Abstract
Prader-Willi syndrome (PWS) and Angelman syndrome (AS) result from the loss of function of imprinted genes in human chromosome 15q11–q13. The central part of mouse chromosome 7 is homologous to human 15q11–q13, with conservation of both gene order and imprinted features. We report here the characterization of a transgene insertion (Epstein–Barr virus Latent Membrane Protein 2A, LMP2A) into mouse chromosome 7C, which has resulted in mouse models for PWS and AS dependent on the sex of the transmitting parent. Epigenotype (allelic expression and DNA methylation) and fluorescence in situ hybridization analyses indicate that the transgene-induced mutation has generated a complete deletion of the PWS/AS-homologous region but has not deleted flanking loci. Because the intact chromosome 7, opposite the deleted homolog, maintains the correct imprint in somatic cells of PWS and AS mice and establishes the correct imprint in male and female germ cells of AS mice, homologous association and replication asynchrony are not part of the imprinting mechanism. This heritable-deletion mouse model will be particularly useful for the identification of the etiological genes and mechanisms, phenotypic basis, and investigation of therapeutic approaches for PWS.
Keywords: imprinting, epigenetic, failure-to-thrive, methylation, non-Merdeliar
Prader-Willi and Angelman syndromes (PWS and AS) are distinct neurobehavioral disorders caused by a loss of function of imprinted genes in human chromosome 15q11–q13 (1). The most common molecular defect leading to PWS and AS is an identical 4-megabase deletion of the entire imprinted domain (1, 2). However, the deletion is always paternal in origin for PWS and maternal in origin for AS. Maternal uniparental disomy (UPD) is also frequent in PWS, with paternal UPD or mutations in the UBE3A gene occurring rarely in AS patients (1, 3). As expected from the inheritance patterns, UBE3A is imprinted in man and mouse with maternal expression only in specific brain regions (1, 4). In contrast, PWS patients with inheritance consistent with a single gene mutation are not found, suggesting that PWS is a contiguous gene syndrome resulting from the loss of function of several paternally expressed, imprinted genes (1).
Additional molecular complexity in these syndromes involves rare PWS and AS patients with biparental inheritance but an abnormal epigenotype (1). Some of these imprinting mutation patients have a microdeletion located at the 5′ end of the SNURF–SNRPN bicistronic gene in PWS (1, 5, 6) and 30 kilobases (kb) upstream of this for AS (1, 7). These microdeletions define an imprinting center (IC), which is thought to control erasure and resetting of the chromosome 15q11–q13 imprint in the germ line, leading to transmission of the “grandparental” imprint to a child with PWS or AS (1).
The genes in human chromosome 15q11–q13 have homologs in mouse chromosome 7C, with conservation of both gene order and imprinted features (1). Previously, the breeding of mice with balanced translocations was used to generate animals with maternal UPD for 7C, but these mice died within 1 week postnatally from failure-to-thrive (8), whereas paternal UPD for chromosome 7A–C gave a more subtle phenotype characterized by AS-like features and severe late-onset obesity (9). In the latter mice, some phenotypic features may result from UPD for an imprinted region in proximal chromosome 7 (10). This problem has been overcome with the establishment of AS mouse models with null mutations in the Ube3a gene (ref. 4; K. C. Goss, J. C. Schryver, and D. K. Johnson, unpublished data), each with a mild neurological and behavioral phenotype and similar characteristics as AS patients. Finally, a 42-kb deletion of the Snrpn locus resulted in mice with an imprinting mutation and a phenotype equivalent to those with maternal UPD, indicating conservation of the IC mechanism in mice (11).
Here we describe a mouse line in which an Epstein–Barr virus Latent Membrane Protein 2A (LMP2A) transgene insertion has created a deletion of the entire PWS/AS-homologous region. Paternal transmission of the transgene results in loss of paternally derived gene expression in this domain, whereas maternal transmission results in loss of imprinted expression of the AS gene, Ube3a. This PWS mouse model is unique in being genetically sustainable, thereby providing an important resource to dissect the specific genes and mechanisms responsible for PWS. Furthermore, segregation and imprint analyses indicate that homologous association and replication asynchrony are not part of the imprinting mechanism for this imprinted domain.
MATERIALS AND METHODS
Transgenic Mice.
The SacI–KpnI fragment of the Eμ-LMP2A vector (pRL209), in which the Ig heavy chain promoter and enhancer controls the LMP2A gene, was microinjected into (B6 × SJL) F1 single-cell fertilized eggs as described (12). The LMP2A transgenic animals were bred to C57BL/6 mice (The Jackson Laboratory) for over four generations at Northwestern University and subsequently to CD-1 mice (Charles River Breeding Laboratories) for over four generations at Case Western Reserve University. Transgenics were detected by PCR (all PCR primer sequences and conditions here and below are available from the corresponding author). All animal experiments were in accordance with university animal welfare guidelines.
Southern Analyses.
For differential methylation of Snurf–Snrpn, tail genomic DNA was double digested with EcoRI and MluI, electrophoresed on a 0.8% agarose gel, Southern blotted, and hybridized with a PCR-amplified probe from a Snurf–Snrpn exon 1 subclone (ref. 13; see above). Methylation of Zfp127 and Ndn were similarly assayed by using digestion with XbaI and EagI, and hybridization with a 1.3-kb EcoRI–NotI Zfp127 genomic fragment (14) or Ndn genomic PCR probe (see above). For methylation analysis of the LMP2A transgene, DNA was digested with HpaII or BamHI and HpaII, and hybridization was with a 2.0-kb EcoRI fragment from the pRL34 LMP2A cDNA (12). For dosage analyses, DNA was digested with BamHI, EcoRI, or PstI and Southern blots were probed with Herc2 cDNA probes (2), LMP2A as above, or a control probe for a unique gene from chromosome 6A (unpublished data).
Expression Analyses.
Total RNA was extracted from freshly dissected brain or cerebellum with RNAzolB (Cinna/Biotecx Laboratories, Friendswood, TX), of which 5 μg underwent reverse transcription (RT) by using SuperScript II (GIBCO/BRL) with random hexamers as primers, and 0.04 of the RT reaction used for 25-μl PCR amplifications of Zfp127, Ndn, Snurf exons 1–3, Snrpn exons 9–10, Ipw, Ube3a, and Herc2 (see above).
Fluorescence in Situ Hybridization (FISH).
Spleens were dissected from TgPWS/AS(del) mice and wild-type littermates, and slides were prepared by using standard procedures (available from the corresponding author). Slides were hybridized with biotin- or digoxigenin-labeled probe (150 ng) with 1 μg of mouse Cot-1 DNA as competitor and washed by using standard methods, bound probe was detected with fluorescein-labeled anti-digoxigenin and Texas red avidin, and chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI), before mounting slides in antifade solution (Oncor). At least 20 metaphases were analyzed in each experiment. The probes used include Snurf–Snrpn bacterial artificial chromosome (BAC) 397F16 (13); Zfp127 λ44 and λ46 (14); BAC BH6, which maps between Ube3a and Gabrb3 (M. Dhar and D. K. Johnson, personal communication); a 6.9-kb 3′ Herc2 cDNA probe (cDNA coordinates 8, 302–15, 247) (2); BAC 220N6 for Mlsn1 (15); Igf1r genomic subclones of 5.4 kb and 7.2 kb, from a 17-kb λ bacteriophage (16); and the LMP2A cDNA for the transgene array (see above; ref. 12). Because the injected transgene fragment contains a 93-nt overlap with the pBS KS(II) vector but not the pUC18, pCR2.1, or BAC vectors, cDNA and phage subclones were transferred to the pUC18 vector for FISH analysis.
RESULTS
Phenotype of Epstein–Barr Virus LMP2A Transgenic Mice.
Nine lines of transgenic mice with lymphocyte cell lineage expression of LMP2A were generated (12). With the exception of the line described in this study, derived from a female founder (13F′), the other eight independent transgenic lines exhibited no gross developmental phenotype except in lymphocyte cell lineages, which is described elsewhere (12). Only the line from 13F′, which we have named TgPWS/AS(del) based on data presented below, showed an imprinted phenotype by mating and an imprinted epigenotype (see below).
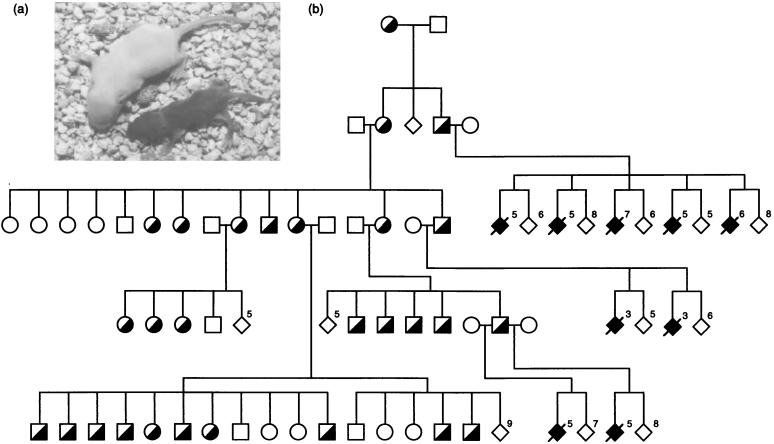
The TgPWS/AS(del) line showed imprinted inheritance, with a lethal phenotype only appearing after paternal, but not maternal, transmission (Fig. 1 a and b). After paternal inheritance of the transgene, all pups appeared normal at birth. However, each day thereafter, the transgenic mice failed to maintain the same growth rate as their wild-type littermates and died within 1 week (Fig. 1a). These mice appeared dehydrated, although milk was observed in their stomachs and survival beyond 48 hours indicates that they were adsorbing some nutrients. They also showed reduced movement compared with their wild-type littermates, and they had irregular respiratory rates. After maternal transmission, all mice were viable and fertile, with no obvious phenotypic effects.
Figure 1.
Imprinted phenotype (a) and inheritance (b) of TgPWS/AS(del) mice. (a) Representative 5-day-old littermates in which the wild-type animal (albino coat color) on left weighs 6.2 g, whereas a PWS pup inheriting the transgene paternally (dark coat color) weighs only 2.3 g. (b) Filled symbols represent the severe failure-to-thrive phenotype seen in transgenic offspring after paternal inheritance only (PWS model), whereas half-filled symbols represent animals inheriting the transgene mutation maternally (a genetic model of AS). This represents only one branch of the complete pedigree. The number of animals of a particular genotype in a given litter are indicated. Diamonds show animals of both or unidentified sex.
Molecular Characterization of Transgenic Animals.
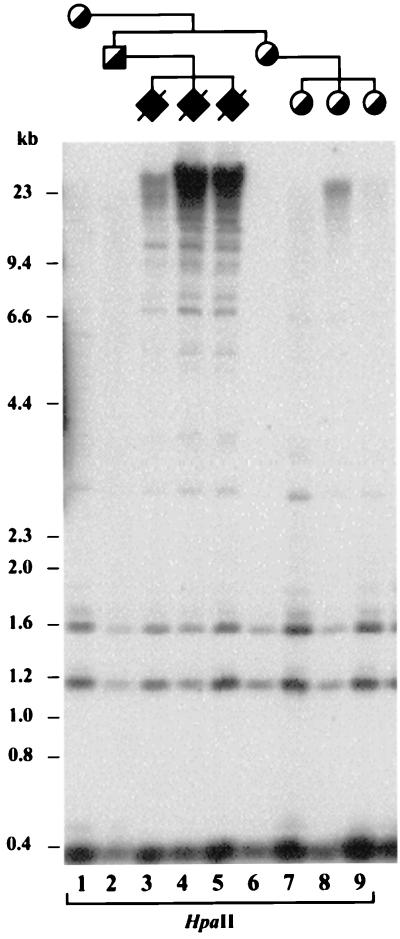
DNA methylation and expression analyses were first performed on the LMP2A transgene to determine whether it was imprinted. Digestion of DNA from transgenic mice with HpaII (Fig. 2) or BamHI and HpaII (data not shown) and hybridization with a transgene probe shows that after paternal inheritance, the transgene is largely methylated, but after maternal inheritance, the transgene is mostly unmethylated, and this pattern is observed through multiple generations. Gene expression analysis of the transgene by RT-PCR showed that LMP2A is expressed after both maternal and paternal inheritance (data not shown).
Figure 2.
DNA methylation analysis of the Epstein–Barr virus LMP2A transgene. Paternal inheritance results in transgene DNA that is largely undigested by the methylation-sensitive restriction enzyme HpaII, whereas maternal inheritance of the transgene results in DNA that is mostly digested by HpaII (the 0.4- and 1.2-kb lower bands are unmethylated). Symbols are as for Fig. 1.
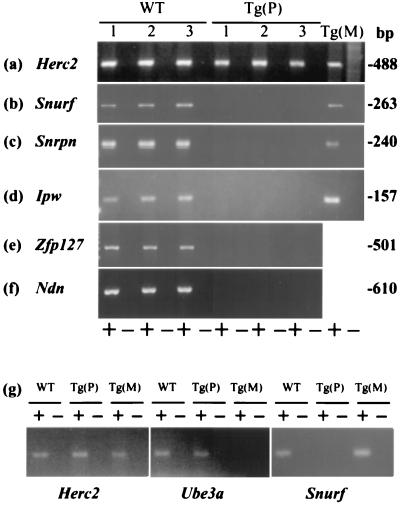
The transgene insertion site was mapped to mouse chromosome 7B/C with FISH by using the transgene and a chromosome 7-specific paint as probes on metaphase chromosomes from transgenic animals (data not shown). No karyotypic abnormality was noted by this analysis. This information, coupled with the imprinted phenotype similar to that seen in mice with maternal UPD or an imprinting mutation for chromosome 7C (8, 11) led us to explore the possibility that the transgene had altered gene expression in the PWS-homologous region. Zfp127 (14), Ndn (17, 18), Snurf (6), Snrpn (13, 19), and Ipw (20) are imprinted genes normally expressed from the paternal allele only, whereas Herc2, a control gene, is not imprinted (2). By using RT-PCR on RNA isolated from brain, it can be seen that animals inheriting the transgene maternally and wild-type littermates show normal expression of the control (Fig. 3a) and each of these imprinted genes (Fig. 3 b–f). In contrast, expression of each of the imprinted genes is undetectable for mice inheriting the transgene paternally (Fig. 3 b–f), although the control gene Herc2 is expressed robustly (Fig. 3a).
Figure 3.
Expression analyses by RT-PCR. The nonimprinted control gene Herc2 is expressed in all animals (a and g). Mice inheriting the transgene paternally [Tg(P)] show a complete lack of expression of any of the five known paternally expressed genes in chromosome 7C (b–g), whereas wild-type littermates (WT) and mice inheriting the transgene maternally [Tg(M)] express each of these imprinted genes (total brain RNA was used for a–f and cerebellum RNA for g). In contrast, the Ube3a gene shows virtually no expression in cerebellum from Tg(M) mice compared with abundant expression in Tg(P) and WT cerebellum (g). A + or − indicates that reverse transcriptase was or was not added to the RT reaction, respectively.
Three alternative models might explain the loss of paternal imprinted gene expression from this chromosomal region. (i) The transgene insertion may be associated with a deletion of all paternally expressed genes in the PWS-homologous region, because the integration of a transgene into an endogenous chromosome can be accompanied by chromosomal rearrangements (21, 22). (ii) The transgene insertion might interfere with the IC mechanism, as for PWS imprinting mutations (1, 5, 11), so that mice inheriting the transgene paternally have two copies of each gene but each copy has a maternal epigenotype. (iii) The transgene insertion might block paternal gene expression through the formation of heterochromatin, as occurs in repeat-induced silencing (23).
A Transgene-Induced Deletion Mouse Model of PWS.
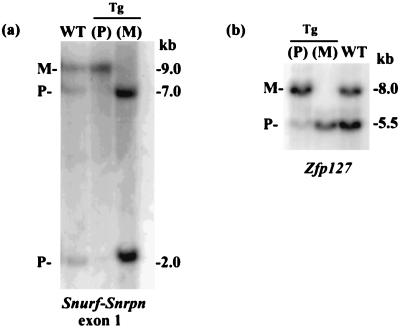
To distinguish between these models, DNA methylation studies were performed for Snurf–Snrpn, Ndn, and Zfp127. The latter two genes map about 1 megabase distal of Snurf–Snrpn and are part of the coordinated imprint switch mediated by the IC (1, 5, 11). Analysis of DNA from wild-type animals, double digested with EcoRI–MluI with a 5′ Snurf probe, yields a methylated 9-kb fragment corresponding to the maternal allele and unmethylated 7-kb and 2-kb fragments corresponding to the paternal allele (13). In mice inheriting the transgene paternally, the unmethylated bands are completely absent (Fig. 4a). Similarly, in mice inheriting the transgene maternally, the methylated band is completely absent (Fig. 4a). Likewise, analyses of DNA digested with XbaI–EagI and hybridized with a 5′ Zfp127 probe (Fig. 4b) or 5′ Ndn probe (data not shown) demonstrate that mice inheriting the transgene paternally only show the methylated band, whereas maternal inheritance of the transgene yields only the unmethylated band. The finding of monoallelic DNA methylation on paternal and maternal transmission for these three loci proves that the transgene insertion had in fact created a large deletion on integration that encompasses each of these paternally expressed genes within chromosome 7C. Had it disrupted the IC imprint switch element alone or silenced paternal gene expression by a position effect or by heterochromatization, these events would have fixed that chromosome with a maternal imprint so that maternal transmission of the transgene would have resulted in a normal, biallelic methylation pattern at nondeleted loci.
Figure 4.
DNA methylation analyses. (a) Digestion of DNA with EcoRI and MluI and hybridization with a Snurf–Snrpn exon 1 probe identifies bands corresponding to the methylated maternal (M) and unmethylated paternal (P) alleles in wild-type mice. Paternal or maternal inheritance of the transgene yields only the maternal or paternal methylation pattern, respectively. (b) Similar results are seen for Zfp127 (XbaI–EagI), mapping 1 megabase distal of Snrpn.
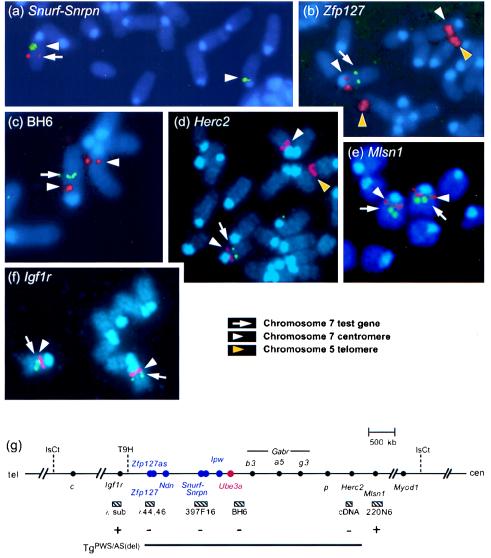
FISH studies were carried out to define the extent of the deletion in the PWS/AS-homologous domain. By using BAC 397F16, which spans the Snurf–Snrpn gene (13) coupled with a probe specific for the centromere of chromosome 7 (24), we confirmed that Snurf–Snrpn was deleted in transgenic mice (Fig. 5a). Use of the centromere probe combined with two overlapping phage spanning Zfp127 (14) confirmed that the deletion extends distally at least as far as this imprinted locus (Fig. 5 b and g). Proximally, hybridization of BAC BH6, which maps just proximal of Ube3a, demonstrated a single set of signals, indicating that Ube3a is also deleted in transgenic mice (Fig. 5 c and g). Similarly, FISH using a 3′ Herc2 cDNA probe, a large gene that maps at the boundary of the PWS/AS domain (2), detects signals on both chromosome 7B/C homologs in wild-type animals (data not shown) but only on a single chromosome 7 in TgPWS/AS(del) mice (Fig. 5d). Consistent with these results, Southern blot analysis with a 614-bp Herc2 3′-untranslated region probe detected band signals of 50% intensity in transgenic mice compared to wild-type littermates, whereas a control probe detected equal intensity signals in all lanes. Similar results were obtained for all 5′ Herc2 cDNA probes (data not shown), indicating that the entire gene is deleted (Fig. 5g).
Figure 5.
Mapping the extent of the deletion by FISH. (a) Metaphase chromosome spreads of mice heterozygous for the transgene were hybridized with a mouse chromosome 7 centromere-specific probe (also hybridizes to telomere of chromosome 5) coupled with the Snurf–Snrpn BAC 397F16. The single set of red signals indicates that this locus is deleted in transgenic animals. Similar FISH analysis demonstrates that the deletion encompasses Zfp127 (green signal) (b), BAC BH6 located just proximal of Ube3a (green signal) (c), and the 3′ end of the Herc2 gene (green signal) (d). In each case, the test probe is deleted from one chromosome 7 homolog. In contrast, two sets of green signals are seen for the Mlsn1 BAC 220N6 (e) and the Igf1r gene (f), indicating that these two loci are intact on the TgPWS/AS(del) chromosome. (g) Summary map showing that the deletion (thick line) associated with transgene insertion encompasses the imprinted and nonimprinted domains of mouse chromosome 7C that are syntenic to the PWS/AS-homologous domain in human 15q11–q13. Loci immediately proximal (Mlsn1), syntenic to human 15q13–q14, and distal (Igf1r), syntenic to human 15q25–qter, are intact. Dashed lines, translocations; cen, centromere; tel, telomere; sub, subclones; hatched boxes, FISH probes; + or −, intact or deleted FISH probe; blue or red, paternal- or maternal-only.
We next examined genes mapped outside the PWS/AS region. The melastatin (Mlsn1) gene maps just proximal to p, and the human homolog maps to 15q13–q14 distal to the PWS/AS deletions (15). In contrast to the results above for genes in the PWS/AS interval, the Mlsn1 BAC 220N6 (15) showed a strong FISH signal on both chromosome 7B/C homologs (Fig. 5e). Whereas no imprinted or physically linked genes are known distal of Zfp127, the next known genetically linked gene is Igf1r (16). FISH analysis with two genomic subclones demonstrated that this locus is also intact on both the TgPWS/AS(del) and normal chromosome 7 homologs (Fig. 5f). Combined, our data demonstrate a deletion of all loci from the mouse PWS/AS-homologous region but not closely flanking proximal or distal genes (summarized in Fig. 5g).
A Deletion Mouse Model of AS.
Because Ube3a is imprinted only in some regions of mouse and human brain (1, 4), we dissected cerebellum and demonstrated that expression of Ube3a is virtually absent in mice with maternal inheritance of the transgene compared with robust levels in both wild-type littermates and mice inheriting the transgene paternally (Fig. 3g). In contrast, paternally expressed (Snurf) and nonimprinted (Herc2) control genes behave as expected (Fig. 3g). These results confirm the FISH studies that showed Ube3a to be deleted.
Breeding of the transgene through females (AS mice) to albino CD-1 mice also allowed an estimation of the genetic distance between the transgene, detected by PCR, and the albino (c) mutation, detected by coat-color phenotype (similar analyses of paternal segregation could not be accurately evaluated because of some early postnatal loss of PWS mice). In this cross, the AS dams are heterozygous at both the transgene and c. Of 76 segregants, 11 recombinants were detected (3 albino AS-transgenic and 8 dark nontransgenic), whereas 65 were nonrecombinant (27 dark AS-transgenic mice and 38 albino nontransgenic). Therefore, the transgene (equivalent to the distal deletion endpoint) maps 14.5 ± 4 centimorgans proximal of c.
DISCUSSION
In this report we have described a mouse model of PWS and AS in which a chromosomal deletion was mediated by insertion of tandem copies of a transgene. This transgenic model is equivalent to the majority (70%) of PWS and AS patients who have a 4-megabase deletion of human chromosome 15q11–q13 (1, 2). Previous mouse models of PWS have maternal UPD or an imprinting mutation (8, 11). All three PWS mouse models share a very similar phenotype, with severe failure-to-thrive and early postnatal lethality, as expected for PWS (1, 6). Although our transgenic PWS mice live several days longer than those with UPD or an imprinting mutation, this may be caused by the genetic background. Maternal transmission of the transgene yields viable and fertile mice that represent a genetic mouse model of AS; these animals are expected to have an equivalent phenotype to Ube3a-null mice, in which the mild neurological and behavioral phenotype is only observed on careful experimental analysis (ref. 4; K. C. Goss, J. C. Schryver, and D. K. Johnson, unpublished data). These phenotypic comparisons provide strong evidence that, other than the imprinted genes as shown here, no other flanking dominant genes are deleted. Because our TgPWS/AS(del) line is maintained in the hemizygous state, recessive genes within the deletion are not unmasked, although the deletion does include several genes associated with recessive phenotypes (Gabrb3, p, Herc2; refs. 1, 2, 25, and 26).
Although the exact breakpoints have not been identified, the deletion is unlikely to be much larger than defined in this work (Fig. 5g) because (i) the deletion is not cytogenetically visible; (ii) the transgene (distal deletion breakpoint) maps 14.5 ± 4 centiMorgans from the albino (c) locus, in perfect agreement with the observed female recombination rate between p and c (27); (iii) the deletion does not include dominant genes that alter the phenotype from that seen in other mouse models of PWS or AS (see above); (iv) the deletion does not include at the proximal end a gene (Mlsn1) whose human ortholog in 15q13–q14 maps distal of the PWS/AS deletions (15); and (v) the deletion does not distally include the next known flanking gene, Igf1r. These considerations further attest to the specificity of the TgPWS/AS(del) mice as an animal model of PWS and AS.
The TgPWS/AS(del) deletion encompasses all genes whose homologs are deleted in PWS and AS patients with a common 4-megabase deletion (1, 2). It is believed that the AS phenotype of patients with UPD is milder than that of deletion patients (28). Some milder clinical features have been described in PWS patients with UPD compared with deletion patients (29), although in both syndromes, the cardinal features are present in all patients. Nevertheless, these studies suggest that hemizygosity for some nonimprinted genes may contribute to the clinical presentation of AS and PWS. The transgene-deletion AS and PWS mouse models described here, compared with mouse models based on UPD (8, 9), imprinting mutations (11), or gene mutation (4), may be useful for investigation of the contribution of nonimprinted genes to the AS and PWS phenotypes.
The transgene-deletion mouse model of PWS and AS represents a powerful model for rapidly determining the imprinted status of any gene mapped to mouse chromosome 7C or conserved human 15q11–q13 genes, including many expected to be identified in this region by the Human Genome Project. Most mouse genes are assessed for imprinting by using F1 crosses between strains or subspecies of mice, but the identification of transcribed and genomic polymorphisms can be a rate-limiting step. In contrast, analysis of imprinting by using tissues from mice with the TgPWS/AS(del) mutation inherited maternally or paternally is simple and rapid, as described here for DNA methylation and gene expression and elsewhere for imprinted protein expression (6).
A curious finding of this study is that the transgene is methylated after paternal but not maternal inheritance, in contrast to the vast majority of transgenes, which are methylated only after maternal inheritance (30–32). Because only one of nine independent transgenic lines demonstrated this imprinted epigenotype, it is unlikely to be due to the sequence-context of the construct often seen for other transgenes (25, 31, 32). One possibility is that the LMP2A transgene inserted into or near an endogenous locus that is normally imprinted and differentially methylated and has thereby acquired a similar pattern of methylation. Although our studies have shown that the transgene has deleted all of the known imprinted genes (1, 6, 14) in the region, the extent of the imprinted domain is unknown distal of Zfp127. In the syntenic human region, the ortholog of the Zfp127 gene is estimated to be no more than 50–200 kb from the end of the imprinted domain (refs. 1 and 2; unpublished data); therefore, additional imprinted genes may be present at the distal end of the mouse imprinted domain that could represent a transgene insertion target. Nevertheless, despite having a methylation imprint, the transgene is not functionally imprinted, probably because of the insertion of multiple copies of the transgene into the chromosome (estimated copy number ≥45; unpublished data), with many copies highly methylated after paternal inheritance but several remaining unmethylated and retaining the ability to be transcribed.
An important implication of the heritability of our deletion mouse model regards the mechanism of germ-line imprint establishment and its maintenance in somatic cells. Several studies have identified replication timing asynchrony (33, 34) and homologous association (35) of imprinted chromosome regions. However, transmission of the TgPWS/AS(del) mutation maternally or paternally does not interfere with proper imprint establishment on the intact chromosome 7 homolog in the germ line of AS mice or with imprint maintenance (in PWS and AS mice) in somatic cells postfertilization. Because the TgPWS/AS(del) mutation deletes the entire imprinted domain, including the IC, homologous association and replication asynchrony cannot be required for either imprint establishment or maintenance. This establishes that the imprinting mechanism for this region is purely cis acting (but does not exclude a role in cis for replication timing).
Our TgPWS/AS(del) line has a unique advantage over other PWS mouse models. Mice with maternal UPD are generated at low frequency and high cost, because of the breeding scheme requiring chromosome missegregation (8). Because of the neonatal lethality of PWS mice, transmission of a targeted IC mutation from male chimeras only occurs until the chimera stops breeding or dies, and transmission from female chimeras has not been observed (11). In contrast, our TgPWS/AS(del) PWS model is maintained by maternal transmission, producing viable and fertile transgenic N1 offspring at 50% frequency that on subsequent paternal transmission give rise to the PWS mouse model phenotype in half of the paternally derived N2 offspring. Thus, this mouse line serves as a sustainable resource for future experiments aimed at detailed pathological analyses and the genetic dissection of the specific genes contributing to the PWS phenotype. This may be accomplished by additional transgenic experiments in which imprinted genes from the PWS region are added back to the TgPWS/AS(del) mice, potentially correcting the failure-to-thrive and any subsequent PWS mouse phenotypes. Genetic and nongenetic therapeutic approaches in PWS that use this PWS deletion mouse model may also be assessed in the future.
Acknowledgments
We thank M. Schaldach and Dr. T. A. Gray for help with animal care; Drs. M. Dhar, D. K. Johnson, and A. Efstratiadis for reagents; and Drs. H. F. Willard and E. M. Rinchik for critical comments. This work was funded by donations from anonymous Prader-Willi Syndrome Association members and grants from the National Institutes of Health (HD31491 and HD36079) to R.D.N. National Institutes of Health Training Grants supported, in part, J.M.G. (GM08613), M.M. (AI07476), and R.G.C. (CA09560). J.D.T. is supported by the U.S. Department of Energy Lawrence Livermore National Laboratory Contract W7405-ENG-48. R.L. is supported by National Cancer Institute Grants CA62234 and CA73507 and is a scholar of the Leukemia Society of America.
ABBREVIATIONS
- AS
Angelman syndrome
- BAC
bacterial artificial chromosome
- FISH
fluorescence in situ hybridization
- IC
imprinting center
- PWS
Prader-Willi syndrome
- RT
reverse transcription
- UPD
uniparental disomy
- kb
kilobase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Nicholls R D, Saitoh S, Horsthemke B. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 2.Ji Y, Walkowicz M J, Buiting K, Johnson D K, Tarvin R, Rinchik E M, Horsthemke B, Stubbs L, Nicholls R D. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- 3.Malzac P, Webber H, Moncla A, Graham J M, Kukolich M, Williams C, Pagon R A, Ramsdell L A, Kishino T, Wagstaff J. Am J Hum Genet. 1998;62:1353–1360. doi: 10.1086/301877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y-h, Armstrong D, Albrecht U, Atkins C M, Noebels J L, Eichele G, Sweatt J D, Beaudet A L. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 5.Ohta T, Gray T A, Rogan P K, Buiting K, Gabriel J M, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll D J, et al. Am J Hum Genet. 1999;64:397–413. doi: 10.1086/302233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray T A, Saitoh S, Nicholls R D. Proc Natl Acad Sci USA. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohta T, Buiting K, Kokkonen H, McCandless S, Heeger S, Driscoll D J, Cassidy S B, Horsthemke B, Nicholls R D. Am J Hum Genet. 1999;64:385–396. doi: 10.1086/302232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cattanach B M, Barr J A, Evans E P, Burtenshaw M, Beechey C V, Leff SE, Brannan C I, Copeland N G, Jenkins N A, Jones J. Nat Genet. 1992;2:270–274. doi: 10.1038/ng1292-270. [DOI] [PubMed] [Google Scholar]
- 9.Cattanach B M, Barr J A, Beechey C V, Martin J, Noebels J, Jones J. Mamm Genome. 1997;8:472–478. doi: 10.1007/s003359900479. [DOI] [PubMed] [Google Scholar]
- 10.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li L-L, Tada M, Suzuki R. Nat Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 11.Yang T, Adamson T E, Resnick J L, Leff S, Wevrick R, Francke U, Jenkins N A, Copeland N G, Brannan C I. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 12.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 13.Gabriel J M, Gray T A, Stubbs L, Saitoh S, Nicholls R D. Mamm Genome. 1998;9:788–793. doi: 10.1007/s003359900868. [DOI] [PubMed] [Google Scholar]
- 14.Jong M T C, Carey A H, Caldwell K, Lau M H, Handel M A, Driscoll D J, Stewart C L, Rinchik E M, Nicholls R D. Hum Mol Genet. 1999;8:795–803. doi: 10.1093/hmg/8.5.795. [DOI] [PubMed] [Google Scholar]
- 15.Hunter J J, Shao J, Smutko J S, Dussault B J, Nagle D L, Woolf E A, Holmgren L M, Moore K J, Shyjan A W. Genomics. 1998;54:116–123. doi: 10.1006/geno.1998.5549. [DOI] [PubMed] [Google Scholar]
- 16.Liu J-P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 17.MacDonald H R, Wevrick R. Hum Mol Genet. 1997;6:1873–1878. doi: 10.1093/hmg/6.11.1873. [DOI] [PubMed] [Google Scholar]
- 18.Jay P, Rougeulle C, Massacrier A, Monda A, Mattei M-G, Malzac P, Roeckel N, Taviaux S, Lefranc J-L B, Cau P, et al. Nat Genet. 1997;17:357–363. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- 19.Leff S E, Brannan C I, Reed M L, Özçelik T, Francke U, Copeland N G, Jenkins N A. Nat Genet. 1992;2:259–264. doi: 10.1038/ng1292-259. [DOI] [PubMed] [Google Scholar]
- 20.Wevrick R, Francke U. Hum Mol Genet. 1997;6:325–332. doi: 10.1093/hmg/6.2.325. [DOI] [PubMed] [Google Scholar]
- 21.Woychik R P, Alagramam K. Int J Dev Biol. 1998;42:1009–1017. [PubMed] [Google Scholar]
- 22.Morgan D, Turnpenny L, Goodship J, Dai W, Majumder K, Matthews L, Gardner A, Schuster G, Vien L, Harrison W, et al. Nat Genet. 1998;20:149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]
- 23.Garrick D, Fiering S, Martin D, Whitelaw E. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 24.Rounds D, Brueckner M, Ward D. Genomics. 1995;29:612–622. doi: 10.1006/geno.1995.9958. [DOI] [PubMed] [Google Scholar]
- 25.Culiat C T, Stubbs L J, Woychik R P, Russell L B, Johnson D K, Rinchik E M. Nat Genet. 1995;11:344–346. doi: 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- 26.Homanics G E, DeLorey T M, Firestone L L, Quinlan J J, Handforth A, Harrison N L, Krasowski M D, Rick C E M, Korpi E R, Mäkelä R, et al. Proc Natl Acad Sci USA. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams R W, Holdener B C, Angel J M, Oakey R, Hunter K W. Mamm Genome. 1998;8:S136–S159. doi: 10.1007/s003359900652. [DOI] [PubMed] [Google Scholar]
- 28.Smith A, Marks R, Haan E, Dixon J, Trent R J. J Med Genet. 1997;54:426–429. doi: 10.1136/jmg.34.5.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cassidy S B, Forsythe M, Heeger S, Nicholls R D, Schork N, Benn P, Schwartz S. Am J Med Genet. 1997;68:433–440. [PubMed] [Google Scholar]
- 30.Reik W, Howlett S K, Surani M A. Development (Cambridge, U.K.) 1990. Suppl., 99–106. [PubMed] [Google Scholar]
- 31.Chaillet J R. Mutat Res. 1994;307:441–449. doi: 10.1016/0027-5107(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 32.Chaillet J R, Bader D S, Leder P. Genes Dev. 1995;9:1177–1187. doi: 10.1101/gad.9.10.1177. [DOI] [PubMed] [Google Scholar]
- 33.Kitsberg D, Selig S, Brandeis M, Simon I, Driscoll D J, Nicholls R D, Cedar H. Nature (London) 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- 34.LaSalle J M, Lalande M. Nat Genet. 1995;9:386–394. doi: 10.1038/ng0495-386. [DOI] [PubMed] [Google Scholar]
- 35.LaSalle J M, Lalande M. Science. 1995;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]