Abstract
With cDNA microarrays, it is now possible to compare the expression of many genes simultaneously. To maximize the likelihood of finding genes whose expression is altered under the experimental conditions, it would be advantageous to be able to select clones for tissue-appropriate cDNA sets. We have taken advantage of the extensive sequence information in the dbEST expressed sequence tag (EST) database to identify a neural crest-derived melanocyte cDNA set for microarray analysis. Analysis of characterized genes with dbEST identified one library that contained ESTs representing 21 neural crest-expressed genes (library 198). The distribution of the ESTs corresponding to these genes was biased toward being derived from library 198. This is in contrast to the EST distribution profile for a set of control genes, characterized to be more ubiquitously expressed in multiple tissues (P < 1 × 10−9). From library 198, a subset of 852 clustered ESTs were selected that have a library distribution profile similar to that of the 21 neural crest-expressed genes. Microarray analysis demonstrated the majority of the neural crest-selected 852 ESTs (Mel1 array) were differentially expressed in melanoma cell lines compared with a non-neural crest kidney epithelial cell line (P < 1 × 10−8). This was not observed with an array of 1,238 ESTs that was selected without library origin bias (P = 0.204). This study presents an approach for selecting tissue-appropriate cDNAs that can be used to examine the expression profiles of developmental processes and diseases.
The epidermal melanocyte is a distinct cell type derived from the pluripotent neural crest. Several human genetic disorders (Piebaldism, albinism, Waardenburg’s syndrome, melanoma) are caused by mutations in genes that control melanocyte differentiation, survival, or function (1, 2). The emerging technology of cDNA microarrays provides a powerful tool for identifying genes whose expression is altered in disease states (3, 4, 5). This method involves spotting thousands of cDNA clones (probes) on a solid support, hybridizing the array with two labeled mRNA samples (targets) and comparing the relative expression of these clones between the two mRNA samples (6). For microarray studies, emphasis is often placed on the selection of mRNA samples being compared. However, it is also important that the cDNA clones analyzed are likely to be expressed within the tissue/model system being investigated. Until all genes are available, a tissue-appropriate cDNA probe set for microarray analysis would be advantageous for dissecting the transcriptional regulation of neural crest melanocyte (NC-M) derivatives. We have used a database analysis approach to identify a set of expressed sequence tag (EST) clusters that are derived primarily from NC-M tissues. Using cDNA microarray analysis, we confirmed that cDNA probes selected by this approach are differentially expressed in NC-M derivatives relative to non-neural crest derived samples. This NC-M cDNA microarray will be useful for the identification of genes that have altered expression in neural crest disorders.
MATERIALS AND METHODS
BLAST Analysis and Percent Melanocyte (%Mel) Calculation.
The 22,889 ESTs from library 198 (Soares melanocyte 2NbHM, RNA from normal foreskin melanocytes) were assembled into 10,826 contigs with sequence alignment, editing, and assembly programs phred and phrap (ref. 14; http://bozeman.genome.washington.edu/). Contig redundancy was reduced by clustering nonoverlapping sequence contigs using the EST clone identification number, which is common for the nonoverlapping 5′ and 3′ sequence reads for a single EST cDNA clone. The consensus sequence from each cluster was compared with dbEST sequences in parallel by using a blastn–dbEST query (Oct. 4, 1998). For each query-identified EST (P < 10−100), the source library was identified from dbEST. By using this information, %Mel was calculated for each cluster as (the total number of ESTs from library 198)/(the total number of ESTs in dbEST independent of the library origin) ×100. The %Mel for genes listed in Table 1 was derived from the appropriate cluster for each gene.
Table 1.
Neural crest–melanocyte gene set
| Gene | Human disease | OMIM | EST, no.
|
%Mel | ||
|---|---|---|---|---|---|---|
| In 198 | In libraries other than 198 | Total in dbEST | ||||
| KIT | Piebaldism | 164920 | 1 | 0 | 1 | 100 |
| DCT | 191275 | 3 | 2 | 5 | 60 | |
| EDNRB | WS-4 | 131244 | 7 | 2 | 9 | 77.8 |
| ERBB3 | 190151 | 5 | 13 | 18 | 27.8 | |
| L1CAM | 308840 | 2 | 0 | 2 | 100 | |
| LYST | CHS | 214500 | 2 | 1 | 3 | 66.7 |
| MART1 | U06452* | 5 | 0 | 5 | 100 | |
| MDA-7 | U16261* | 1 | 0 | 1 | 100 | |
| MLSN1 | 603576 | 15 | 6 | 21 | 71.4 | |
| MET | 164860 | 5 | 32 | 37 | 13.5 | |
| MITF | WS-2a | 156845 | 5 | 2 | 7 | 71.4 |
| MSG1 | 300149 | 57 | 22 | 79 | 72.2 | |
| MYO5A | Griscelli’s syndrome | 160777 | 1 | 0 | 1 | 100 |
| NES | 600915 | 1 | 0 | 1 | 100 | |
| OCA2 | OCA2 | 203200 | 2 | 2 | 4 | 50 |
| PAX3 | WS-1,3 | 193500 | 5 | 0 | 5 | 100 |
| PMEL17 | 155550 | 10 | 8 | 18 | 55.6 | |
| SLUG | 602150 | 13 | 10 | 23 | 56.5 | |
| SOX10 | WS-4 | 602229 | 7 | 21 | 26 | 26.9 |
| TYRP1 | OCA3 | 203290 | 3 | 8 | 11 | 27.2 |
| TYR | OCA1 | 203100 | 7 | 1 | 8 | 87.5 |
GenBank accession no.
Slide Preparation.
The microarray slides for the 1,238 EST microarray and Mel1 set were prepared as described (3, 15). EST clone inserts for the Mel1 set were amplified with PCR primers AEK M13F-1 CTGCAAGGCGATTAAGTTGGGTAAC and AEK M13R-1 GTGAGCGGATAACAATTTCACACAGGAAACAGC.
Microarray Probe and Hybridization.
RNA was prepared similar to that of Khan et al. (5) with the exception being that, after the initial RNA isolation (Qiagen RNeasy kit), a subsequent extraction step with RNAsol (Tel-Test, Friendswood, TX) was performed. The fluorescence-labeled cDNA probes were prepared as described (5). UACC383 and MNT-1 were Cy5 (Amersham Pharmacia) fluor-labeled and independently compared with Cy3 (Amersham Pharmacia) fluor-labeled 293T RNA. Hybridizations for Mel1 array were performed in duplicate. Hybridizations were carried out at 65°C for 16 hr. Slides were washed at room temperature in 0.5× SSC/0.1% SDS for 3 min, followed by a 3-min wash in 0.6× SSC. Slides were immediately spun dry.
Cell Culture.
All cells were grown to 80–95% confluence. 293T cells were grown in DMEM containing 10% FBS, 2 mM l-glutamine, and 100 units/ml each penicillin and streptomycin. MNT1 cells (16) were grown in DMEM, 20% FBS, 10% AIM-5 medium (Life Technologies, Grand Island, NY), 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 1 M Hepes, 2 mM l-glutamine, and 100 units/ml each penicillin and streptomycin. UACC 383 cells (University of Arizona Comprehensive Tissue Culture Core Facility, Tucson, AZ) were grown in RPMI medium 1640, 10% FBS, 2 mM glutamine, and 100 units/ml each penicillin and streptomycin.
Image Acquisition and Analysis.
Fluorescence intensities at the immobilized probes were determined from images taken with a custom confocal microscope equipped with laser excitation sources and interference filters appropriate for the Cy3 and Cy5 fluors. Separate scans were taken for each fluor at a resolution of 225 μm2 per pixel and 65,536 gray levels. Image segmentation to identify areas of hybridization, normalization of the intensities between the two fluor images, and calculation of the normalized mean fluorescent values at each target were as described (5, 17). Normalization between the images was used to adjust for the different efficiencies in labeling and detection with the two different fluors. This was achieved by manual matching of the detection sensitivities to bring a set of 88 internal control genes (http://www.nhgri.nih.gov) to nearly equal intensity followed by computational calculation of the residual scalar required for optimal intensity matching for this set of genes.
RESULTS
Within dbEST, there are over 1 × 106 ESTs derived from over 750 libraries obtained from a variety of tissue sources (7). The cDNA sequences for 21 genes (NC-M set, Table 1), previously shown to be expressed during NC-M development, were used to screen dbEST to identify all representative ESTs for each of the 21 genes. Analysis of the NC-M set determined that the ESTs from these 21 genes were most frequently found in library 198, Soares melanocyte 2NbHM. This result suggested that library 198 is a valid source from which to identify genes involved in the regulation of NC-M development. For each gene, we calculated the fraction of ESTs from library 198 divided by the total number of ESTs from dbEST (%Mel, Table 1). The average %Mel for the NC-M genes analyzed was 69.74 ± 28.51% even though library 198 represents <3% of the ESTs within the database.
To determine whether the skewing of ESTs to library 198 is unique to NC-M genes, we next analyzed a characterized set of genes that have demonstrated expression in many diverse tissues. A set of 46 control genes was selected under the following criteria: (i) previous characterization by microarray analysis to demonstrate similar levels of expression in a diverse set of tissues (http://genome.nhgri.nih.gov/melanocyte/) (5) and (ii) at least one EST identified by blastn analysis of dbEST derived from library 198. The calculation of %Mel was performed as described for the 21 NC-M genes. In contrast to the NC-M genes, the ESTs from the 46 gene control set had a %Mel of 9.07 ± 10.26%, which is significantly different from the NC-M control genes (P = 1.2 × 10−10) (Fig. 1A). This result suggests that the %Mel criteria could be used to select a set of cDNA clones from library 198 for microarray analysis that would be expressed preferentially in NC-M derived tissues.
Figure 1.
Distribution of %Mel in control and array sets. (A) Distribution of the NC-M set (filled bars) is compared with the 46-gene set (open bars). (B) The distribution of the 6,507 clusters derived from library 198 is shown in relation to the Mel1 subset of 852 cDNA clones, which were selected for microarray analysis. The %Mel (the number of library 198 ESTs for a given cluster/the total number of ESTs for that cluster) was calculated from blastn analysis of cluster consensus sequence against dbEST (see Materials and Methods). Histograms depict the percentage of clusters (y axis) for a given range of %Mel (x axis).
To test this, a dbEST library distribution analysis was performed with all ESTs in library 198 to select a subset of clones with a high %Mel for use in subsequent cDNA microarray analysis. To reduce redundant ESTs representing the same gene, the 22,889 ESTs from library 198 were first assembled into 6,507 clusters; each cluster theoretically represented an individual transcription unit. The sequences composing each cluster were compared with sequences in dbEST, and the %Mel was calculated (Fig. 1B). The average %Mel for the 6,507clusters from library 198 was 36.7 ± 35.6%. Of the 6,507 clusters, 46% fell within the range of the 46-gene control set (from 0–20%Mel), and 36% fell within the range of the NC-M gene set (40–100%Mel). Based on this distribution, a subset of library-198 clusters (852 cDNA clones, Mel1 set) were selected that have an average %Mel = 79.5% ± 34.8%, similar to the distribution found for the NC-M control genes. The %Mel distribution for the Mel1 set compared with the 6,507 clusters is depicted in Fig. 1B.
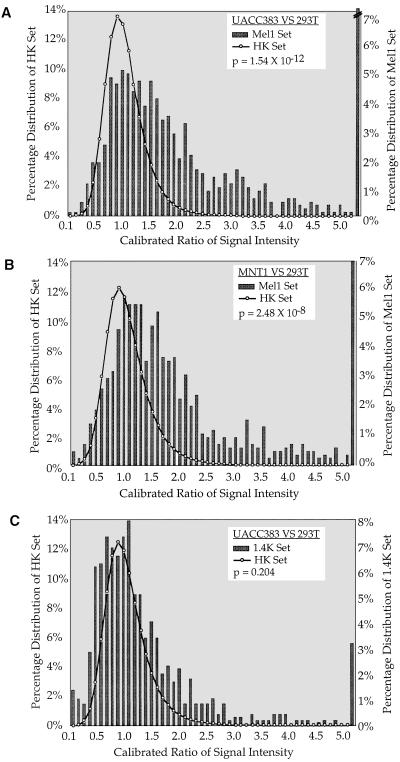
To confirm that the Mel1 cDNA set is enriched for cDNA clones differentially expressed in NC-M derivatives, the Mel1 set was analyzed by using cDNA microarray analysis, comparing two melanoma cell lines relative to a non-NC-M cell line. Calibrated intensity ratios and hybridization profiles were obtained for the Mel1 array comparing the two melanoma cell lines (MNT-1 and UACC 383) independently, relative to the kidney epithelial cell line 293T. A control set of 88 genes previously characterized by microarray analysis as demonstrating similar expression in a diverse set of tissues was used to normalize for labeling efficiencies between cohybridized cDNA samples (5). As anticipated, known NC-M genes showed a higher level of expression in both melanoma cell lines relative to 293T cells (Fig. 2). To determine whether the Mel1 set demonstrated a similar expression profile as the NC-M genes, we compared the distribution of the expression profile of the entire Mel1 array to the distribution of 88 internal control genes (5). Two cDNA sample pairs were analyzed in duplicate: MNT-1 vs. 293T and UACC 383 vs. 293T. The expression profiles of the Mel1 set demonstrated a differential expression profile shifted in the melanoma direction (UACC 383, P = 1.54 × 10−12; MNT-1, P = 2.48 × 10−8, Fig. 3 A and B).
Figure 2.
NC-M gene expression. Color-coded representation of relative intensity values for nine representative NC-M genes (above bar) and three representative internal controls (below bar) for hybridizations comparing UACC383 or MNT-1 cDNA relative to 293T cDNA. The expression ratios depicted in red indicate a higher expression level in the melanoma cell line listed, and those depicted in green indicate a reduced expression in the melanoma cell line relative to the 293T cell line. Black denotes calibrated intensity ratio value near 1. The ratio color scale is noted below, with the saturation of the red or green increasing in proportion to the ratio. Values for the individual calibrated ratios are shown to the right.
Figure 3.
Distribution of relative intensities for Mel1 and 1.4K array. Histogram displaying the relative intensities of the 88 internal control set in comparison to the relative intensities for Mel1 array (A and B) and 1.4K array (C). Representative hybridizations are shown from A. UACC383 melanoma cDNA (Cy5-labeled) vs. 293T kidney cDNA (Cy3-labeled) (C) and MNT-1 melanoma cDNA (Cy5-labeled) vs. kidney 293T cDNA (Cy3-labeled) (B). For each slide, the EST probe intensities (scale of 0–65, 535 fluorescence units) were normalized for labeling efficiency based on the calculation of a normalization constant based on the intensities of 88 internal control ESTs (16). Previous experiments have shown that a single scalar can bring the intensity ratios of this set of control genes close to 1 when comparing a wide range of cell lines (5). This normalization constant is used to calculate the calibrated ratio for the intensity of each EST. Normalization constants are 1.070 (A), 1.078 (B), and 0.839 (C). Comparison of the shift in intensity profiles between the 88 control genes and the Mel1 (A and B) or 1.4K (C) was calculated by using the Mann–Whitney U test (18). Bars represent the percentage of cDNA probes (y axis) within a specific calibrated ratio range (x axis). The broken bar (in A) shows that the actual percentage (15%) is above the range of the figure. The distribution of the internal control set calibrated ratios is represented by the solid line overlying the histogram of the Mel1 set cDNA probes.
The UACC 383 melanoma and 293T kidney samples were also compared by using another microarray containing 1,238 human ESTs that were (i) previously characterized by microarray analysis (5), (ii) selected based on the identification of ESTs corresponding to characterized genes in GenBank (8), and (iii) selected without a tissue-specific bias. Again, the same 88 control genes were used to normalize the two labeled cDNA samples. In contrast to the shift observed in the expression profile with the Mel1 array, there is no significant shift in the expression profile with the 1,238 EST nontissue bias array (P = 0.204, Fig. 3C). These results are consistent with our previous observations that most genes represented in the set of 1,238 ESTs do not demonstrate significant variations in calibrated intensity-ratio values over a wide variety of tissues (unpublished data). Taken together, our results demonstrate that a database-analysis approach could be used to identify a set of cDNA probes that are significantly up-regulated in NC-M derivatives compared with non-neural crest-derived samples.
DISCUSSION
When using cDNA microarrays for expression profile analysis, it is important to carefully consider the thousands of EST clones that are arrayed on to the glass slide and to carefully consider the RNAs that are being compared. Human cDNA microarray sets described to date represent a subset of the genes expressed in the human genome. They have provided valuable tools in analyzing the expression profiles of tumor cell samples (3, 5), of fibroblast cells in response to serum (9), and of inflammatory disease (4). These human EST sets available are not yet as complete as those from organisms whose entire genome has been sequenced (10–12). Even with the entire human genome sequence in hand [as anticipated it will be in 2003 (13)], it will still require a tremendous amount of work to identify all of the predicted expressed sequences and the various splice forms. Additionally, the number of ESTs that are placed on cDNA microarrays will, in the near term, represent a compromise between the maximal numbers of ESTs one’s equipment will allow one to place on the slide, the costs of acquiring and preparing ESTs for spotting, and the reduction in the number of arrays that can be produced as the number of genes included gets larger. The Mel1 cDNA set will not contain all genes that are essential for proper melanocyte function, e.g., those genes with more diverse expression patterns. However, genes that are broadly expressed are available on the generalized cDNA sets (9). Selection of tissue-appropriate EST array sets will complement the more general chips currently being analyzed by providing smaller, more readily produced arrays that contain genes that are likely to be expressed in the specific RNA source.
We have demonstrated that one can use a data-mining approach to select a tissue-appropriate set of cDNA clones for microarray analysis. Similar approaches may be useful for selecting sets of cDNA probes for other tissues. However, the informatic selection must be combined with appropriate expression analyses to determine the validity of the cDNA set. We have used dbEST to identify a set of cDNA probes appropriate for the analysis of NC-M development by using cDNA microarrays.
Given that many of the NC-M control genes (Table 1) are expressed at embryonic stages of NC-M development, this set should provide a useful reagent for the analysis of the patterns of transcriptional regulation of NC-M development. In addition, this set will be useful for the characterization of altered expression patterns occurring in disease states such as melanoma.
Acknowledgments
We would like to acknowledge John Leuder for technical assistance with RNA preparation and Jane Ellison for sequencing. We thank V. Hearing and the Varmus laboratory for cell lines MNT-1 and 293T, respectively. We also thank for U. Teichmann, L. Biesecker, P. Swartzberg, R. Nussbaum, H. Arnheiter, and members of the Pavan laboratory for thought-provoking discussions.
ABBREVIATIONS
- NC-M
neural crest-derived melanocyte
- dbEST
expressed sequence tag database
- %Mel
percent melanocyte
References
- 1.Barsh G S. Trends Genet. 1996;8:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 2.Jackson I J. Hum Mol Genet. 1997;10:1613–1624. doi: 10.1093/hmg/6.10.1613. [DOI] [PubMed] [Google Scholar]
- 3.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 4.Heller R A, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley D E, Davis R W. Proc Natl Acad Sci USA. 1997;6:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan J, Simon R, Bittner M, Chen Y, Leighton S B, Pohida T, Smith P D, Jiang Y, Gooden G C, Trent J M, Meltzer P S. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 6.Duggan D J, Bittner M, Chen Y, Meltzer P, Trent J M. Nat Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 7.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 8.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 9.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 10.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1997;94:13057–62. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;131:359–367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 12.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;5:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Collins F S, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- 14.Ewing B, Hillier L, Wendl M, Green P. Genome Res. 1998;3:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 15.Shalon D, Smith S J, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 16.Piantelli M, Maggiano N, Ricci R, Larocca L M, Capelli A, Scambia G, Isola G, Natali P G, Ranelletti F O. J Invest Dermatol. 1995;2:248–253. doi: 10.1111/1523-1747.ep12317599. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Dougherty E R, Bittner M L. Biomed Optics. 1997;2:364–374. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- 18.Sokal R R, Rohlf F J. Biometry: The Principles and Practice of Statistics in Biological Research. Third Ed. New York: Freeman; 1995. pp. 427–431. [Google Scholar]