Abstract
ATP hydrolysis and polypeptide binding, the two key activities of Hsp70 molecular chaperones, are inherent properties of different domains of the protein. The coupling of these two activities is critical because the bound nucleotide determines, in part, the affinity of Hsp70s for protein substrate. In addition, cochaperones of the Hsp40 (DnaJ) class, which stimulate Hsp70 ATPase activity, have been proposed to play an important role in promoting efficient Hsp70 substrate binding. Because little is understood about this functional interaction between domains of Hsp70s, we investigated mutations in the region encoding the ATPase domain that acted as intragenic suppressors of a lethal mutation (I485N) mapping to the peptide-binding domain of the mitochondrial Hsp70 Ssc1. Analogous amino acid substitution in the ATPase domain of the Escherichia coli Hsp70 DnaK had a similar intragenic suppressive effect on the corresponding I462T temperature-sensitive peptide-binding domain mutation. I462T protein had a normal basal ATPase activity and was capable of nucleotide-dependent conformation changes. However, the reduced affinity of I462T for substrate peptide (and DnaJ) is likely responsible for the inability of I462T to function in vivo. The suppressor mutation (D79A) appears to partly alleviate the defect in DnaJ ATPase stimulation caused by I462T, suggesting that alteration in the interaction with DnaJ may alter the chaperone cycle to allow productive interaction with polypeptide substrates. Preservation of the intragenic suppression phenotypes between eukaryotic mitochondrial and bacterial Hsp70s suggests that the phenomenon studied here is a fundamental aspect of the function of Hsp70:Hsp40 chaperone machines.
Members of the ubiquitous 70-kDa heat shock protein (Hsp70) class of molecular chaperones are critical in cellular responses to stresses that cause protein denaturation as well as for normal cellular growth because of their roles in protein folding, protein translocation across membranes, and assembly and disassembly of oligomeric structures (1–3). The ability of Hsp70s to act in these diverse cellular processes is determined by virtue of their ability to bind to exposed hydrophobic segments of unfolded or partially folded polypeptide chains, preventing nonproductive association of hydrophobic regions and thus facilitating proper folding (4). Hsp70 chaperone activity is driven by an ATPase activity that regulates cycles of polypeptide binding and release (5). These two crucial activities are localized to different domains: ATPase activity to the 44-kDa N-terminal domain, substrate binding to the adjacent 18-kDa domain. Structures of both of these highly conserved domains have been solved separately with x-ray crystallography (6, 7). However, there is little information available concerning how the two domains communicate with one another. Nevertheless, there is little doubt that such communication is essential, because nucleotide binding profoundly affects interaction with protein substrates, and addition of peptide substrates stimulates the ATPase activity of Hsp70s.
Hsp70s can be viewed as alternating between two forms: the ATP-bound form, which has low affinity for substrates, and the ADP-bound form, which has a relatively high affinity for substrates (8). Although conformational differences between the ATP- and ADP-bound state are detectable, the structural nature of these differences remains obscure (9, 10). Nucleotide hydrolysis, the rate-limiting step of the ATPase cycle, leads to conformation changes in both domains and thus acts to convert Hsp70 from a less to a more stably binding form in regards to protein substrates (11, 12).
The intrinsic ATPase activity of Hsp70s is much too weak to be stoichiometrically coupled to the substrate binding–release cycle (8). Consequently, cochaperones of the DnaJ (Hsp40) and GrpE families function to increase ATP turnover, allowing productive chaperone action (13). The DnaJ proteins specifically catalyze the nucleotide hydrolysis step, whereas GrpE and related proteins stimulate nucleotide release (11, 14, 15). Thus, DnaJs promote a more stable interaction with protein substrates, whereas GrpE facilitates substrate release by stimulating release of ADP and allowing rebinding of the typically more abundant ATP. DnaJ proteins are found in eukaryotes in every cellular compartment where an Hsp70 has been identified, as well as in prokaryotes, whereas GrpE-like proteins seem to be restricted to prokaryotes and mitochondria.
In vivo, Hsp70s function in concert with members of the DnaJ and GrpE families as “chaperone machines.” The best characterized chaperone machine is that of Escherichia coli, which is composed of DnaK, DnaJ, and GrpE. This machinery is required for the replication of λ bacteriophage and the refolding of host proteins after a thermal stress (16, 17). Ssc1, Mdj1, and Mge1 comprise the analogous chaperone machine in Saccharomyces cerevisiae mitochondria. The majority of proteins that function in yeast mitochondria are synthesized as precursors in the cytosol and translocated across the mitochondrial membranes. Ssc1 is an integral part of the mitochondrial translocation machinery (18). Ssc1 binds to incoming preproteins and, in an ATP-dependent manner, aids preprotein movement across the membranes, making translocation unidirectional (19). Ssc1 also functions in the refolding of newly translocated proteins; both Mge1 and Mdj1 assist in this process (20, 21).
DnaJs are multidomain proteins defined by the presence of a J domain. The J domain is required for the ATP-dependent interaction of DnaJ with Hsp70s and for stimulation of Hsp70 ATPase (22). NMR analysis reveals that this J domain interaction occurs via the ATPase domain of DnaK (23). Recent studies indicate an interaction between the J domain and a channel at the base of the ATPase domain of DnaK (24, 25). A second site near, or coincident with, the peptide-binding pocket of DnaK is also suggested by these and other studies, although the relevance of this binding site in vivo is unclear (26–28). However, the precise elements of the DnaK–DnaJ interaction are still largely undefined.
Proper functioning of the Hsp70 chaperone machine requires productive contacts between the domains of Hsp70 as well as with cochaperones, which may interact across both domains. Although much work has been carried out with purified wild-type (wt) proteins, the functional interplay among these different domains/proteins that allows efficient action in vivo remains to be established. We believe that analysis of mutant proteins will aid in our understanding of this complex macromolecular system by allowing the testing of models concerning the consequences of the interactions between components. With this idea in mind, we undertook an analysis of amino acid alterations in the ATPase domain of Hsp70 that could suppress the defect caused by an amino acid change in the peptide-binding domain. In vitro analysis revealed that a peptide-binding domain mutant protein is defective in interaction with both DnaJ and peptides. A second amino acid change that partially restored the ability of the protein to function in vivo also resulted in partial restoration of the ability of DnaJ to stimulate the ATPase activity of the mutant, suggesting that restoring the functional interaction of DnaJ may enable this mutant to function in vivo.
MATERIALS AND METHODS
Media, Plasmids, and Strains.
Media. LB medium was prepared as described (29) and supplemented with 100 μg/ml ampicillin, 100 μg/ml kanamycin, and/or 25 μg/ml chloramphenicol where appropriate. Yeast YPD (rich glucose media) and minimal media were prepared as described (30).
Plasmids.
pRS314-SSC1 (ori ampR TRP1 SSC1) was described previously (15) and contains wt SSC1. pQE60 (ori ampR) is a vector control (Qiagen, Chatsworth, CA). pBB46 (ori ampR dnaK) is a DnaK expression plasmid derived from pQE60 having dnaK under control of an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter (27).
S.
cerevisiae strain DG252 [trp1–1 ura3–1 leu2–3, 112 his3–11, 15 ade2–1 can1–100 GAL2+ met2-Δ1 lys2-Δ2 ssc1ΔClaI∷LEU2 pGAL1:SSC1(URA3)]. Chromosomal copy of SSC1 is deleted for the internal 1.4-kilobase ClaI fragment and marked with LEU2. Viability is maintained by the pGAL1:SSC1(URA3) plasmid containing wt SSC1 gene under control of the GAL1 promoter (18).
E.
coli. E. coli strain BB205 [ΔdnaK52∷camR sidB1 pBB20 (R6K ori kanR lacIq)] (27) is a derivative of MC4100 deleted for dnaK.
Site-Directed Mutagenesis.
Sequential rounds of PCR with two complementary degenerate oligonucleotide primers were used to create mutations at designated positions (31). Regions subjected to PCR were either replaced by the wt gene or sequenced to verify that no additional mutations were created. In the process of constructing SSC1 D107A, initial experiments identified a second mutation at a position encoding I485N, which was created as a by-product of Taq-dependent PCR. Phenotypic analysis revealed I485N had a lethal phenotype, which was then further investigated.
Random Mutagenesis and Selection of SSC1 Suppressor Mutants.
The first 1,606 nt of SSC1 were amplified by PCR for mutagenesis and gave a mutagenesis rate of 0.185% or ≈1 mutation per 500 bp, as determined by sequencing. Reactions contained 1 ng of pRS314-SSC1 template DNA, 5 pmol of each primer, 10 mM Tris⋅HCl (pH 9.0), 1.5 mM MgCl2, 25 mM KCl, 0.25 mM each dNTP, and 2.5 units of Taq polymerase (Promega). PCR products were digested with PstI and SalI and ligated into pRS314-SSC1-I485N lacking the same fragment; thus, the first 1,045 nt of SSC1 were randomly mutagenized to create a library. This library was transformed into yeast strain DG252, and growth at 23°C on glucose minimal media was selected.
In Vivo Precursor Processing.
Experiments were performed essentially as described (32), with the omission of tunicamycin. Briefly, strains were grown at 23°C overnight, shifted to 37°C for 15 minutes, and labeled with [35S]methionine/cysteine for 2 min. Labeling was stopped by addition of methionine, cysteine, and cycloheximide to 0.5 mg/ml. At the indicated time points, samples were withdrawn, processed, and immunoprecipitated with antibodies to Hsp60 or the F1β subunit of the F1F0 ATP synthase as described. Samples were analyzed by SDS/PAGE and visualized by using a PhosphorImager (Molecular Dynamics).
Protein Purification.
wt or mutant DnaK was expressed from plasmid pNRK416 in a BL21(DE3) dnaKJ− strain and purified as described (33). DnaJ was overproduced as described (26) and purified by DEAE-Sepharose and hydroxylapatite chromatography as described (34). Protein concentrations were determined by Bradford assay by using the Bio-Rad protein assay reagent with ovalbumin as a standard. Spectrophotometric measurement of DnaK concentration indicated ovalbumin gave artificially high values, and thus DnaK concentrations obtained from Bradford assays were corrected to reflect this discrepancy.
Single-Turnover ATPase Assays.
For complex formation, DnaK (16 μg) was incubated with 100 μCi (1 Ci = 37 GBq) of [α-32P]ATP (DuPont NEG-003H, 3,000 Ci/mmol) in buffer A [25 mM Hepes⋅KOH, pH 7.5/100 mM KCl/11 mM Mg(OAc)2] containing 100 nM ATP for 15 minutes at 30°C. Complex was separated from free ATP on a Nick column (Amersham Pharmacia), adjusted to 10% glycerol, and frozen at −70°C. Single-turnover assays were performed as described (15) at 30°C in buffer A containing 0.2 μM DnaJ or 5 μM peptide p5 (CALLLSAPRR). DnaK was present at ≈0.35 μM. The rate of ATP hydrolysis was determined by fitting to a first-order rate equation by nonlinear regression analysis by using prism version 2.0 (GraphPad, San Diego, CA). Data shown are the results from two identical experiments with SEM given. Similar results were obtained with different protein preparations. Similar results were obtained in experiments by using 50% lower DnaK concentration, ensuring that DnaK saturation was achieved. Wt, I462T, and D79A/I462T proteins each ran as monomers as determined by HPLC gel filtration by using a Superose 12 column.
Fluorescence Anisotropy Peptide-Binding Assay.
Peptide APPY (CALLQSRLLLSAPRRAAATARY) was labeled with fluorescein (F-APPY) as described (35). F-APPY (35 nM) was incubated in buffer B [50 mM Hepes⋅KOH, pH 7.6/50 mM KCl/10 mM Mg(OAc)2] with 3 nM–10 μM wt or mutant DnaK overnight at 4°C in the dark to reach equilibrium. Measurements were made with the Beacon 2000 Fluorescence Polarization system (Panvera, Madison, WI) at 25°C with excitation at 490 nm and emission at 535 nm. Data were fit to a quadratic single site binding equation (35) by using Microsoft excel.
Surface Plasmon Resonance Analysis.
Surface plasmon resonance was used to detect DnaK–DnaJ interaction with a BIAcore 2000 apparatus (BIAcore, Uppsala, Sweden) at the University of Wisconsin-Madison Biophysical Instrumentation Facility. All experiments were performed at 25°C in buffer B with 0.003% P20 surfactant (BIAcore). A fusion of biotin carboxyl carrier protein to the C terminus of DnaJ was used to biotinylate DnaJ by expression in E. coli. Expression of this fusion protein rescues growth of a strain lacking dnaJ, indicating that biotin carboxyl carrier protein does not alter DnaJ function (24). Approximately 1,000 response units of biotinylated DnaJ biotin carboxyl carrier protein (as crude cell-free extract) were immobilized on a streptavidin-crosslinked SA sensor chip surface according to the manufacturer’s protocols, corresponding to ≈1.0 ng/mm2. This method of coupling avoids heterogeneity of immobilization inherent in chemical crosslinking of DnaJ. NaCl (10 μl, 2 M) was then injected to remove nonspecifically bound proteins. wt or mutant DnaK was preincubated in buffer B containing 1 mM ATP for 5 min before injection for 3.5 minutes at 15 μl/min. Routinely, DnaK samples were injected in multichannel mode first over a control flow cell treated with 10 μl of 50 μg/ml biotin and then over a second flow cell having immobilized biotinylated DnaJ biotin carboxyl carrier protein, and background binding to biotin was then subtracted from each signal.
Tryptophan Fluorescence.
Measurements were performed on a QuantaMaster1 fluorescence spectrometer from Photon Technology International (South Brunswick, NJ). Excitation was at 290 nm and emission was recorded from 310 to 410 nm with a 4-nm slit width. Spectra were determined at 0.65 μM DnaK in buffer B at 25°C; samples were preincubated with either 1 mM ATP or ADP (final concentration) for 5 min. To determine peak maxima, data were fit by nonlinear regression analysis to an asymmetric curve described on the left by a Gaussian peak function and on the right by a mixed Gaussian/Laurentzian function by using Microsoft excel.
RESULTS
A Mutation in the ATPase Domain Suppresses the Lethality and Translocation Defect of a Peptide-Binding Domain Mutant.
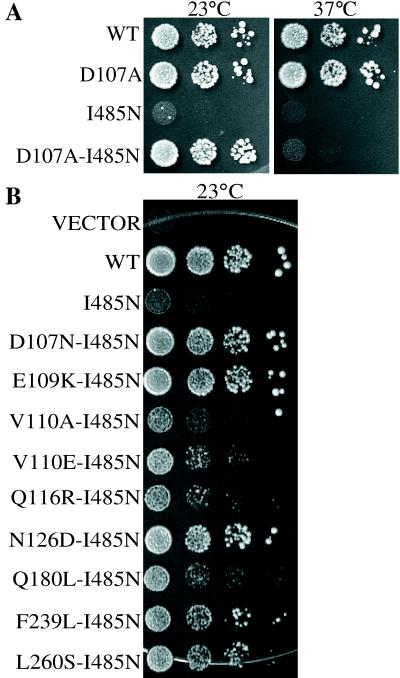
To gain a better understanding of the interplay between domains of Hsp70, we analyzed a gene encoding mitochondrial Hsp70 having two mutations, one causing an amino acid change in the ATPase domain and the second causing an amino acid change in the peptide-binding domain (see Materials and Methods). The peptide-binding domain mutation (I485N), which results in exchange of an asparagine for an isoleucine at position 485, in the context of an otherwise wt SSC1 gene is a null mutation, resulting in a lethal phenotype (Fig. 1A). However, the presence of a second mutation, D107A, allows growth of the I485N mutant at temperatures of 30°C and lower. The D107A mutation in the context of an otherwise wt SSC1 sequence promotes wt growth at all temperatures. Therefore, a single amino acid change in the ATPase domain resulted in intragenic suppression of a defect caused by an amino acid substitution in the peptide-binding domain.
Figure 1.
Growth phenotype of yeast SSC1 mutants. pRS314-SSC1 carrying either a wt or mutant copy of SSC1 (as indicated) was transformed into strain DG252. Strains were grown in galactose minimal selective media overnight at 23°C, 1:10 serial dilutions were made, and equivalent numbers of cells were plated on rich glucose media and incubated at the indicated temperatures to test the ability of mutant SSC1 genes to support growth. (A) Growth of D107A, I485N, and D107A/I485N. (B) Growth of I485N in the presence of additional ATPase domain mutations.
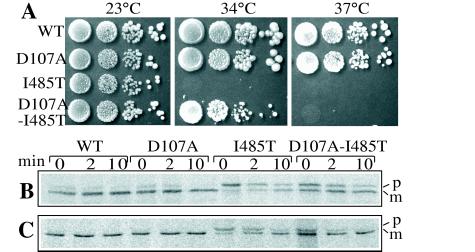
Change of I485 to threonine leads to a temperature-sensitive growth phenotype, suggesting that it has a similar, but milder, affect on Ssc1 structure compared with I485N. This temperature-sensitive phenotype is partially suppressed by the D107A mutation (Fig. 2A) and allowed us to analyze the effects of a mutation at the I485 residue in vivo. Because Ssc1 is the chaperone required for translocation of cytosolic precursor proteins into mitochondria (18), we monitored cleavage of mitochondrial leader sequences by in vivo pulse–chase analysis. Strains were grown at 23°C, briefly shifted to 37°C, and then pulsed with [35S]methionine and cysteine before initiating a chase by addition of unlabeled amino acids. Processing of both F1β and Hsp60 to mature form occurred so rapidly in the wt and D107A strains that nearly all of the protein reached mature form during the pulse labeling (Fig. 2B). However, the time course of processing was much slower in an I485T strain than in wt, with the majority of both F1β and Hsp60 being in the precursor form at the beginning of the chase. The presence of the D107A mutation in the context of I485T allowed more rapid processing. Thus, in vivo precursor-processing phenotypes correlate well with the growth phenotypes, because the D107A mutation partially alleviates I485T growth as well as processing defects.
Figure 2.
Growth and precursor processing defect of SSC1-I485T is partly alleviated by D107A. (A) pRS314-SSC1 plasmids carrying either a wt or mutant copy of SSC1 (as indicated) were transformed into DG252 cells; transformants were streaked on plates containing 5-fluoroorotic acid to select for loss of the URA3-marked plasmid containing wt SSC1. 5-Fluoroorotic acid-resistant cells were grown in YPD overnight, 1:10 serial dilutions were made, and cells were plated on YPD and incubated at the indicated temperatures. (B and C) In vivo pulse–chase analysis of mitochondrial precursor processing. Strains containing wt or mutant copies of SSC1 were pulse labeled with [35S]methionine and cysteine at 37°C for 2 min, at which time cold methionine and cysteine were added, and incubation continued for the indicated time in minutes before harvest and immunoprecipitation with antibodies to Hsp60 (B) or F1β (C). Samples were analyzed by SDS/PAGE and visualized by using a PhosphorImager.
Additional Amino Acid Changes in the ATPase Domain Suppress the I485N Lethal Phenotype.
To determine whether other mutations in the ATPase domain could suppress the phenotype of the I485N mutant, we performed a genetic selection. The region of SSC1 encoding the ATPase domain was mutagenized by PCR and cloned into a vector containing the remainder of the SSC1 gene having the I485N mutation. Mutant genes able to suppress the I485N phenotype were selected by transformation into strain DG252 and plating at the nonpermissive temperature of 23°C. Nine additional intragenic suppressors were analyzed. The nine suppressors represented eight different codons (Fig. 1B). The extent of suppression varied. A second mutation at the D107 residue, D107N, was obtained, which allowed nearly wt growth rates at 23°C, whereas changes at V110 suppressed only weakly. None of the suppressor mutations allowed growth at 37°C (data not shown).
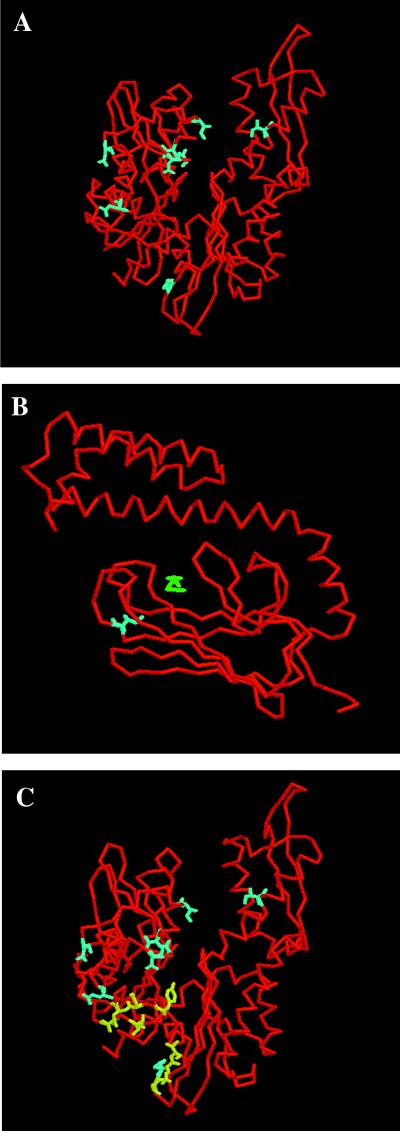
To determine whether the suppressor residues clustered to a particular region of the ATPase domain, we located the residues on the crystal structure of DnaK (Fig. 3 A and B) (36), which is 62% identical to Ssc1 at the amino acid level (37). The residues altered by the suppressor mutations map to a single face of the structure, on the backside of the traditional view in a region toward the center and bottom cleft. Most of the residues are surface exposed.
Figure 3.
Location of residues on DnaK crystal structures (PDB entries 1DKG and 1DKX). ATPase domain (A) is rotated 180° relative to the traditional view; peptide-binding domain (B). Residues described in this study are blue: Ssc1 D107 (DnaK D79), E109 (E81), V110 (V82), Q116 (I88), N126 (N98), Q180 (Q152), F239 (F216), L260 (I237), I485 (I462). Peptide substrate is green. Residues identified in other studies (C) are yellow: DnaK R167, I169, T215 (24); DnaK Y145, N147, D148, E217, V218 (25).
Analogous Mutations in E. coli dnaK Show Similar Intragenic Suppression Phenotype.
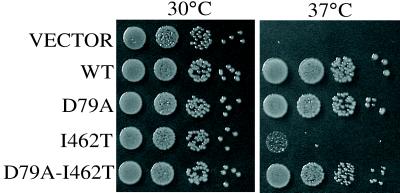
To understand the basis of the defects in the peptide-binding domain mutants as well as the mechanism of suppression, we purified the mutant proteins and carried out biochemical analyses. However, the purification of the mutant Ssc1 proteins proved problematic. We attempted to carry out these studies in a system in which Hsp70 biochemical parameters were well established. Therefore, we synthesized mutations in E. coli dnaK (see Materials and Methods) analogous to the mutations in yeast SSC1. dnaK is required for growth of E. coli at temperatures of 37°C and higher (38). dnaK I462T (= SSC1 I485T) mutant cells grew extremely slowly at 37°C and were nonviable at higher temperatures. To validate this study, we needed to establish that a similar intragenic suppression existed in the E. coli system. Such suppression was indeed the case, as D79A/I462T (= SSC1 D107A/I485T) allowed robust growth at 37°C (Fig. 4). However, suppression was not complete, because growth of the double mutant was severely compromised at higher temperatures (data not shown). This preservation of the intragenic suppression phenotype indicates that DnaK and Ssc1 are structurally similar such that the mechanism for suppression is preserved in the two systems.
Figure 4.
Similar intragenic suppression phenotype is seen for analogous mutations in E. coli dnaK. pQE60 vector or pBB46 containing wt or mutant dnaK under the control of an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter was transformed into strain BB205. Strains were grown overnight at 30°C, a permissive growth temperature for dnaK mutants. Serial dilutions (1:10) were made, equivalent numbers of cells were plated on media containing 15 μM IPTG, and cells were grown overnight at the indicated temperatures.
I462T Is Defective in Interaction with Peptide Substrates.
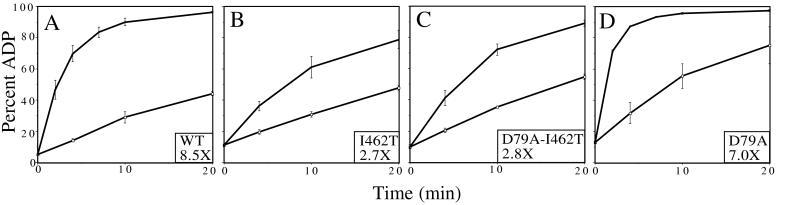
Because binding protein substrates is a fundamental property of Hsp70s, we asked whether the I462T mutant had defects in peptide-binding activities. First, the ability of p5 peptide to stimulate ATPase activity in single-turnover assays was used as an indirect assessment of activity. Single-turnover ATPase assays were performed by using preformed DnaK–[α-32P]ATP complexes in the presence and absence of peptide. The basal rate of ATP hydrolysis by wt DnaK was 0.040 ± 0.007 min−1 (Fig. 5), as previously reported (33, 39). I462T showed no significant alteration in basal activity (0.051 ± 0.014 min−1). Addition of a 15-fold molar excess of peptide p5 over DnaK allowed us to delineate differences in stimulation of wt and mutant DnaK. Whereas wt ATPase activity was stimulated 8.5-fold by p5, I462T was stimulated only 2.7-fold. Similar results were seen with a second unrelated peptide (data not shown).
Figure 5.
Peptide p5 stimulation of DnaK ATPase activity. Single-turnover ATPase assays were performed in the absence (□) or presence (♦) of p5. DnaK concentration is ≈0.35 μM; p5 is 5 μM. Fold stimulation of ATPase activity by p5 is shown in the box. (A) wt DnaK. (B) I462T. (C) D79A-I462T. (D) D79A.
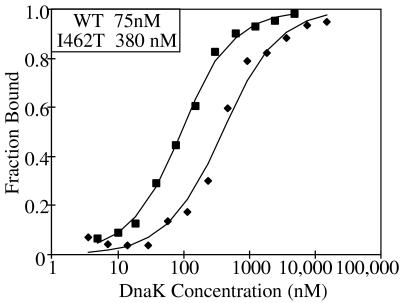
To more directly test the peptide-binding ability of Hsp70, we compared the affinity of wt and I462T for peptide in the absence of ATP by using fluorescence anisotropy. Fluorescein-labeled peptide APPY (F-APPY) was used and found to have a Kd for wt DnaK of 75 nM (Fig. 6), as previously published (35). I462T had an ≈5-fold higher Kd for F-APPY, indicating that this mutant protein interacts less stably with peptide and providing an explanation for the decreased ability of p5 to stimulate the ATPase activity.
Figure 6.
Fluorescence anisotropy assay of peptide binding to wt and I462T DnaK. F-APPY (35 nM) fluorescein-labeled peptide was incubated with the indicated concentrations of wt (■) or I462T (⧫) DnaK overnight to achieve equilibrium. Fluorescence polarization was determined at 25°C with excitation at 490 nm and emission at 535 nm. Data were fitted to a quadratic single-site binding equation to determine the Kd of DnaK for peptide, which is shown in the box. Data is expressed as fraction of DnaK bound to F-APPY. At saturation, the maximum polarization for wt was approximately 240 polarization milliunits, whereas the maximum for I462T was approximately 205 polarization milliunits.
I462T Is Defective in Interaction with DnaJ.
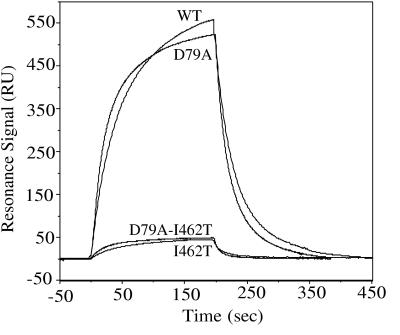
Because DnaJ stimulates the ATPase activity of DnaK and is thought to interact with the peptide-binding domain as well as the ATPase domain, we tested the interaction of I462T with DnaJ (24, 26, 27). The real-time interaction of mutant DnaK with DnaJ was investigated by using BIAcore based on surface plasmon resonance. After immobilization of DnaJ on the sensor chip surface, various concentrations of wt and mutant DnaK were injected over the surface for the association phase; buffer was then injected over the surface to measure dissociation. During kinetic evaluation of BIAcore results, we and others have found that the binding curves do not fit well to a single or double exponential, suggesting that the DnaK–DnaJ interaction measured does not fit a simple model (25, 26). This is not surprising, because DnaJ likely binds to more than one site on DnaK, and the nucleotide status of DnaK also affects binding. Consequently, we use these results only as a qualitative measure of DnaK–DnaJ binding. At 1.0 μM DnaK, about 550 response units of wt DnaK bound DnaJ (Fig. 7). I462T bound to DnaJ with much lower affinity (only ≈50 response units bound), supporting the hypothesis that this point mutant is defective in interaction with DnaJ.
Figure 7.
Surface plasmon resonance analysis of the interaction of immobilized DnaJ with wt and mutant DnaK proteins. DnaK (1 μM) was preincubated with 1 mM ATP for 5 min, and injection of DnaK over the sensor chip began at 0 seconds; buffer was injected at 210 seconds. A response in response units (RU) indicates binding of DnaK. Background binding to a flow cell containing immobilized biotin was subtracted from each sensorgram.
Because DnaJ stimulates the rate of ATP hydrolysis by DnaK (11), we predicted that the DnaJ-binding-defective I462T mutant would be defective in DnaJ stimulation of ATP hydrolysis. To test this idea, single-turnover ATPase assays were performed (Fig. 8). We selected an ≈2:1 ratio of DnaK/DnaJ to allow us to easily compare the degree of DnaJ stimulation of wt and mutant DnaK ATPase. An 8.1-fold stimulation of wt DnaK ATPase activity by DnaJ was observed, but only a 2.0-fold stimulation of I462T ATPase activity was seen. Therefore, in these established in vitro assays, I462T is defective in interaction with DnaJ.
Figure 8.
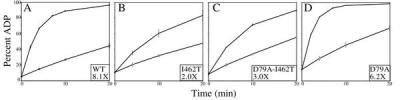
DnaJ stimulation of DnaK ATPase activity. Single-turnover ATPase assays were performed in the absence (□) or presence (♦) of DnaJ. DnaK concentration is ≈0.35 μM; DnaJ is 0.2 μM. Fold stimulation of ATPase activity by DnaJ is shown in the box. (A) wt DnaK. (B) I462T. (C) D79A-I462T. (D) D79A.
DnaK Mutant Proteins Undergo Nucleotide-Dependent Conformation Changes.
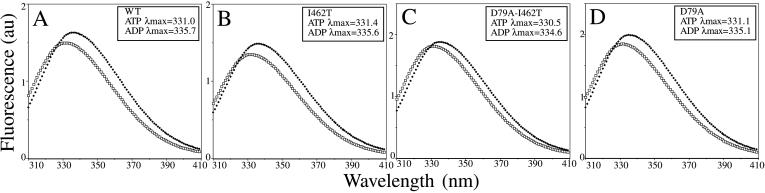
We wanted to gain insight into the means by which D79A suppresses the defect caused by the I462T change. Two general possibilities exist for the mechanism of suppression of a peptide-binding domain alteration by ATPase domain changes: (i) the alterations affect the direct interaction of the two domains or (ii) the alterations affect the interaction of proteins eliciting effects in both domains. To determine whether direct communication between the ATPase domain and the C terminus is affected in the mutants, we monitored intrinsic fluorescence of the single, N-terminally located tryptophan of DnaK. The ATP-induced alteration in the emission spectrum has been shown to require the C terminus of the protein and is thought to be caused by altered domain interactions (9). Each of the mutant proteins, including I462T, showed a blue shift and quenching of fluorescence in the presence of ATP relative to ADP typical of wt protein (Fig. 9). Thus I462T is able to make the conformation changes measurable by this assay, suggesting that neither this amino acid change, nor the D79A change, affect direct interdomain communication.
Figure 9.
Intrinsic tryptophan fluorescence emission spectra of wt and mutant DnaK proteins. DnaK (0.65 μM) was preincubated with 1 mM ADP (♦) or ATP (□). To determine peak maxima (shown in box), data were fit by nonlinear regression analysis to an asymmetric curve described on the left by a Gaussian peak function and on the right by a mixed Gaussian/Laurentzian function by using Microsoft excel. (A) wt DnaK. (B) I462T. (C) D79A-I462T. (D) D79A.
Effect of D79A on the Properties of I462T.
We also tested whether the D79A alteration affected interaction with peptide or DnaJ. No difference in the physical interaction of D79A/I462T with DnaJ in the BIAcore assay or peptide in the fluorescence anisotropy assay compared with I462T was observed (Fig. 7; data not shown). The interaction of D79A was indistinguishable from wild-type (Fig. 7).
Single-turnover ATPase assays revealed that D79A/I462T has a basal ATPase activity similar to wt, whereas D79A has a basal ATPase activity ≈2-fold higher than wt. D79A/I462T stimulation by peptide was similar to that for the I462T mutant protein, showing only a 2.8-fold stimulation, compared with 2.7-fold. Interestingly, DnaJ stimulated D79A/I462T 3.0-fold, a small but reproducible enhancement compared with the 2.0-fold seen for I462T (Fig. 8). This difference in stimulation by DnaJ is the only difference we have observed in biochemical assays comparing I462T and D79A/I462T.
DISCUSSION
Communication between Hsp70 ATPase and peptide-binding domains is fundamental to their function because adenine nucleotides regulate the nature of substrate interactions (8); however, the mechanism of domain communication is ill-defined because of lack of both structural and genetic information. Therefore, we initiated studies to understand the functionally important elements of Hsp70 domain communication in vivo by using two approaches: a genetic analysis of Hsp70 mutants in different domains and an in vitro biochemical analysis of these mutant proteins. The mutation in the region of dnaK encoding the peptide-binding domain discussed in this report alters an absolutely conserved amino acid located in a loop proposed to be critical for maintaining the structure of the peptide-binding pocket, although not directly contacting peptide (7). That an amino acid alteration at this location has an affect on the affinity of interaction of the mutant protein with peptide substrates is not surprising, because other dnaK mutations causing amino acid alterations outside the peptide-binding pocket have similar effects (27). Because we found no alterations in ATP-dependent conformational changes as monitored by tryptophan fluorescence, basal ATPase activity, or ATP-binding properties, we suspect that this alteration in peptide binding is central to the observed phenotypic effect.
The dual DnaJ and peptide-binding defects we observed in I462T are not surprising, because other DnaK mutant proteins have recently been reported to show a correlation between peptide-binding and DnaJ-binding defects (24, 27). These findings support the idea that DnaJ may bind at or near the peptide-binding pocket of DnaK, in addition to its well characterized interaction with the ATPase domain. Whether interaction with the peptide-binding domain is biologically significant or an artifact of in vitro biochemical experiments exacerbated by abnormally high DnaJ levels in the assays is a matter of debate (28, 40). In this view, the J domain interacts with the ATPase domain, whereas another part of DnaJ binds as a substrate in the peptide-binding pocket. However, there is general consensus that stimulation of ATPase activity by binding of DnaJ to the ATPase domain in conjunction with binding of an unfolded protein or peptide substrate in the peptide-binding pocket is central to the chaperone activity of Hsp70s.
The major question raised by the studies reported here is how amino acid changes in the ATPase domain suppress the effect of defects in the peptide-binding domain. Simplistically, one can envision two distinct models as to how communication between domains might be facilitated in vivo: (i) via direct interaction between the ATPase and peptide-binding domains mediated by conformation changes directly affecting interfacing residues in the two domains or (ii) via an indirect communication mediated by a cochaperone that binds to both domains or elicits effects in both domains. ATP-dependent conformational changes assessed by alterations in tryptophan fluorescence were the same for mutant and wt proteins. Although this assay may not detect subtle conformational changes, these results suggest that direct domain communication is not affected in the mutants. Therefore, we focused on the interaction with cochaperones. It is unlikely that the mutants are defective in indirect domain communications mediated by the GrpE cochaperone, because the suppressor residues map to the face of the ATPase domain which does not interact with GrpE, according to the DnaK–GrpE co-crystal structure (Fig. 3A) (36). In addition, results from isolated mitochondria showed wt levels of Mge1 bound to SSC1 mutant proteins, whereas Mdj1, a DnaJ homologue of the mitochondrial matrix, showed decreased interaction (data not shown). Thus, we focused on the interaction of the mutant proteins with the DnaJ cochaperone and peptide substrates.
We observed a small but reproducible reversal of DnaJ stimulation of ATPase activity in the double mutant (3.0-fold) compared with I462T protein (2.0-fold), whereas wt protein was stimulated 8.1-fold under the same conditions. No enhanced stimulation of the double-mutant protein with peptide substrates was observed, however, suggesting a DnaJ-specific effect. No significant recovery of physical binding of D79A/I462T double mutant protein to DnaJ or peptide (data not shown) was seen compared with I462T. But the second amino acid alteration in the ATPase domain need not alter the affinity of DnaK for DnaJ or peptide (“physical interaction”) to affect the ability of DnaJ to stimulate ATPase activity. A defect in the “functional interaction” with DnaJ or peptide could be reversed by allowing recovery of a DnaJ- or peptide-elicited response on binding without changing the affinity of the interaction. Such a reversal could alter the conformation of the ATPase domain and thus the efficacy of the functional interaction.
The overlap in location of our suppressors with DnaK residues published as critical for interaction with DnaJ (Fig. 3C) provides additional support for the view that suppression could be DnaJ-mediated. Suh et al (24) found dnaK mutations at positions encoding R167, I169, and T215, which map to the same face of the ATPase domain, as allele-specific suppressors of the dnaJ D35N mutant. Moreover, R167H bound with higher affinity to mutant DnaJ than to wt DnaJ, strongly implicating this region of the ATPase domain as a site for interaction with DnaJ (Fig. 3C). Gassler et al. (25) created multiple alanine substitutions of conserved ATPase-domain residues and found these to be defective in DnaJ interactions (Fig. 3C). Taken together, the above evidence suggests that this particular face of the ATPase domain is critical for interaction with DnaJ. Some residues on this face may directly interact with DnaJ, and others may be crucial for maintaining the proper Hsp70 structure for optimal in vivo interactions with DnaJ. Substitutions at particular positions could increase or decrease the affinity of the physical interaction with DnaJ. Other substitutions could cause alterations in ATPase domain structure that could either enhance or dampen the ability of DnaJ to elicit interdomain communication.
The results of the experiments reported here suggest a possible biochemical mechanism of suppression. In the absence of cochaperones, ATP hydrolysis is not stoichiometrically coupled to the much faster substrate binding/release cycle (8). DnaJ helps couple these two processes by increasing ATP hydrolysis and thus the amount of DnaK in the ADP form, leading to enhanced interaction with substrates (41). If the interaction of DnaJ with the peptide-binding domain pocket is an artifact of experimental conditions and the critical interaction is with the ATPase domain, we favor the following model. The interaction of I462T with DnaJ in vivo is normal. The decrease in affinity for peptide is the critical defect of I462T, and this leads to a decrease in stable interaction with substrates. One can imagine two ways of overcoming this defect: (i) by directly increasing affinity for substrates or (ii) by increasing the conversion of DnaK to the ADP form. D79A/I462T thus would partially suppress via the second method. According to this model, the effect of DnaJ interaction with the ATPase domain is enhanced, increasing the stimulation of ATP hydrolysis, thus promoting capture of polypeptide substrates.
If in fact the interaction of DnaJ with the peptide-binding domain observed in vitro is important for in vivo chaperone-binding cycles, alternative models need to be considered, because a dual mechanism of DnaJ signaling may exist. In this case, the decrease in peptide-binding activity need not be the critical defect of I462T. In fact, there is no unequivocal information as to the magnitude of a decrease in affinity for unfolded polypeptides tolerated in vivo. Rather, the defective interaction with DnaJ might be the consequential change. Suppression might then be a direct consequence of the partial restoration of the efficacy of DnaJ’s ability to stimulate ATPase activity.
These hypotheses of DnaJ-dependent suppression require more rigorous testing, particularly because the magnitude of both the growth suppression and the enhancement of DnaJ-stimulated ATPase activity is small. It should be noted that the dnaK suppressor was not directly selected, but rather was constructed as a site-directed change based on the results with the yeast mitochondrial Hsp70 Ssc1. Analysis of stronger dnaK ATPase domain suppressors of the I462T growth defect should clarify the mechanism of suppression.
The preservation of the intragenic suppression phenotype between S. cerevisiae SSC1 and E. coli dnaK suggests that the structures of Ssc1 and DnaK are similar. In addition, it suggests that the mechanism of suppression is preserved in the two systems, and implies that our findings apply to the general Hsp70–DnaJ protein interaction in both prokaryotes and eukaryotes.
Acknowledgments
We are grateful to W. Walter for help performing many of the experiments; C. Gross and W. Suh for giving helpful advice, sharing unpublished results, and giving helpful comments on the manuscript; L. Gierasch and D. Montgomery for helpful suggestions and generously providing fluorescein labeled peptide; R. Raines for helpful suggestions and use of the fluorimeter; and C. Whalen and J. Bruno for suggestions for BIAcore experiments. We thank M. Gottesman, B. Burkholder, R. McMacken, and C. Gross for providing strains, and K. Hutchison for creation of spreadsheet programs. This work was supported by National Institutes of Health Training Grants T32-GM07215 (J.E.D.) and 5T32-GM08349 (J.E.D. and C.V.), and National Institutes of Health Grant 5R01-GM27870 (E.A.C.).
ABBREVIATIONS
- hsp
heat shock protein
- wt
wild type
References
- 1.Craig E A, Gambill B D, Nelson R J. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morimoto R I, Tissieres A, Georgopoulos C. The Biology of Heat Shock Proteins and Molecular Chaperones. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 3.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 4.Flynn G, Pohl J, Flocco M, Rothman J. Nature (London) 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 5.Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C. J Biol Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- 6.Flaherty K M, DeLuca-Flaherty C, McKay D B. Nature (London) 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X, Zhao X, Burkholder W, Gragerov A, Ogata C, Gottesman M, Hendrickson W. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid D, Baici A, Gehring H, Christen P. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 9.Buchberger A, Theyssen H, Schroder H, McCarty J S, Virgallita G, Milkereit P, Reinstein J, Bukau B. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- 10.Shi L, Kataoka M, Fink A L. Biochemistry. 1996;35:3297–3308. doi: 10.1021/bi951984l. [DOI] [PubMed] [Google Scholar]
- 11.McCarty J, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 12.Theyssen H, Schuster H P, Packschies L, Bukau B, Reinstein J. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 13.Liberek K, Marszalek J, Ang D, Georgopoulos C. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Packschies L, Theyssen H, Buchberger A, Bukau B, Goody R, Reinstein J. Biochemistry. 1997;36:3417–3422. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- 15.Miao B, Davis J E, Craig E A. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- 16.Liberek K, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1988;85:6632–6636. doi: 10.1073/pnas.85.18.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaitanaris G A, Papavassiliou A G, Rubock P, Silverstein S J, Gottesman M E. Cell. 1990;61:1013–1020. doi: 10.1016/0092-8674(90)90066-n. [DOI] [PubMed] [Google Scholar]
- 18.Kang P J, Ostermann J, Shilling J, Neupert W, Craig E A, Pfanner N. Nature (London) 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 19.Dekker P, Pfanner N. In: Molecular Chaperones and Folding Catalysts. Bukau B, editor. London: Harwood; 1999. pp. 236–262. [Google Scholar]
- 20.Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 21.Laloraya S, Dekker P, Voos W, Craig E, Pfanner N. Mol Cell Biol. 1995;15:7098–7105. doi: 10.1128/mcb.15.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 23.Greene M, Maskos K, Landry S. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suh W-C, Burkholder W, Lu C Z, Zhao X, Gottesman M, Gross C. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gassler C, Buchberger A, Laufen T, Mayer M, Schroder H, Valencia A, Bukau B. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karzai A W, McMacken R. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 27.Burkholder W F, Zhao X, Zhu X, Hendrickson W A, Gragerov A, Gottesman M E. Proc Natl Acad Sci USA. 1996;93:10632–10637. doi: 10.1073/pnas.93.20.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laufen T, Mayer M, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ausubel F, Brent R, Kingston R, Moore D, Seidman J G, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 30.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 31.Cormack B. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. New York: Wiley; 1994. pp. 8.5.7–8.5.9. [Google Scholar]
- 32.Baxter B K, James P, Evans T, Craig E A. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamath-Loeb A, Lu C Z, Suh W-C, Lonetto M, Gross C. J Biol Chem. 1995;271:30051–30059. doi: 10.1074/jbc.270.50.30051. [DOI] [PubMed] [Google Scholar]
- 34.Zylicz M, Yamamoto T, McKittrick, Sell S, Georgopolous C. J Biol Chem. 1985;260:7591–7598. [PubMed] [Google Scholar]
- 35.Montgomery D, Morimoto R, Gierasch L. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 36.Harrison C J, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 37.Boorstein W R, Ziegelhoffer T, Craig E A. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 38.Georgopoulos C, Ang D, Liberek K, Zylicz M. In: Stress Proteins in Biology and Medicine. Morimoto R, Tissieres A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 191–222. [Google Scholar]
- 39.Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misselwitz B, Staeck O, Rapoport T. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 41.Pierpaoli E, Sandmeier E, Baici A, Schonfeld H-J, Gisler S, Christen P. J Mol Biol. 1997;269:757–768. doi: 10.1006/jmbi.1997.1072. [DOI] [PubMed] [Google Scholar]