervos P, Raducha M, Harris H, Kadesch T. Proc Natl Acad Sci USA. 1988;85:6342–6346. doi: 10.1073/pnas.85.17.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen N D, Barton S C, Surani M A H, Reik W. In: Mammalian Development: A Practical Approach. Monk M, editor. Oxford: IRL; 1987. pp. 217–233. [Google Scholar]
- 26.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley A. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: IRL; 1987. pp. 113–151. [Google Scholar]
- 28.Kaufman M H. The Atlas of Mouse Development. London: Academic; 1992. pp. 2–5. [Google Scholar]
- 29.Pomp D, Murray J D, Medrano J F. Mouse Genome. 1991;89:279. [Google Scholar]
- 30.Paldi A, Deltour L, Jami J. Transgenic Res. 1993;2:325–329. doi: 10.1007/BF01976173. [DOI] [PubMed] [Google Scholar]
- 31.DePrimo S E, Stambrook P J, Stringer J R. Transgenic Res. 1996;5:459–466. doi: 10.1007/BF01980211. [DOI] [PubMed] [Google Scholar]
- 32.Brunkow M E, Tilghman S M. Genes Dev. 1991;5:1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- 33.Poirier F, Chan C T, Timmons P M, Robertson E J, Evans M J, Rigby P W J. Development (Cambridge, UK) 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 34.Pachnis V, Belayew A, Tilghman S M. Proc Natl Acad Sci USA. 1984;81:5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrick D, Fiering S, Martin D I, Whitelaw E. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 36.Dorer D R, Henikoff S. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 37.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kass S U, Landsberger N, Wolffe A P. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 39.Thorvaldsen J L, Duran K L, Bartolomei M S. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Araki K, Araki M, Yamamura K. Nucleic Acids Res. 1997;25:868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]