Abstract
Susceptibility to type I diabetes is linked to class II MHC alleles in both mouse and man. However, the molecular mechanisms by which MHC molecules mediate disease susceptibility are unknown. To analyze how I-A alleles predispose to, or prevent, the development of type I diabetes, we have chosen, as the first step, to investigate the immune response to an important islet cell protein in diabetes-susceptible and diabetes-resistant mice. MHC class II alleles conferring susceptibility and resistance to diabetes select completely different sets of immunogenic epitopes from the β islet cell autoantigen glutamic acid decarboxylase 65. Peptide-binding studies, analysis of MHC restriction, and immunization with these peptide epitopes indicate that the two amino acid substitutions within the I-Aβ chain that distinguish a diabetes-susceptibility from a diabetes-resistance allele are sufficient to alter peptide binding and MHC restriction and may also influence antigen presentation and the selection of the T cell repertoire. The data indicate that the molecular mechanisms for class II-mediated selection of immunodominant epitopes are complex and differ for each individual peptide epitope. Further study of the functional characteristics of the response to these epitopes should provide insight into mechanisms of MHC-mediated diabetes susceptibility.
Type I diabetes and several other autoimmune diseases, including rheumatoid arthritis, show strong association and linkage with very specific sequence polymorphisms in MHC class II molecules (1). These polymorphisms are found in several different, susceptible MHC alleles in humans and animals. In nonobese diabetic (NOD) mice, the MHC class II region encodes a unique I-A molecule (I-Ag7) and a nonexpressed I-E molecule (2, 3). The I-Aβg7 allele in NOD mice encodes serine instead of aspartic acid at position 57. Transgenic introduction of a resistant I-Aβ chain, or a mutated I-Ag7 β chain, converts the susceptible into a resistant phenotype. Several groups have produced NOD mice that are transgenic for a “nonsusceptible” (aspartic acid at position 57) MHC class II I-A allele. These I-A transgenic (Tg) mice on the NOD background exhibit markedly decreased levels of insulitis and diabetes (4–6). More specifically, Lund and co-workers (7) have shown that transgenic NOD mice carrying an I-Ag7 allele mutated at position 56 (His to Pro) or 57 (Ser to Asp) are partially or completely protected from both diabetes and insulitis. These results show that I-Aβ position 57 is the major MHC-linked genetic polymorphism determining susceptibility or resistance to type I diabetes.
Although associations and linkage of MHC class I and II genotype with disease susceptibility have been shown in several autoimmune diseases, the mechanisms of susceptibility are not understood. Two T cell receptor (TCR) transgenic models recently have been used to investigate the molecular basis for resistance to type I diabetes. One (8) found that protective MHC class II molecules provide resistance via thymic deletion of diabetogenic T cells. The second study (9) found that resistant class II alleles cause positive selection of regulatory T cells. Both results may be correct, but only for their respective TCRs.
A third study (10) found that one diabetogenic TCR (see ref. 9) was positively selected by diabetes-susceptible (I-Ag7) as well as diabetes-resistant (I-Ag7.PD) class II alleles, and neither allele negatively selected this TCR in the periphery. Again, these results apply only to this TCR.
In any event, these studies investigate protection from diabetes induced by a single transgenic TCR, but do not address how susceptible class II alleles mediate susceptibility. To understand susceptibility, it may be necessary to characterize completely the autoimmune response in type I diabetes. This will require identifying the critical target autoantigens for type I diabetes, their peptide epitopes, and the functional characteristics [e.g., T helper 1 (Th1) vs. Th2] of responding T cells. Do susceptible and resistant alleles select functionally different T cell repertoires specific for autoantigen epitopes? What are the effects of susceptible and resistant alleles on antigen processing/presentation, interaction with CLIP and H-2M molecules, and on proteolytic processing pathways?
To initiate this approach, we designed alternative experiments to investigate the molecular mechanisms of MHC-linked disease susceptibility and protection. NOD wild-type and NOD.PD Tg mice were used as animal models for type I diabetes-susceptible and diabetes-resistant MHC genotypes, respectively. NOD.PD Tg mice express a site-specific mutant I-Aβg7 allele in which amino acids 56 and 57 have been mutated from histidine and serine to proline and aspartic acid (11). The only difference between wild-type NOD mice and NOD.PD mice is at positions 56 and 57 of the transgenic MHC class II I-Aβ chain. NOD.PD Tg mice have been fully backcrossed to NOD mice, and these mice do not develop diabetes.
The first step in this experimental approach is to characterize the immunogenic T cell epitopes of islet cell autoantigens in diabetes-susceptible and diabetes-resistant strains. The islet cell autoantigen chosen in this study is glutamic acid decarboxylase 65 (GAD65), which has been shown to play an important role in the pathogenesis of type I diabetes in humans and in NOD mice (12–15). After identification of the immunogenic epitopes of GAD65, possible mechanisms for control of MHC-linked susceptibility/resistance to diabetes were analyzed by studying MHC restriction, peptide-binding affinity, cytokine response profiles, and T cell responses to immunization with peptides identified in susceptible and resistant mice.
The results show that I-Ag7 molecules select a completely different set of immunogenic epitopes of GAD65 for presentation to CD4 T cells from those identified by the diabetes-resistant I-Ag7.PD molecules. The two amino acid substitutions within the I-Aβ chain that distinguish NOD wild-type mice from NOD.PD Tg mice are sufficient to influence dramatically MHC restriction, peptide binding, selection of the T cell repertoire and/or antigen-presentation pathways. Previous studies suggested that protective class II alleles mediated their effects either by positive selection of regulatory T cells (9) or by thymic deletion of autoreactive T cells (8). The data in the present study are compatible with these previous results and provide libraries of peptide epitopes and peptide-specific T cell hybridomas, which can be used to further analyze mechanisms of MHC-mediated susceptibility to type I diabetes.
MATERIALS AND METHODS
Generation and Screening of NOD.PD Tg Mice.
The construction of the NOD.PD transgene and characterization of the resulting Tg mouse lines have been described in detail elsewhere (11). All Tg mice in this study were heterozygous for the NOD.PD transgene. Outcrosses to BALB/c indicate that I-Ag7.PD is expressed at a level 100–150% that of I-Ag7.
To identify mice carrying the NOD.PD transgene, three oligonucleotide primers spanning the mutated region were designed to screen mice in two separate PCRs. Primer EC-PD4 (5′-CACCAGTTCAAGGGCGAG-3′), which annealed upstream of the mutated site, served as the 5′ primer. Two 3′ primers were created. Primer EC-PD5 (5′-TTGTAGTACTCGGCGTCC-3′) includes a 4-bp sequence discrepancy at the 3′ end that anneals only to the PD transgene but not to the g7 allele, when the proper annealing temperature is used. Only DNA from NOD.PD Tg mice will generate a 161-bp DNA product with primers EC-PD4 and EC-PD5 (annealing temperature 56°C). Primer EC-PD6 (5′-CCGCAGGGAGGTGGGGAC-3′) anneals downstream from EC-PD5. PCRs using EC-PD4 and EC-PD6 (annealing temperature 58°C) will yield a 255-bp PCR product in both Tg and non-Tg mice. The sequence difference between the PD transgene and I-Ag7 creates an EagI site in the PD transgene. Digestion of the 255-bp product from the PD transgene by EagI yields 139- and 116-bp fragments, whereas the 255-bp product from the g7 template remains undigested.
Generation of GAD65-Specific T Cell Hybridomas.
Nine-week-old female NOD (The Jackson Laboratory) and NOD.PD Tg mice were immunized in the hind footpads and at the base of the tail with 50 μg of GAD65 protein in incomplete Freund’s adjuvant (IFA). Ten days later, cells were isolated from popliteal and inguinal lymph nodes, resuspended at 5 × 106 cells per ml in RPMI complete medium containing 1% NOD mouse serum, and restimulated in vitro with 10 μg/ml GAD65. Four days later, the cells were purified by Lympholyte-M (Cedarland Laboratories, Ontario, Canada) separation and cultured with 10 units/ml rIL-2 overnight. The GAD65-activated T cells were fused with the BW 5147 cell line by using 50% polyethylene glycol (Sigma). A detailed protocol for generation of T cell hybridomas has been described elsewhere (16).
Epitope Screening Using a Europium-Based IL-2 Sandwich ELISA.
The epitope mapping of GAD65 involved three consecutive steps (16). T cell hybridomas initially were screened with GAD65 whole protein. The hybridomas that responded to GAD65 subsequently were screened with 10 pools (10–12 peptides in each pool) of 15-mer peptides overlapping by 10 aa, spanning the entire 585-aa sequence to identify the peptide(s) encoding GAD65 epitopes. The epitope specificity of pool-positive hybridomas was decoded by using individual peptides from the regional pools.
A culture condition for screening GAD65-specific T cell hybridomas has been described elsewhere (16). The ability of the hybridomas to respond to antigen (peptide or protein) was assessed by IL-2 production, as detected by sandwich ELISA.
Competitive Inhibition Peptide–MHC Binding Assay.
The peptide–MHC-binding assay used in this study was modified from the protocol originally developed by Nepom and coworkers (17). M12.C3.g7 and M12.C3.g7.PD B cell transfectants express exclusively I-Ag7 and I-Ag7.PD molecules, respectively. These two cell lines had been subcloned and sorted by fluorescent cell sorter to obtain the same levels of surface expression of I-Ag7 and I-Ag7.PD molecules. M12.C3.g7 and M12.C3.PD transfectants were fixed with 0.5% paraformaldehyde. The cells then were washed and the cell pellets were resuspended in 200 μl of citrate phosphate binding buffer, pH 4.5. Various concentrations of unlabeled inhibitory peptides then were added to the cell suspensions. After 4 hr of incubation at 37°C, a biotinylated reference peptide was added and the mixture was incubated for 18–24 hr at 37°C. The biotinylated peptides used as reference peptides were known to bind well to their target MHC molecules (18). The reference peptide for binding to M12.C3.g7 (I-Ag7) transfectants was λ repressor 12–24 peptide. The reference peptide for binding to M12.C3.g7.PD (I-Ag7.PD) transfectants was ovalbumin 323–339 peptide. After incubation overnight, the cells were washed with Hanks’ balanced salt solution and lysed with 100 μl of 1% Nonidet P-40 with protease inhibitors. The cell lysates were transferred to ELISA plates precoated with I-Ag7-specific mAb (OX-6) (PharMingen) and incubated overnight at 4°C. After washing, europium-labeled streptavidin was added at 100 μl per well and incubated for 1 hr at room temperature. The level of fluorescence was measured with an LKB-Wallac fluorescence plate reader (Wallac, Gaithersburg, MD).
Measurement of Antigen-Specific Cytokine Expression by ELISA.
Wild-type NOD and NOD.PD mice were immunized with 50 μg of GAD65 protein in IFA in the hind footpads and the base of the tail. Ten days later, draining lymph node cells were isolated and stimulated in vitro with 20 μg/ml GAD65, a “g7” or “PD” peptide pool, or individual “g7” or “PD” peptides (20 μM/peptide) for 72 hr. Culture supernatants were harvested, and cytokine (IFN-γ and IL-4) production was determined by immunoassay by using purified capture and biotinylated detection monoclonal pairs (PharMingen).
RESULTS
Mapping of Immunogenic T Cell Epitopes of GAD65 in NOD Wild-Type Mice and Characterization of the Core Sequences of These Epitopes.
Nine-week-old NOD mice were immunized with GAD65 protein in IFA to generate T cell hybridomas for epitope mapping. The immunogenic epitopes of GAD65 identified in NOD mice and the frequency of GAD65 peptide-specific T cell hybridomas are shown in Table 1. Five immunogenic epitopes (p206–220, p221–235, p286–300, p401–415, and p561–575) were identified. p206–220, p221–235, and p286–300 are the three most frequent epitopes among the five identified. Forty percent of GAD65-specific T cell hybridomas recognize p206–220, and 39% respond to p221–235. Unimmunized NOD mice 9 weeks of age also were used to perform epitope mapping. In 780 screened T cell hybridomas, two and one of the hybridomas were specific for p286–300 and p206–220, respectively. None of the epitopes identified in this study overlap with the three GAD65 epitopes (p247, p509, and p524) previously described by Kaufman et al. (13).
Table 1.
Immunogenic T cell epitopes of GAD65 identified in NOD mice
| Peptide | No. of T cell hybridomas | % (n = 74) |
|---|---|---|
| 206–220 | 30 | 41 |
| 221–235 | 29 | 39 |
| 286–300 | 7 | 9 |
| 401–415 | 3 | 4 |
| 561–575 | 5 | 7 |
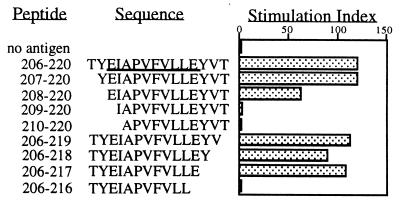
Fine mapping of peptide epitopes identified in NOD mice was performed by using variant peptides that were truncated gradually from the N and C termini of the epitopes. Three p206–220-specific and three p286–300-specific T cell hybridomas were used for characterizing the core sequences of their respective peptide epitopes. Five p221–235-specific T cell hybridomas were used for identifying their core sequence. The same core epitope was identified for all the hybridomas of a given specificity. The length of the peptide core of p206–220 and p221–235 needed for optimal T cell response is 10 residues, defined as 208–217 (Fig. 1) and 223–232 (data not shown). The peptide core for p286–300 is shorter, and a seven-residue peptide (p289–295) is able to induce a T cell response (data not shown).
Figure 1.
Identification of the peptide core sequence to p206–220-specific T cell hybridomas. T cell hybridomas were stimulated with 5 μg/ml wild-type or variant peptides for 48 hr. The ability of the hybridomas to respond to antigen was assessed on the basis of the level of IL-2 production, as detected by ELISA. The stimulation index is the ratio of IL-2 level from culture supernatants with peptides to that of culture supernatants without peptides. The underlined sequence represents the peptide core sequence of that epitope.
Mapping of Immunogenic T Cell Epitopes of GAD65 in NOD.PD Tg Mice and Identification of MHC Restriction of These T Cell Hybridomas.
The generation and screening of T cell hybridomas from NOD.PD mice (diabetes-resistant strain) were accomplished by using a protocol identical to that described in the previous section. Nine-week-old NOD.PD mice were immunized with GAD65 in IFA to generate GAD65-specific T cell hybridomas. Table 2 shows the number and frequency of GAD65-specific T cell hybridomas generated from NOD.PD mice. Eight immunogenic epitopes of GAD65 were identified in NOD.PD mice. Five of these T cell epitopes had been found previously in wild-type NOD mice. However, three of the epitopes (p456–470, p331–345, and p551–565) are newly identified epitopes. P456–470 is the dominant epitope of GAD65 derived from NOD.PD mice. Fifty-two percent of GAD65-specific T cell hybridomas derived from NOD.PD mice responded to p456–470.
Table 2.
Immunogenic T cell epitopes of GAD65 identified in NOD.PD transgenic mice
| Peptide | No. of T cell hybridomas | % (n = 81) |
|---|---|---|
| 456–470 | 42 | 52 |
| 331–345 | 5 | 6 |
| 551–565 | 8 | 9 |
| 206–220 | 3 | 4 |
| 221–235 | 2 | 2 |
| 286–300 | 11 | 13 |
| 401–415 | 3 | 4 |
| 561–575 | 7 | 8 |
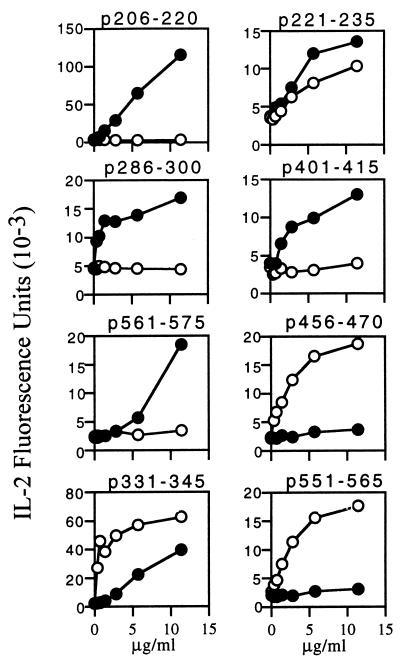
Because endogenous I-Ag7 molecules also are expressed on the surface of antigen presentation cells (APC) in NOD.PD Tg mice, it is essential to determine the MHC restriction of the T cell hybridomas derived from NOD.PD mice. To determine MHC restriction, we tested the response of individual T cell hybridomas to a specific peptide presented by M12.C3.g7 or M12.C3.g7.PD B cell transfectants. M12.C3.g7 and M12.C3.g7.PD transfectants express exclusively I-Ag7 and I-Ag7.PD molecules, respectively. If T cell hybridomas responded more strongly to their specific antigen presented by I-Ag7 or I-Ag7.PD molecules, these T cell hybridomas were considered to be restricted to I-Ag7 or to I-Ag7.PD molecules. The results of these experiments with hybridomas from NOD.PD mice are shown in Fig. 2. For the five epitopes, p206–220, p221–235, p286–300, p401–415, and p561–575, the T cell hybridomas gave a much stronger response to peptide plus I-Ag7 and gave a weak or no response to peptide plus I-Ag7.PD. We therefore refer to these five I-Ag7-restricted T cell epitopes as “g7” epitopes. For the other three epitopes (p456–470, p331–345, and p551–565), T cell hybridomas had much stronger responses to peptide plus I-Ag7.PD, but not I-Ag7 molecules. These three I-Ag7.PD-restricted T cell epitopes therefore are designated as “PD” epitopes. Several T cell hybridomas with the same peptide specificity were tested for MHC restriction and uniformly gave the same pattern for all eight epitopes. The degree of responses to p221–235 showed some extent of variation between tested T cell hybridomas. However, these p221–235-specific hybridomas consistently gave a stronger response to the peptide plus I-Ag7 than I-Ag7.PD. These g7-restricted and PD-restricted T cell epitopes of GAD65, with their amino acid sequences, are listed in Table 3.
Figure 2.
MHC restriction of GAD65-specific T cell hybridomas. T cell hybridomas were stimulated with various concentrations of specific peptides presented by M12.C3.g7 (●) or M12.C3.g7.PD (○) cells for 48 hr. The ability of the hybridomas to respond to antigen was assessed on the basis of the level of IL-2 production. The x axis represents the concentration of peptide, and the y axis represents the arbitrary fluorescence units obtained from the IL-2 ELISA.
Table 3.
g7 and PD epitopes and their amino acid sequences
| Epitope | Peptide | Sequence |
|---|---|---|
| g7 | 206–220 | TYELAPVFVLLEYVT |
| 221–235 | LKKMRFIIGWPGGSG | |
| 286–300 | KKGAAAIGIGTDSVI | |
| 401–415 | PLOCSALLVREEGLM | |
| 561–575 | ISNPAATHQDIDFLI | |
| PD | 456–470 | WLMWRAKGTTGFEAH |
| 331–345 | LVSATAGTTVYGAFD | |
| 551–565 | GDKVNFFRMVISNPA |
The Binding of g7 and PD Peptide Epitopes of GAD65 to I-Ag7 and I-Ag7.PD MHC Class II Molecules.
If allele-specific peptide binding to MHC is found for diabetes-susceptible (g7) and diabetes-resistant (PD) epitopes, it would indicate that epitope selection may play a key role in determining MHC-linked disease susceptibility and resistance. If not, other mechanisms may be involved in the control of MHC-mediated disease susceptibility.
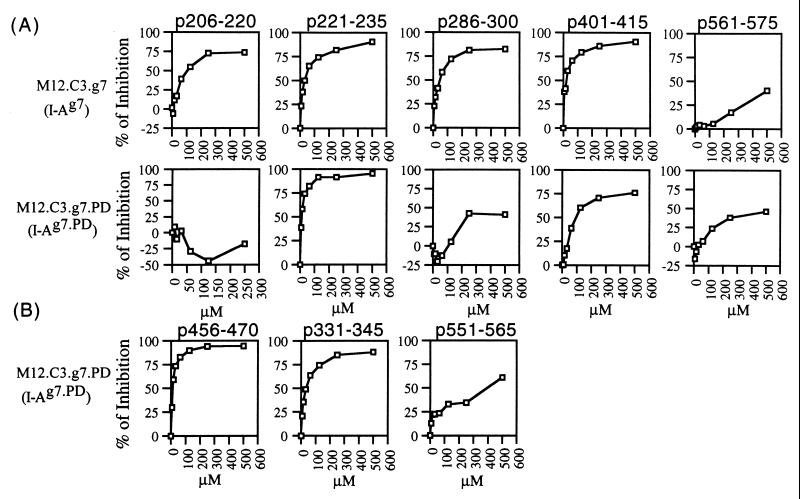
To test the hypothesis of selective binding of T cell epitopes, we measured the relative binding avidity of each g7 and PD epitope to I-Ag7 and I-Ag7.PD molecules by using a competitive peptide–MHC-binding assay. The binding results for the five g7 epitopes showed that some g7 peptides preferentially bound to I-Ag7 molecules, whereas others did not show such preferential binding (Fig. 3A). Three g7 epitopes, p206–220, p286–300, and p401–415, bound preferentially to I-Ag7 molecules. Peptide 206–220 bound exclusively to I-Ag7 molecules. Peptide 286–300 and p401–415 bound more strongly to I-Ag7 than I-Ag7.PD molecules. The other g7 peptides (p221–235 and p561–575) did not bind preferentially to either I-Ag7 or I-Ag7.PD molecules. Peptide 221–235 bound strongly to I-Ag7 and I-Ag7.PD molecules, and p561–575 bound weakly to both I-Ag7 and I-Ag7.PD molecules.
Figure 3.
The binding of g7 (A) and PD (B) epitopes to I-Ag7 and I-Ag7.PD molecules. M12.C3.g7 and M12.C3.g7.PD cells were used, respectively, as binding targets in this competitive peptide/MHC-binding assay. The inhibitory peptides are shown on the top of each pair of experiments. The x axis represents various concentrations (μM) of inhibitory peptides, and the y axis represents the percentage of inhibition, which was calculated by the following formula: % of inhibition = [(R − R1)/R] × 100%, where R = signal from assays containing only reference peptide, and R1 = signal from assays containing reference peptide and inhibitory peptide.
In contrast to the binding characteristics of the g7 epitopes, all three PD epitopes preferentially bound to I-Ag7.PD molecules (Fig. 3B). Peptides 456–470 and p331–345 bound much more strongly to I-Ag7.PD molecules than to I-Ag7. Peptide 551–565 bound exclusively to I-Ag7.PD molecules. The IC50 for all three PD epitopes binding to M12.C3.g7 are more than 500 μM (data not shown in Fig. 3B). Table 4 shows the level of relative binding capacity for g7 and PD epitopes, based on the approximate IC50 value for each epitope.
Table 4.
Relative binding capacity of GAD65 immunogenic epitopes to I-Ag7 and I-Ag7.PD class II molecules
| Peptide | Immunogenic epitope of | Binding
|
|
|---|---|---|---|
| To I-Ag7 | To I-Ag7.PD | ||
| 206–220 | g7 | +++ | − |
| 221–235 | g7 | ++++ | ++++ |
| 286–300 | g7 | ++++ | + |
| 401–415 | g7 | ++++ | +++ |
| 561–575 | g7 | + | + |
| 456–470 | PD | + | ++++ |
| 331–345 | PD | + | ++++ |
| 551–565 | PD | − | ++ |
++++, 50% inhibition obtained with 0–50 μM inhibitory peptide; +++, 50% inhibition obtained with 51–250 μM inhibitory peptide; ++, 50% inhibition obtained with 251–500 μM inhibitory peptide; +, 50% inhibition obtained with greater than 501 μM inhibitory peptide; −, no inhibition detected.
Cytokine Production by T Cells Stimulated with g7 and PD Epitopes.
I-Ag7 and I-Ag7.PD molecules identify completely different sets of immunodominant GAD65 epitopes to T cells, but the functional significance of this difference is not clear. To evaluate the qualitative aspects of GAD65-specific immune responses, NOD wild-type and NOD.PD mice were immunized with 50 μg of GAD65 in IFA, and cytokine production was determined 72 hr after restimulation in vitro of T cells from draining lymph nodes with either native antigen, a peptide pool, or individual peptide epitopes.
The cytokine response data (data not shown) demonstrated that large amounts of IFN-γ were secreted by T cells derived from both NOD wild-type and NOD.PD Tg mice when stimulated with GAD65 protein. T cells in NOD mice produced more IFN-γ when stimulated with g7 but not PD peptide pools. Peptide 206–220-specific T cells secreted the highest concentration of IFN-γ among the other g7 epitope-specific T cells derived from NOD mice. T cells in NOD.PD mice produce more IFN-γ when stimulated with PD but not g7 peptide pools. P456–470-specific T cells secreted the highest concentration of IFN-γ among the other PD epitope-specific T cells derived from NOD.PD mice.
Small amounts of IL-4 were secreted by T cells derived from both NOD wild-type and NOD.PD Tg mice when stimulated with GAD65 protein. T cells derived from NOD.PD mice produce slightly more IL-4 than those derived from NOD mice. IL-4 could not be detected when NOD- and NOD.PD-derived T cells were stimulated with peptide pools and individual peptide epitopes.
Cross-Immunization of Wild-Type NOD Mice with PD Epitopes.
“Cross-immunization” in this study refers to the injection of PD epitopes into NOD wild-type mice. The purpose of this experiment was to determine whether T cells specific for PD epitopes exist in the periphery of wild-type NOD mice. This procedure might allow us to test the hypothesis of T cell repertoire selection we proposed previously. NOD mice were immunized with two PD epitopes (p456–470 or p331–345) emulsified in complete Freund’s adjuvant (CFA) to generate PD epitope-specific, I-Ag7-restricted T cell hybridomas. As a positive control, NOD mice were immunized with a g7 epitope (p206–220) in CFA to generate p206–220-specific, I-Ag7-restricted T cell hybridomas. The results of cross-immunization experiments are summarized in Table 5. Eighty-nine and 91% of screened T cell hybridomas derived from NOD mice immunized with PD epitopes respond to their priming peptides, p456–470 and p331–345, respectively. Although the PD epitope-specific, I-Ag7-restricted T cells exist in the periphery of NOD mice, all of them failed to respond to GAD65 protein at levels as high as 50 μg/ml. In the control group, 85% of screened T cell hybridomas derived from NOD mice primed with p206–220 responded to the given peptide. Unlike PD epitope-specific, I-Ag7-restricted T cell hybridomas, the majority (83%) of the g7 epitope-specific, I-Ag7-restricted T cell hybridomas responded well to 10 μg/ml GAD65 protein.
Table 5.
Cross-immunization experiments
| Immunizing peptide | % of peptide-positive hybridomas | % of peptide-specific hybridomas responding to GAD65 protein |
|---|---|---|
| 456–470 (PD epitope) | 448/405 = 89% | 0/41 = 0% |
| 331–345 (PD epitope) | 458/504 = 91% | 0/48 = 0% |
| 206–220 (g7 epitope) | 144/168 = 85% | 40/48 = 83% |
NOD mice were immunized with two PD epitopes (p456–470 or p331–345), 50 μg emulsified in complete Freund’s adjuvant (CFA), to generate PD epitope-specific. I-Ag7 restricted T cell hybridomas. In another group, NOD mice were immunized with a g7 epitope (p206–p220), 50 μg in CFA, to generate p206–220-specific, I-Ag7-restricted T cell hybridomas. The percentage of peptide-positive hybridomas represents the ratio of the number of peptide-responding T cell hybridomas to the total number of screened hybridomas. The percentage of peptide-specific hybridomas responding to GAD65 protein represents the ratio of the number of peptide-responding T cell hybridomas responding to GAD65 to the total number of peptide-responding T cell hybridomas that were screened.
DISCUSSION
The experiments reported here demonstrate that I-Ag7 and I-Ag7.PD molecules identify completely different epitopes of GAD65 to T cells. These studies in I-Aβ Tg mice clearly demonstrate that a 2-aa difference in the α-helical domain of the β-chains of I-Ag7 and I-Ag7.PD can dramatically affect the specificity of autoreactive T cell responses to the islet cell antigen GAD65. These differences may be central to our understanding of the disease susceptibility mediated by I-Ag7 and the protective effect mediated by I-Ag7.PD alleles.
N- and C-terminal truncation data demonstrate the minimum number of residues required for T cell responses. Although the truncation experiments do not identify the potential sites for T cell and MHC contact with peptides, this information helps us predict a potential peptide-binding motif for I-Ag7 molecules. I-Ag7-binding (19) and HLA-DRB1*0405-binding (20–22) motifs have been identified by acid elution of peptides from MHC molecules. HLA-DRB1*0405 encodes a β-chain lacking an aspartic acid at position 57, similar to the I-Ag7 allele in NOD mice. In comparing the immunogenic peptides identified in our study with the I-Ag7 and DRB1*0405 peptide-binding motifs, none of these peptides completely fit the characterized DRB1*0405 and I-Ag7 motifs. However, it is not unusual for the amino acid sequence of immunogenic epitopes not to correlate with the sequence of predicted motifs. Because the motifs are based on peptide elution studies and thus select for abundant peptides, it is likely that the motifs correspond to peptides that bind with high affinity. It is possible that some autoreactive T cells specific for high-affinity, motif-fitting GAD65 peptides may have been eliminated in the thymus by negative selection. As a result, these peptides may not be detected as immunogenic for peripheral T cells.
The data shown in Fig. 3 and Table 4 demonstrate that peptide binding alone cannot account for the immunodominance of different epitopes of GAD65 identified by I-Ag7 and I-Ag7.PD molecules. Peptide 206–220, the most dominant epitope selected by I-Ag7 molecules, is not the epitope that has the highest binding affinity among the g7 epitopes. In addition, two g7 epitopes are able to bind well both I-Ag7 and I-Ag7.PD molecules. Even though these two sets of epitopes show some degree of preferential binding to their I-A molecules, only p206–220 (g7 epitope) and p551–565 (PD epitope) show exclusive binding to I-Ag7 and I-Ag7.PD molecules, respectively. These observations imply that other mechanisms are involved in the selection of distinct T cell epitopes by susceptible and resistant MHC alleles.
It has been proposed that the development of insulin-dependent diabetes is controlled by the Th1 vs. Th2 phenotype of autoreactive Th cells: Th1 cells would promote diabetes, whereas Th2 cells would protect from disease (23, 24). However, cytokine experiments reveal that there are no qualitative differences in the cytokine profiles of T cells derived from NOD and NOD.PD mice after stimulation with GAD65. T cells from both strains produce Th1-like cytokines. It is surprising that a large amount of Th1 cytokine (IFN-γ) was produced by GAD65-specific T cells in diabetes-resistant NOD.PD mice. One explanation may be that although GAD65-specific T cells secrete inflammatory Th1 cytokines in NOD.PD mice, other islet antigen-specific T cells may secrete Th2-like cytokines that delay or block the process of type I diabetes. The age of the mice is also critical for the disease process. T cells of the same antigen specificity may have different cytokine profiles at different ages. Furthermore, immunogenic epitopes from autoantigens have been shown to differ in mice of different ages because of epitope spreading (13). It is also possible that distinct T cell epitopes of islet cell antigens may elicit different effector functions.
Cross-immunization experiments with the PD epitope p456–470 demonstrated that this peptide is also immunogenic in wild-type NOD mice if the peptide epitope is used for immunization and T cell stimulation. However, I-Ag7-restricted T cell hybridomas specific for p456–470 (which were not identified in NOD wild-type mice after immunization with native protein) responded only to synthetic peptide, not the protein antigen. In identical studies of the second PD epitope, p331–345, similar findings were observed. Why do PD peptide-specific, I-Ag7-restricted T cell hybridomas respond only to GAD65 peptide, but not whole protein? Three possibilities may explain this phenomenon. First, the PD epitopes may be functionally recessive (cryptic) in NOD mice. In other words, these two PD epitopes may not be processed and presented efficiently on the surface of I-Ag7-expressing APC in NOD wild-type mice. Thus, antigen-specific T cells never have the opportunity to meet the processed peptide antigens. It is evident that the hierarchy of display of peptide determinants on a protein antigen can differ greatly in different alleles of MHC molecules, as well as in the same MHC molecule on APC in different locations (25, 26).
Second, differences may exist in the frequency of peripheral TCR in NOD wild-type and NOD.PD Tg mice that are specific for the naturally processed peptide/MHC complex (derived from protein antigen), the synthetic peptide/MHC complex, or both. Viner et al. (27) showed that the interaction of free peptides with class II MHC molecules can generate complexes that are antigenically dissimilar to those resulting from intracellular processing of intact antigens. It is possible that the T cells responding to synthetic peptide antigens are different from T cells that are able to recognize naturally processed epitopes. This phenomenon has been described as type A (response to peptide and protein antigen) and type B (exclusively to peptide) T cells by Unanue and his colleagues (28). The third possibility is that these PD epitope-specific, I-Ag7-restricted T cells have a very low binding affinity to PD epitopes on I-Ag7 molecules. Therefore, the T cells responding to PD epitopes in NOD mice can be detected only after administration of a large amount of peptides in adjuvant or aqueous form (data not shown).
Based on the results of this study, it is possible that selective binding of T cell peptide epitopes, the ability to process/present antigen, and selection of a distinct T cell repertoire may all be involved in the control of MHC-linked susceptibility. Susceptibility alleles (I-Ag7) predispose to type I diabetes by allowing binding and presentation of pathogenic self-peptides as well as allowing the development of an autoreactive T cell repertoire in the periphery. In contrast, the lack of pathogenic T cell responses in resistant strains (individuals) is due to the absence or suppression of one or more of these three capabilities. Furthermore, the control mechanisms involved in the selection of autoantigen T cell epitopes are complex and appear to differ for each peptide epitope. For p206–220, peptide binding plays an important role in recognition of this epitope by T cells in diabetes-susceptible mice. On the other hand, p221–235 binds equally well to I-Ag7 and I-Ag7.PD molecules. Therefore, other mechanisms also can play critical roles in MHC control of disease susceptibility. These include on/off rate of the peptide/MHC complex, the half-life of this complex, and the effect of H-2M molecules on antigen presentation by susceptible and resistant alleles. All of these factors can have major effects on peptide/MHC complex surface expression and on Th1/Th2 differentiation.
The present experiments cannot resolve these three possibilities. To further analyze this problem, it will be necessary to measure on/off rates of individual peptide epitopes for each MHC allele; the effect of H-2M molecules on antigen presentation; and the time course of the peptide-specific T cell response and its functional characteristics, by using tetramers of I-Ag7- and I-Ag7.PD-specific peptide complexes, to trace each antigen-specific T cell. Generation of TCR Tg mice for the three g7- and two PD-dominant epitopes will permit further characterization of the response to these five epitopes. The knowledge from these studies will help us understand how I-A alleles predispose to or prevent the development of type I diabetes.
Acknowledgments
We thank Drs. O. Kanagawa and E. Unanue (Washington University) for kindly providing M12.C3.g7 and M12.C3.g7PD B cell transfectants. C.-C.C. was supported by an American Diabetes Association mentor-based fellowship. This work was supported by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases.
ABBREVIATIONS
- GAD65
glutamic acid decarboxylase 65
- TCR
T cell receptor
- APC
antigen presentation cells
- Tg
transgenic
- Th
T helper
- IFA
incomplete Freund’s adjuvant
- NOD mice
nonobese diabetic mice
References
- 1.Castano L, Eisenbarth G S. Annu Rev Immunol. 1990;8:647–680. doi: 10.1146/annurev.iy.08.040190.003243. [DOI] [PubMed] [Google Scholar]
- 2.Hattori M, Buse J B, Jackson R A, Glimcher L, Dorf M E, Minami M, Makini S, Moriwaki K, Kuzuya H, Imura H, et al. Science. 1986;231:733–736. doi: 10.1126/science.3003909. [DOI] [PubMed] [Google Scholar]
- 3.Acha-Orbea H, McDevitt H O. Proc Natl Acad USA. 1987;84:2435–2439. doi: 10.1073/pnas.84.8.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slattery R M, Kjer-Nielsen L, Allison J, Charlton B, Mandel T E, Miller J F A P. Nature (London) 1990;345:724–726. doi: 10.1038/345724a0. [DOI] [PubMed] [Google Scholar]
- 5.Miyazaki T, Uno M, Kikutani H, Kishimoto T, Kimoto M, Nishimoto H, Miyazaki J, Yamamura K. Nature (London) 1990;345:722–724. doi: 10.1038/345722a0. [DOI] [PubMed] [Google Scholar]
- 6.Singer S M, Tisch R, Yang X-D, McDevitt H O. Proc Natl Acad Sci USA. 1993;90:9566–9570. doi: 10.1073/pnas.90.20.9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lund T, O’Reilly L, Hutchings P, Kanagawa O, Simpson E, Gravely R, Chandler P, Dyson J, Picard J K, Edwards A, et al. Nature (London) 1990;345:727–729. doi: 10.1038/345727a0. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt D, Verdaguer J, Averill N, Santamaria P. J Exp Med. 1997;186:1059–1075. doi: 10.1084/jem.186.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luhder F, Katz J, Benoist C, Mathis D. J Exp Med. 1998;187:379–387. doi: 10.1084/jem.187.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanagawa O, Vaupel B A, Xu G, Unanue E R, Katz J D. J Immunol. 1998;161:4489–4492. [PubMed] [Google Scholar]
- 11.Singer S M, Tisch R T, Yang X D, Sytwu H K, Liblau R, McDevitt H O. Diabetes. 1998;47:1570–1577. doi: 10.2337/diabetes.47.10.1570. [DOI] [PubMed] [Google Scholar]
- 12.Tisch R, Yang X-D, Singer S M, Liblau R S, Fugger L, McDevitt H O. Nature (London) 1993;366:72–75. doi: 10.1038/366072a0. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman D L, Clare-Salzler M, Tian J, Forsthuber T, Ting G, Robinson P, Atkinson M A, Sercarz E E, Tobin A, Lehmann P V. Nature (London) 1993;366:69–72. doi: 10.1038/366069a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tisch R, Liblau R S, Yang X D, Liblau P, McDevitt H O. Diabetes. 1998;47:894–899. doi: 10.2337/diabetes.47.6.894. [DOI] [PubMed] [Google Scholar]
- 15.Tian J, Clare-Salzler M, Herschenfeld A, Middleton B, Newman D, Mueller R, Arita S, Evans C, Atkinson M A, Mullen Y, et al. Nat Med. 1996;2:1348–1353. doi: 10.1038/nm1296-1348. [DOI] [PubMed] [Google Scholar]
- 16.Chao C C, McDevitt H O. Immunogenetics. 1997;46:29–34. doi: 10.1007/s002510050238. [DOI] [PubMed] [Google Scholar]
- 17.Buckner J, Kwok W W, Nepom B, Nepom G T. J Immunol. 1996;157:4940–4945. [PubMed] [Google Scholar]
- 18.Buus S, Sette A, Colon S M, Miles C, Grey H M. Science. 1987;235:1353–1358. doi: 10.1126/science.2435001. [DOI] [PubMed] [Google Scholar]
- 19.Reich E P, von Grafenstein H, Barlow A, Swenson K E, Williams K, Janeway C A. J Immunol. 1994;152:2279–2288. [PubMed] [Google Scholar]
- 20.Ramensee H-C, Friede T, Stevanovic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita S, Takahashi K, Motoki M, Komoriya K, Ikagawa S, Nishimura Y. J Exp Med. 1994;180:873–883. doi: 10.1084/jem.180.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinouchi R, Kobayashi H, Sato K, Kimura S, Katagiri M. Immunogenetics. 1994;40:376–378. doi: 10.1007/BF01246679. [DOI] [PubMed] [Google Scholar]
- 23.Katz J D, Wang B, Haskins K, Benoist C, Mathis D. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 24.Katz J D, Benoist C, Mathis D. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- 25.Sercarz E E, Lehmann P, Ametani A, Benichou G, Miller A, Moudgil K. Annu Rev Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 26.Schneider S C, Sercarz E E. Hum Immunol. 1997;54:148–158. doi: 10.1016/s0198-8859(97)00079-7. [DOI] [PubMed] [Google Scholar]
- 27.Viner N J, Nelson C A, Deck B, Unanue E R. J Immunol. 1996;156:2365–2368. [PubMed] [Google Scholar]
- 28.Nelson C A, Unanue E R. Immunol Rev. 1996;151:81–105. doi: 10.1111/j.1600-065x.1996.tb00704.x. [DOI] [PubMed] [Google Scholar]