Abstract
The transgenic expression of a toxin gene or a thymidine kinase gene under the control of cell type-specific promoter/enhancer has been shown to be useful for removing a specific cell population in mice. However, this approach requires extensive analysis of the control elements for gene expression in the preparation of the transgenic constructs, and furthermore, the toxin gene might be expressed ectopically because of random integration, resulting in aberrant depletion of unrelated cells. To avoid such difficulties with the transgenic approach, we established a method for the specific depletion of a cell population by replacing a uniquely expressed gene in the population with the diphtheria toxin gene by using homologous recombination. The NKR-P1 gene, a specific cell surface marker of natural killer (NK) cells, was selected as the target gene for depleting NK cells. In chimeric mice reconstituted with embryonic stem cells in which the NKR-P1 gene was replaced by the toxin gene, NKR-P1+ cells were almost completely depleted, and NK cell function was abrogated in the embryonic stem cell-derived lymphoid cells. Other cell lineages developed normally. These results show that all NK cells express NKR-P1, that NKR-P1+ cells do not influence the development of T and B cells, and further, that this technology of cell targeting is a fast and powerful method of generating mice lacking any chosen cell population.
The currently established gene targeting technology has provided significant insights into the in vivo function of particular genes by depleting the gene product in a wide range of biological materials (1–3). In contrast, to understand the in vivo function of a specific cell population, one of the approaches is the depletion of the population instead of a molecule. The application of antibodies specific for a cell surface molecule of a cell population has been widely used to remove this specific population. However, this approach often resulted in depleting the population only to a limited extent (4–6). To deplete a cell population in vivo more specifically, transgenic approaches have been examined, in which either a toxin or a thymidine kinase (TK) gene was expressed under the control of cell type-specific promoter/enhancer (7–13). Diphtheria toxin A fragment (DT-A) and ricin A have been used as toxin genes, and herpes simplex virus 1 TK has often been used for cell depletion in an inducible manner. Depletion of the cell population expressing the toxin or TK gene in such transgenic mice seemed to be successful but was often incomplete. More importantly, there were significant phenotypic differences in several lines of transgenic mice expressing the same transgene. These data suggest that the toxin gene might be expressed ectopically in unrelated tissues/cells because of random integration, resulting in aberrant depletion of unrelated cells. These awkward results were attributed to the transgenic mouse approach, not to the use of a toxin gene because of the following characteristics of the DT gene. The expression of a single molecule of DT-A has been shown to be enough to kill cells (14). Because DT-A does not enter living cells in the absence of the DT-B fragment (15), it has been shown that the bystander effect of DT-A alone on unrelated cells is negligible. In spite of these obvious limitations of the transgenic approach, an improved approach such as the use of homologous recombination to target the toxin or TK gene into the specific site has not been reported. Therefore, we have attempted to establish a method by which a specific cell population expressing a particular gene is depleted by specific integration of the DT gene at the initiation site of the gene by using homologous recombination.
NKR-P1 is a type II membrane glycoprotein with a C type lectin domain in the extracellular region (16, 17). Murine NKR-P1 is known to be a hallmark of surface markers for natural killer (NK) cells (18) and contains three homologous genes: NKR-P1A, NKR-P1B, and NKR-P1C. Conventional T cells, except for a small number of NK1.1+ T cells (19), B cells, and nonlymphoid cells, do not express NKR-P1. The expression of NKR-P1C (CD161, NK1.1) depends on the strain of mice: NK cells from C57BL/6 express NKR-P1C, and those from BALB/c mice do not.
In this study, the NKR-P1 gene was used as the target gene for specific depletion of NK cells by replacing the NKR-P1 gene with the DT gene. As a result, all NK cells and NK function were depleted in the cells derived from embryonic stem (ES) cells. These results show that all NK cells express NKR-P1 and that the approach we used is a successful and convenient method for depleting a specific cell population in vivo.
MATERIALS AND METHODS
Construction of Targeting Vector and Production of NKDT Chimeric Mice.
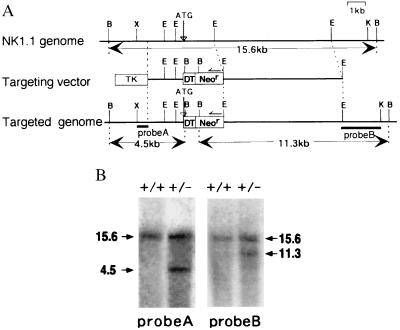
The genomic NK1.1 gene was isolated from a genomic library of C57BL/6 origin by using a full-length NK1.1 cDNA probe. The initiation codon of NK1.1 was replaced by that of DT-A (7) by recombinant PCR. The targeting vector consisted of a long arm [a 7-kilobase (kb) EcoRI fragment containing all exons except exon 1], a short arm (a 2.1-kb PCR product linked to DT-A), DT-A, neor from pKJ2 (20), and an HSV-tk gene from pIC19R/MC1-TK (21), as shown in Fig. 1A.
Figure 1.
Generation of NKDT chimeric mice. (A) A partial map of the NK1.1 gene locus and the targeting construct. The targeting vector in pBluescript II SK (Stratagene) was constructed by replacing the 3′ downstream from the NK1.1 initiation codon with the DT-A (DT) and neor (Neor) genes. Restriction-enzyme-sensitive sites are indicated as follows: B, BamHI; E, EcoRI; K, KpnI; X, XhoI. (B) Southern blot analysis of the represented ES clone with specific homologous recombination. The genomic DNA was digested with BamHI and hybridized with probe A (5′) and probe B (3′) as indicated in A. Molecular sizes (kb) of the bands are indicated at the margin. +/+: wild-type, +/−: heterozygotes.
Because NK1.1 is expressed on NK cells from C57BL/6 but not BALB/c or 129 mice, the ES cell line BL6/III derived from C57BL/6 mice (22) was used for gene targeting. The linearized targeting vector was introduced into BL6/III ES cells by electroporation. After selection in the presence of 150 μg/ml G418 (GIBCO) and 2 μM ganciclovir as well as screening the colonies by PCR and Southern blotting, two clones with specific homologous recombination were obtained. The chimeric mice were generated by microinjection of the ES cells into BALB/c blastocysts (23).
Flow Cytometric Analysis.
Cells of thymus, spleen, peripheral blood, and bone marrow were isolated from 9- to 18-week-old mice, stained with fluorescence- or biotin-conjugated antibodies, and analyzed with a fluorescence-activated cell sorter (FACScan; Becton Dickinson). In some experiments, T cells were enriched by depletion of B cells by using anti-mouse Ig-coupled magnetic beads as described (13). The following antibodies were used in this study: anti-NK1.1 (PK136), anti-pan-NK (DX5), anti-HSA (M1/69), anti-CD4 (RM4-5), anti-CD8α (53-6.7), anti-CD3ɛ (2C11), anti-B220 (RA3-6B2), anti-Mac-1 (M1/70), anti-Gr-1 (RB6-8C5), anti-red cell (TER119), anti-H-2Kb (AF6-88.5), and anti-H-2Dd (34-2-12). These antibodies were all purchased from PharMingen.
NK Cytotoxic Assay.
Splenocytes were depleted of B cells by anti-mouse IgG-coupled magnetic beads as described (13) and stained with H-2Kb and H-2Dd (or NK1.1). H-2Kb+ cells of chimeric mice and NK1.1+ cell-depleted cells of C57BL/6 mice were sorted by FACStar plus. These cells were cultured in the presence of IL-2 (1,000 units/ml) for 5 days to induce NK cells. A cytotoxic assay was carried out as reported (24). Target cells were labeled with PKH-2 green fluorescence dye (Sigma) and mixed with graded numbers of effector cells and incubated for 4 h. Then, the proportion of dead cells among PKH-2-stained cells was determined by propidium iodide staining and flow cytometric analysis.
T Cell Proliferation Assay.
FACS-sorted T cells (n = 2 × 104) were stimulated with 10 μg/ml of immobilized anti-TCRβ mAb (H57-597) for 48 h and pulsed with 18.5 mBq of [3H]thymidine. The [3H]thymidine incorporated in the cells was measured with a liquid scintillation counter.
RESULTS AND DISCUSSION
We isolated an ≈16-kb genomic DNA fragment containing the whole NKR-P1C cDNA from a C57BL/6 genomic library. A targeting vector was constructed as shown in Fig. 1A. The NK1.1 gene was replaced with DT-A gene at its initiation codon. Because NK1.1 is expressed on NK cells from C57BL/6 mice but not on those from 129 mice, we used the BL6/III ES cell line (22) derived from C57BL/6 mice to generate targeted ES cells. Two independent ES clones possessing specific homologous recombination in one allele of the NK1.1 gene were obtained (Fig. 1B). Two clones of the ES cells in which the NKR-P1 gene was replaced with the DT-A gene (NKDT ES cells) and two parental ES clones (control ES cells) were used for injection into BALB/c blastocysts, and 24 chimeric mice (17 and 7 mice, respectively, for each ES clone) were generated and analyzed. Because both NKDT ES clones showed similar results, data from only one clone are presented here.
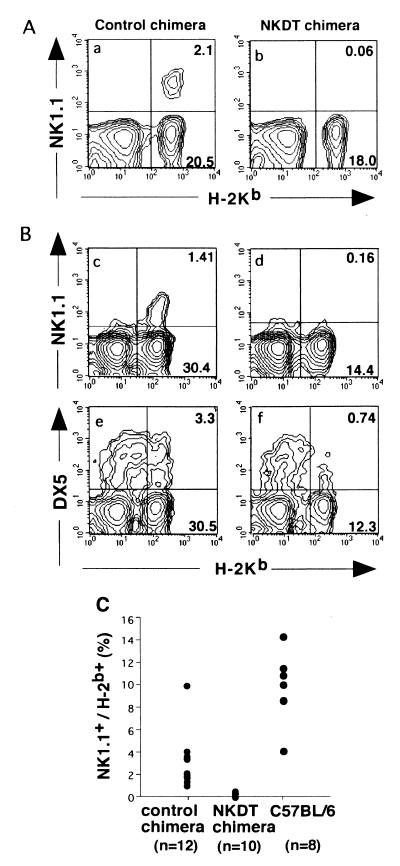
H-2 markers were used to distinguish ES-derived cells from cells with a host origin from chimeric mice. Lymphoid cells derived from ES cells express H-2b, whereas those from host cells express H-2d; thus, we analyzed the generation, cell surface expression, and function of the cells expressing H-2Kb. When peripheral blood cells from chimeric mice were analyzed, 2–30% of lymphoid cells expressed H-2Kb (Fig. 2C), and the proportion of H-2Kb-positive cells almost correlated with the chimerism as judged from hair color (data not shown). Then, to examine whether the toxin-mediated depletion of NK1.1+ cells occurred, the expression of NK1.1 on H-2Kb-expressing cells was analyzed.
Figure 2.
FACS profiles of NK cells in chimeric mice. Peripheral blood cells (A) and splenocytes (B) from chimeric mice reconstituted with NKDT ES cells (NKDT chimera) and control ES cells (control chimera) were stained with PK136 (anti-NK1.1) or DX5 (anti-NK cells) and anti-H-2Kb and anti-HSA mAbs. Expression of H-2Kb and NK1.1 in HSA− cells was shown. NK1.1 expression was analyzed in all 24 chimeric mice. (C) Proportion of NK1.1+ cells in peripheral blood cells derived from ES cells in NKDT and control chimeric mice as well as wild-type C57BL/6 mice.
Although a significant proportion of NK1.1+ cells was detected in control ES-derived cells, NK1.1+ cells were not detectable at all in NKDT ES-derived H-2Kb+ cells in both peripheral blood (Fig. 2A) and spleen (Fig. 2B). Fig. 2C summarizes the proportion of NK1.1+ cells among H-2Kb+ cells in each mouse. The proportion of NK1.1+ cells derived from NKDT ES cells was significantly lower than that from control ES cells. To exclude the possibility that the NKDT chimera just abolished the expression of NK1.1 and that the NK cells remained without NK1.1 expression, we analyzed the expression of DX5, another widely used marker for NK cells (25), although its expression is not completely exclusive for NK cells. DX5+ cells were also strongly reduced in splenocytes derived from NKDT ES cells (Fig. 2B). Taken together, these results indicate that the failure of NK1.1+ cells was caused, not by the unexpected impairment of NK1.1 gene expression by the replacement with DT-A in one allele, but by the removal of NK1.1+ cells by the expression of DT-A.
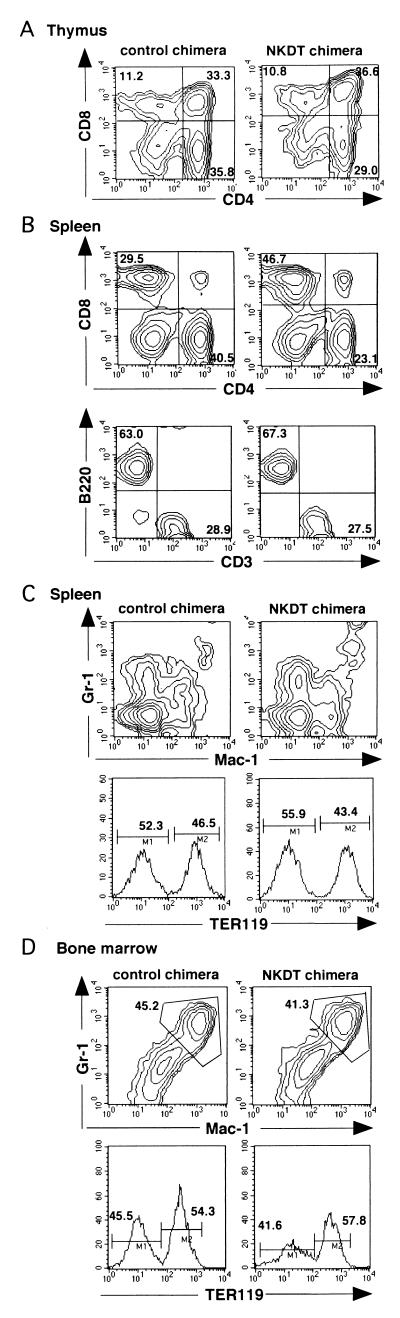
We next determined the effect of the DT-A gene inserted into the NKR-P1 gene for the development of cell populations other than NK cells. When CD4 and CD8 expressions on thymocytes were analyzed, no significant difference was observed between cells derived from NKDT ES cells and control ES cells (Fig. 3A).In contrast to thymocytes, the ratio of CD4+ vs. CD8+ T cells was slightly reduced in splenic T cells derived from NKDT ES cells compared with the ratio from control ES cells, in spite of the fact that the ratio of T to B cells was not altered (Fig. 3B). Furthermore, the developments of the macrophage/granulocyte lineage detected by the expression of Mac-1 and Gr-1 and the developments of the erythrocyte lineage detected by TER119 expression were almost the same in spleen and bone marrow (Fig. 3 C and D). These results show that the effect of the replacement of NK1.1 and the expression of DT-A were restricted in the depletion of cells expressing NK1.1 and did not induce any apparently aberrant deletion of other cells.
Figure 3.
Appearance of hematopoietic cells from chimeric mice. (A and B) Generation of T and B lymphocytes in chimeric mice. Thymocytes (A) and B cell-depleted T cell-enriched splenocytes (B, Upper) from chimeric mice were stained with anti-CD4 and anti-CD8 mAbs, whereas whole splenocytes were stained with anti-CD3 and anti-CD45 (B220) mAbs (B, Lower) in the presence of anti-H-2Kb mAb. Expression of CD4 and CD8 or CD3 and CD45 in H-2Kb+ cells is shown. (C) Appearance of macrophages/granulocytes and red cells in chimeric mice. Splenocytes and bone marrow cells were stained with anti-Gr-1 and anti-Mac-1 mAbs or with TER119 mAb (anti-red cell). Expression of Gr-1 and Mac-1 or TER119 in H-2Kb+ cells is shown. These FACS profiles represent the analyses of the four mice showing high chimerism among the nine mice analyzed.
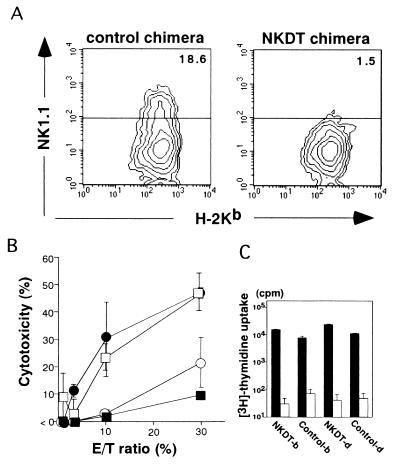
Next, we investigated whether NK cells were depleted functionally by analyzing the NK cell cytotoxic activity of NKDT ES-derived cells. These cells were purified from splenocytes of NKDT and control chimeric mice by flow cytometry after staining for H-2Kb and Dd and expanded for 5 days in the presence of IL-2. Normal C57BL/6 splenocytes as a positive control and NK1.1+ cell-depleted C57BL/6 splenocytes as a negative control were also prepared by cell sorting. Fig. 3A shows the NK1.1 expression on H-2b+ cells derived from ES cells after a 5-day culture. Although both control and NKDT ES-derived cells contained similar numbers of H-2Kb cells, only control ES-derived cells—not NKDT ES-derived cells—contained NK1.1+ cells (Fig. 4A). The cytotoxicity of these cells against an NK-sensitive target cell line, YAC-1, was then analyzed (Fig. 4B). Cells derived from control ES cells showed cytotoxicity comparable to that of cells from normal C57BL/6 mice. In contrast, the cytotoxicity by NKDT ES-derived cells was mostly background, the same level observed with NK1.1+ cell-depleted splenocytes. The observation that FACS-analyzed, NKDT-derived H-2b+ cells had T cell proliferation similar to other control cells upon anti-TCRβ Ab-stimulation (Fig. 4C) proved that all sorted cells are functional and that the failure of natural killing was caused by the specific lack of NK cells. The results of both cell staining and functional analysis indicate that NK1.1+ cells were depleted completely by the homologous recombination of the NKDT gene in the NKR-P1 gene locus.
Figure 4.
NK function of ES-derived splenocytes from chimeric mice. (A) Generation of NK cells from NKDT and control chimeric mice. ES-derived H-2b-positive splenocytes were isolated by flow cytometry and cultured for 5 days in the presence of 1,000 units/ml IL-2. Cultured cells were stained with anti-NK1.1 mAb and anti-H-2Kb mAbs, and the expressions of NK1.1 and H-2Kb are shown. (B) NK activity of cultured splenocytes from chimeric mice. H-2Kb-positive splenocytes cultured as described above from NKDT chimeric (■), control chimeric (□), normal (●), and NK1.1+ cell-depleted (○) splenocytes. Graded numbers of these effector cells were mixed with YAC-1 target cells at the indicated effector-to-target cell ratios, and cytotoxicity was measured as described in Materials and Methods. Data are presented as the means ± SD from experiments performed in triplicate. (C) The proliferative response of FACS-analyzed T cells from chimeric mice. H-2b+ and H-2d+ cells from lymph nodes of both control and NKDT chimeric mice were stimulated with immobilized anti-TCRβ mAb (black bars) or not (open bars); [3H]thymidine was pulsed, and the incorporated [3H]thymidine was measured. Data are presented as means ± SD from experiments performed in triplicate. NK function was analyzed in six individual NKDT mice. Data represent analyses of mice with high chimerism.
In the present study, we successfully demonstrated the generation of mice lacking a particular cell population by replacing the uniquely expressed gene in this population with the DT-A gene. Analysis of various hematopoietic lineage cells indicated that only NK cells were depleted, and the development and function of all other types of cells among the cells differentiated from NKDT ES cells remained intact. Because NK cells and NK function were abrogated completely, our results showed that most NK cells were NK1.1+ and that there were likely no NK1.1− NK cells, consistent with previous data on NK cell depletion by anti-NK1.1 Ab injection (5). Recently, it was suggested that NK1.1+ cells may be important precursor cells for conventional T cells, just as they are for NK cells (26). However, our data clearly showed that such NK1.1+ cells do not play a crucial role in T cell development, because thymocytes and T cells were functionally developed in the complete absence of NK1.1+ cells. The only significant difference we observed was the ratio of CD4+ vs. CD8+ splenic T cells. Although the exact reason for this alteration is unclear, NK cells or NK1.1+ T cells may have some regulatory function for the generation of mature CD4/CD8 T cells. A recent report suggested a possible NK function in the regulation of CD4+ T cell development (27). Further analysis will be required to determine the developmental relationship between NK1.1+ cells and NK1.1− conventional T cells.
This technology that enables site-specific replacement of a toxin gene by homologous recombination provides three major advantages over other cell-depletion methods. First, the technology overcomes the previous problem with toxin-transgenic mice in which the ectopic expression of toxin might result in unexpected effects. Second, the technology does not require the extensive characterization of the promoter/enhancer of the gene. Third, because most genes do not show allelic exclusion, the result of the toxin expression is obvious in chimeric mice or heterozygous mice, precluding the need to generate germ-line-transmitted homozygous mice. Although it has been shown recently that some genes, such as IL-2, have allelic exclusion (28), our results indicated that NKR-P1 is not allelically excluded. Furthermore, this method is able to distinguish whether the gene, under allelic exclusion, is strictly controlled or not.
Therefore, this technology—cell targeting—seems to be a convenient and reliable method for generating mice lacking any specific cell population. It is important to note that the neor gene was inserted together with the DT-A gene into the present targeting construct and remained in the targeted allele. We cannot rule out the possibility that the expression of DT-A could be affected in the presence of the neomycin-resistant (neor) gene, as some unexpected effects caused by the insertion of the neor gene have been reported (29, 30). Accordingly, the present technology can be improved by removing the neor gene by Cre/lox P-mediated recombination to assure the physiological expression of the DT-A gene. Furthermore, when cell-targeting technology is applied to inducible genes, it might be possible to induce specific cell depletion in a developmental stage-specific or stimulation-dependent fashion. Because the TK gene has been used successfully for transgenic mice with inducible, albeit less than complete (10, 13), cell depletion, the TK gene can also be used for conditional depletion of a specific cell population in a cell-targeting system.
Acknowledgments
We thank Dr. S. Y. Park, Dr. S. Okada, and Dr. H. Ohno for experimental help and discussion, Ms. M. Sakuma and Ms. R. Shiina for technical assistance, and Ms. H. Yamaguchi for secretarial assistance. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture, Japan.
ABBREVIATIONS
- NK
natural killer
- DT
diphtheria toxin
- DT-A
diphtheria toxin A chain
- TK
thymidine kinase
- ES
embryonic stem
- kb
kilobase
- FACS
fluorescence-activated cell sorter
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Frohman M A, Martin G R. Cell. 1989;56:145–147. doi: 10.1016/0092-8674(89)90887-8. [DOI] [PubMed] [Google Scholar]
- 2.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn R, Schwenk F. Curr Opin Immunol. 1997;9:183–188. doi: 10.1016/s0952-7915(97)80133-1. [DOI] [PubMed] [Google Scholar]
- 4.Cobbold S P, Jayasuriya A, Nash A, Prospero T D, Waldmann H. Nature (London) 1984;312:548–551. doi: 10.1038/312548a0. [DOI] [PubMed] [Google Scholar]
- 5.Koo G C, Dumont F J, Tutt M, Hackett J, Jr, Kumar V. J Immunol. 1986;137:3742–3747. [PubMed] [Google Scholar]
- 6.Ritz J, Schlossman S F. Blood. 1982;59:1–11. [PubMed] [Google Scholar]
- 7.Palmiter R D, Behringer R R, Quaife C J, Maxwell F, Maxwell I H, Brinster R L. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 8.Breitman M L, Clapoff S, Rossant J, Tsui L C, Glode L M, Maxwell I H, Bernstein A. Science. 1987;238:1563–1565. doi: 10.1126/science.3685993. [DOI] [PubMed] [Google Scholar]
- 9.Landel C P, Zhao J, Bok D, Evans G A. Genes Dev. 1988;2:1168–1178. doi: 10.1101/gad.2.9.1168. [DOI] [PubMed] [Google Scholar]
- 10.Borrelli E, Heyman R, Hsi M, Evans R M. Proc Natl Acad Sci USA. 1988;85:7572–7576. doi: 10.1073/pnas.85.20.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyman R A, Borrelli E, Lesley J, Anderson D, Richman D D, Baird S M, Hyman R, Evans R M. Proc Natl Acad Sci USA. 1989;86:2698–2702. doi: 10.1073/pnas.86.8.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minasi L E, Kamogawa Y, Carding S, Bottomly K, Flavell R A. J Exp Med. 1993;177:1451–1459. doi: 10.1084/jem.177.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamogawa Y, Minasi L A, Carding S R, Bottomly K, Flavell R A. Cell. 1993;75:985–995. doi: 10.1016/0092-8674(93)90542-x. [DOI] [PubMed] [Google Scholar]
- 14.Yamaizumi M, Mekada E, Uchida T, Okada Y. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 15.Pappenheimer A M., Jr Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 16.Giorda R, Rudert W A, Vavassori C, Chambers W H, Hiserodt J C, Trucco M. Science. 1990;249:1298–1300. doi: 10.1126/science.2399464. [DOI] [PubMed] [Google Scholar]
- 17.Lanier L L, Chang C, Phillips J H. J Immunol. 1994;153:2417–2428. [PubMed] [Google Scholar]
- 18.Hackett J, Jr, Tutt M, Lipscomb M, Bennett M, Koo G, Kumar V. J Immunol. 1986;136:3124–3131. [PubMed] [Google Scholar]
- 19.Arase H, Arase N, Ogasawara K, Good R A, Onoe K. Proc Natl Acad Sci USA. 1992;89:6506–6510. doi: 10.1073/pnas.89.14.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boer P H, Potten H, Adra C N, Jardine K, Mullhofer G, McBurney M W. Biochem Genet. 1990;28:299–308. doi: 10.1007/BF02401419. [DOI] [PubMed] [Google Scholar]
- 21.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 22.Ledermann B, Burki K. Exp Cell Res. 1991;197:254–258. doi: 10.1016/0014-4827(91)90430-3. [DOI] [PubMed] [Google Scholar]
- 23.Park S Y, Ueda S, Ohno H, Hamano Y, Tanaka M, Shiratori T, Yamazaki T, Arase H, Arase N, Karasawa A, et al. J Clin Invest. 1998;102:1229–1238. doi: 10.1172/JCI3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arase H, Arase N, Saito T. J Exp Med. 1995;181:1235–1238. doi: 10.1084/jem.181.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore T A, von Freeden-Jeffry U, Murray R, Zlotnik A. J Immunol. 1996;157:2366–2373. [PubMed] [Google Scholar]
- 26.Carlyle J R, Zuniga-Pflucker J C. Immunity. 1998;9:187–197. doi: 10.1016/s1074-7613(00)80601-9. [DOI] [PubMed] [Google Scholar]
- 27.Fort M M, Leach M W, Rennick D M. J Immunol. 1998;161:3256–3261. [PubMed] [Google Scholar]
- 28.Hollander G A, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Simpson S, Terhorst C, Wishart W, Golan D E, Bhan A K, et al. Science. 1998;279:2118–2121. doi: 10.1126/science.279.5359.2118. [DOI] [PubMed] [Google Scholar]
- 29.Olson E N, Arnold H H, Rigby P W, Wold B J. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 30.Shivdasani R A, Fujiwara Y, McDevitt M A, Orkin S H. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]