Short abstract
Our aim was to identify specifically expressed genes using RNA arbitrarily primed (RAP)-polymerase chain reaction (PCR) for differential display in patients with rheumatoid arthritis (RA). In RA, amplification of a distinct PCR product suitable for sequencing could be observed. Sequence analysis identified the PCR product as highly homologous to a 434 base pair segment of the human centromere kinesin-like protein CENP-E. Differential expression of CENP-E was confirmed by quantitative reverse transcription PCR, immunohistochemistry and in situ hybridization. CENP-E expression was independent from prednisolone and could not be completely inhibited by serum starvation. RAP-PCR is a suitable method to identify differentially expressed genes in rheumatoid synovial fibroblasts. Also, because motifs of CENP-E show homologies to jun and fos oncogene products and are involved in virus assembly, CENP-E may be involved in the pathophysiology of RA.
Keywords: arthritis, centromere, differential display, immunohistochemistry, in situ hybridization, RNA fingerprinting
Abstract
Introduction:
Articular destruction by invading synovial fibroblasts is a typical feature in rheumatoid arthritis (RA). Recent data support the hypothesis that key players in this scenario are transformed-appearing synovial fibroblasts at the site of invasion into articular cartilage and bone. They maintain their aggressive phenotype toward cartilage, even when first cultured and thereafter coimplanted together with normal human cartilage into severe combined immunodeficient mice for an extended period of time. However, little is known about the upregulation of genes that leads to this aggressive fibroblast phenotype. To inhibit this progressive growth without interfering with pathways of physiological matrix remodelling, identification of pathways that operate specifically in RA synovial fibroblasts is required. In order to achieve this goal, identification of genes showing upregulation restricted to RA synovial fibroblasts is essential.
Aims:
To identify specifically expressed genes using RNA arbitrarily primed (RAP)-polymerase chain reaction (PCR) for differential display in patients with RA.
Methods:
RNA was extracted from cultured synovial fibroblasts from 10 patients with RA, four patients with osteoarthritis (OA), and one patient with psoriatic arthritis. RAP-PCR was performed using different arbitrary primers for first-strand and second-strand synthesis. First-strand and second-strand synthesis were performed using arbitrary primers: US6 (5' -GTGGTGACAG-3') for first strand, and Nuclear 1+ (5' -ACGAAGAAGAG-3'), OPN28 (5' -GCACCAGGGG-3'), Kinase A2+ (5' -GGTGCCTTTGG-3')and OPN24 (5' -AGGGGCACCA-3') for second-strand synthesis. PCR reactions were loaded onto 8 mol/l urea/6% polyacrylamide-sequencing gels and electrophoresed.Gel slices carrying the target fragment were then excised with a razor blade, eluated and reamplified. After verifying their correct size and purity on 4% agarose gels, the reamplified products derived from the single-strand confirmation polymorphism gel were cloned, and five clones per transcript were sequenced. Thereafter, a GenBank® analysis was performed. Quantitative reverse transcription PCR of the segments was performed using the PCR MIMIC® technique.In-situ expression of centromere kinesin-like protein-E (CENP-E) messenger (m)RNA in RA synovium was assessed using digoxigenin-labelled riboprobes, and CENP-E protein expression in fibroblasts and synovium was performed by immunogold-silver immunohistochemistry and cytochemistry. Functional analysis of CENP-E was done using different approaches (eg glucocorticoid stimulation, serum starvation and growth rate analysis of synovial fibroblasts that expressed CENP-E).
Results:
In RA, amplification of a distinct PCR product suitable for sequencing could be observed. The indicated complementary DNA fragment of 434 base pairs from RA mRNA corresponded to nucleotides 6615-7048 in the human centromere kinesin-like protein CENP-E mRNA (GenBank® accession No. emb/Z15005).The isolated sequence shared greater than 99% nucleic acid (P = 2.9e-169) identity with the human centromere kinesin-like protein CENP-E. Two base changes at positions 6624 (A to C) and 6739 (A to G) did not result in alteration in the amino acid sequence, and therefore 100% amino acid identity could be confirmed. The amplification of 10 clones of the cloned RAP product revealed the presence of CENP-E mRNA in every fibroblast culture examined, showing from 50% (271.000 ± 54.000 phosphor imager arbitrary units) up to fivefold (961.000 ± 145.000 phosphor imager arbitrary units) upregulation when compared with OA fibroblasts. Neither therapy with disease-modifying antirheumatic drugs such as methotrexate, gold, resochine or cyclosporine A, nor therapy with oral steroids influenced CENP-E expression in the RA fibroblasts. Of the eight RA fibroblast populations from RA patients who were receiving disease-modifying antirheumatic drugs, five showed CENP-E upregulation; and of the eight fibroblast populations from RA patients receiving steroids, four showed CENP-E upregulation.
Numerous synovial cells of the patients with RA showed a positive in situ signal for the isolated CENP-E gene segment, confirming CENP-E mRNA production in rheumatoid synovium, whereas in OA synovial tissue CENP-E mRNA could not be detected. In addition, CENP-E expression was independent from medication. This was further confirmed by analysis of the effect of prednisolone on CENP-E expression, which revealed no alteration in CENP-E mRNA after exposure to different (physiological) concentrations of prednisolone. Serum starvation also could not suppress CENP-E mRNA completely.
Discussion:
Since its introduction in 1992, numerous variants of the differential display method and continuous improvements including RAP-PCR have proved to have both efficiency and reliability in examination of differentially regulated genes. The results of the present study reveal that RAP-PCR is a suitable method to identify differentially expressed genes in rheumatoid synovial fibroblasts.
The mRNA, which has been found to be upregulated in rheumatoid synovial fibroblasts, codes for a kinesin-like motor protein named CENP-E, which was first characterized in 1991. It is a member of a family of centromere-associated proteins, of which six (CENP-A to CENP-F) are currently known. CENP-E itself is a kinetochore motor, which accumulates transiently at kinetochores in the G2 phase of the cell cycle before mitosis takes place, appears to modulate chromosome movement and spindle elongation,and is degraded at the end of mitosis. The presence or upregulation of CENP-E has never been associated with RA.
The three-dimensional structure of CENP-E includes a coiled-coil domain. This has important functions and shows links to known pathways in RA pathophysiology. Coiled-coil domains can also be found in jun and fos oncogene products, which are frequently upregulated in RA synovial fibroblasts. They are also involved in DNA binding and transactivation processes resembling the situation in AP-1 (Jun/Fos)-dependent DNA-binding in rheumatoid synovium. Most interestingly, these coiled-coil motifs are crucial for the assembly of viral proteins, and the upregulation of CENP-E might reflect the influence of infectious agents in RA synovium. We also performed experiments showing that serum starvation decreased, but did not completely inhibit CENP-E mRNA expression. This shows that CENP-E is related to, but does not completely depend on proliferation of these cells. In addition, we determined the growth rate of CENP-E high and low expressors, showing that it was independent from the amount of CENP-E expression. supporting the statement that upregulation of CENP-E reflects an activated RA fibroblast phenotype. In summary, the results of the present study support the hypothesis that CENP-E, presumably independently from medication, may not only be upregulated, but may also be involved in RA pathophysiology.
Introduction
Inflammation, altered cellular and humoral immune response, and synovial hyperplasia are typical findings in rheumatoid synovium pathophysiology [1]. On the other hand, there is increasing evidence that T-cell independent pathways, such as upregulation of proto-oncogenes, production of growth factors and the release of matrix-degrading enzymes, lead to progressive destruction of the affected joints [2]. Recent data [3] support the hypothesis that key players in this scenario are transformed-appearing synovial fibroblasts at the site of invasion into articular cartilage and bone. They maintain their aggressive phenotype toward cartilage even when first cultured and thereafter coimplanted together with normal human cartilage into severe combined immunodeficiency mice for an extended period.
To inhibit this progressive growth without interfering with pathways of physiological matrix remodelling, identification of pathways that operate specifically in rheumatoid arthritis (RA) synovial fibroblasts, and not in synovial fibroblasts of other origin [eg those of osteoarthritis (OA)], is required. To achieve this goal, identification of genes that show upregulation that is restricted to RA synovial fibroblasts is essential. Various strategies have been developed to examine tissue-specific gene differences in gene expression. Among these, the differential display approach of RNA arbitrarily primed (RAP)-polymerase chain reaction (PCR) [4] has been proved to be both efficient and reliable in numerous experimental settings [5].
In this study, we analyzed the differential expression of RNAs of RA fibroblasts versus OA fibroblasts derived from synovium of patients undergoing reconstructive surgery or synovectomy.
Materials and methods
Synovial tissue and cell culture
Synovial fibroblasts and tissue were obtained from synovial biopsies of a total of 14 patients (Table 1). Ten patients who met the criteria of the American College of Rheuma-tology for RA [6] were undergoing joint surgery (synovec-tomy or joint replacement by prosthesis implantation), and four patients with long-term OA were undergoing joint surgery (prosthesis implantation) because of severe articular dysfunction. In two patients with RA, fibroblast cultures were obtained not only from the proliferating synovium, but also separately from synovium that was extracted from visible, deep intraosseus invasion areas.
Table 1.
Clinical data of patients
| Patient | Age | Male/female | ESR (mm/1st hour) | WBC (leucocytes/nl) | Medication |
| RA02 | 58 | F | 15 | 16 | M, O |
| RA03 | 59 | F | 85 | 16.5 | C, O |
| RA04 | 37 | M | 26 | 8.8 | G, O |
| RA06 | 55 | M | 37 | 7.8 | M, O |
| RA08 | 66 | F | 45 | 8.7 | - |
| RA09 | 55 | F | 29 | 14.5 | - |
| RA10 | 46 | F | 25 | 11.7 | G, O |
| RA17 | 68 | M | 13 | 6.7 | O |
| RA21 | 31 | M | 26 | 9.7 | G |
| RA22 | 70 | F | 20 | 12.5 | G, O |
| OA02 | 76 | F | 9 | 10.4 | - |
| OA03 | 74 | F | 20 | 8.1 | - |
| OA05 | 65 | M | 13 | 7.1 | - |
| OA06 | 74 | M | 14 | 5.5 | - |
Abbreviations of disease-modifying antirheumatic drugs: C, cyclosporin A; G, gold; M, methotrexate; O, oral steroids; R, resochine; S, sulphasalazine.
Culture of synovial fibroblasts was performed as described recently [3]. In brief, after enzymatic digestion, fibroblasts were grown in culture flasks in Dulbeccos Modified Eagles Medium-Cellgro (Mediatech, Washing-ton, DC, USA) containing 10% fetal calf serum (Gibco Life Technologies, Grand Island, NY, USA) and cultured for four passages. To exclude contamination, synovial fibroblasts were stained for fibroblast markers by immunocytochemistry [> 95% could be stained positively for the fibroblast enzyme prolyl-4-hydroxylase, and none were positive for the macrophage marker CD68 or the neutrophil marker cathepsin G (data not shown)] and tested for mycoplasmas. At 70–80% confluency, cells were harvested and DNA was extracted as outlined below and stored at -70°C. Synovial tissue used for immunohistochemical analysis was immediately snap frozen and stored at -70°C.
RNA extraction
Total cellular RNA was extracted using the RNeasy spin column purification kit (Qiagen, Hilden, Germany). To remove contaminating genomic DNA, the total RNA was treated with DNase I (0.2U/μl; Boehringer Mannheim, Hannheim, Germany). RNA concentrations were measured spectrophotometrically at 260 nm and adjusted, and equal aliquots were then electrophoresed on 1% agarose gels stained with ethidium bromide to compare large and small ribosomal RNAs qualitatively and to exclude degradation. When starting with fresh RNAs, we performed one RAP-PCR, leaving out the reverse transcriptase as a control for DNA contamination.
RNA arbitrarily primed polymerase chain reaction of total cellular RNA
RAP-PCR of total cellular RNA was performed as previously described [4]. In brief, three different concentrations of RNA (500, 250 and 100 ng) were used as templates for each experiment. First-strand synthesis was carried out using 2 μmol/l first-strand arbitrary primer (for sequence see below), and second-strand synthesis was performed using 4 μmol/l arbitrary second primer and subsequently cycled through 35 low-stringency cycles. Arbitrary primers used were US6 (5' -GTGGTGACAG-3') for first-strand synthesis, and Nuclear 1+(5' -ACGAAGAAGAG-3'), OPN28 (5' -GCACCAGGGG-3'), kinase A2+ (5' -GGTGCCTTTGG-3') and OPN24 (5' -AGGGGCACCA-3') for second-strand synthesis. PCR products were loaded onto 8 mol/l urea/6% polyacrylamide sequencing gels. Electrophoresis was performed for 4–6 h at 50 W in 1 × Tris-borate EDTA buffer. Gels were then transferred to 3MM Whatman paper, dried under vacuum at 80°C, and directly exposed to Kodak BioMax™ autoradiography film (Kodak, Stuttgart, Germany) at room temperature for 12–72 h, depending on the intensity of radiation of the amplified fragments.Several luminescence labels (autoradiogram markers; Stratagene, San Diego, CA, USA) were attached to the gel to facilitate alignment of the autoradiograms with the gels for subsequent isolation of differentially displayed gene fragments.
Isolation and purification of differentially amplified polymerase chain reaction products
Gel slices carrying the target fragment were then excized with a razor blade and placed in 50 μl TE buffer (10 mmol/l Tris-HCl, 1 mmol/l EDTA, pH8.0). DNA was eluted by incubating at 65°C for 3 h. Eluates were taken for reamplification of the gene fragment using the primers of the original fingerprint and the conditions outlined above for 20 cycles. PCR products were routinely checked by denaturing polyacrylamide gel electrophoresis, loading the reamplified product next to the original fingerprint to verify its size and purity. To exclude contamination of nondifferentially regulated gene products of similar size to that of the regulated transcripts, thereafter we used native polyacrylamide gels to separate the sequences of the reamplified mixture based on single-strand confirmation polymorphism (SSCP) as described previously [7]. After second identification of the gene segment using this procedure, it was cut from the SSCP gel and reamplified a second time.
Cloning, Southern blot and sequencing
After verifying its correct size and purity on 4% agarose gels, the reamplified products derived from the SSCP gel were cloned into PCR®-II Topo using the TOPO-TA-Cloning® Kit DUALPromoter (Invitrogen, De Schelp, The Nether-lands). After blue-white screening of the clones, 10 white colonies and one blue colony were picked and suspended in 50 μl water. Aliquots of these bacterial suspensions were checked by high-stringency PCR for the presence and the correct length of inserts using the T7 and the M13 (or M20) reverse sequencing primers. Clones of the correct length were subsequently grown overnight in 5ml LB medium containing 50 μg/ml of ampicillin for plasmid isolation. Five clones per transcript were sequenced with the Applied Biosystems 373 automatic sequencer using the Perkin Elmer (Norwalk, CT, USA) DNA sequencing kit.The databases of the National Center for Biological Information were screened to align the obtained sequences with known complementary DNA clones, genomic clones and cloned expressed sequence tags.
If sequences were multiply represented and confirmed within the majority of the clones resulting from one RAP-PCR product, inserts were reamplified and used as probes against Southern blots of the original fingerprint gel. This procedure confirms that selected sequences were in fact differentially amplified in the original fingerprint gels. For this, DNA from RAP-PCR fingerprint gels was transferred to nylon membranes (Duralon-UV, Stratagene, San Diego, CA, USA) by capillary action in a 10 × saline sodium citrate (SSC) buffer overnight. After ultraviolet cross-linking, blots were prehybridized and hybridized using established protocols [8].
Confirmation of differential expression by quantitative reverse transcription polymerase chain reaction
Quantitative reverse transcription PCR was performed using the PCR MIMIC®technique (Clontech, Palo Alto, CA, USA). In this competitive PCR method, one set of primers is used to amplify both the target complementary DNA and the nonhomologous internal standard. As the amplified PCR fragments are designed to be different in length, they can be distinguished by gel electrophoresis. Using this method, relative transcript abundances of the different fibroblast populations could be compared after standardization of the coamplified highly abundant internal standard DNA. Radioactive PCR products amplified using centromere kinesin-like protein-E (CENP-E)-specific primers were evaluated by phosphor imaging densitometry (Phosphor Imager; Molecular Dynamics, Sunnyvale, CA, USA) and subsequent data analysis was performed using the Ambis software (Imagequant; Molecular Dynamics). Each PCR amplification was performed in quadruplicate, and relative amount was calculated as phosphor imager arbitrary units (PAU) of CENP-E amplicons of the RA and the OA fibroblast cultures.
Mean ± standard error of the mean was calculated for each of the individual RA fibroblast cultures and compared with the CENP-E production of all OA synovial fibroblast cultures. Statistical analysis was performed using the Mann-Whitney test for nonpaired parameters. P < 0.05 was regarded as statistically significant.
In-situ expression of CENP-E messenger RNA in rheumatoid arthritis synovium
Plasmids containing the isolated CENP-E gene segment were extracted and purified using Nucleobond-AX-Columns (Macherey-Nagel, Düren, Germany) and linearized to permit generation of antisense and sense riboprobes. In-situ hybridization was performed as published recently [3,9]. In brief, antisense and sense RNA probes were transcribed by T3 and T7 RNA polymerase (Stratagene). Probes were labelled with digoxigenin-uridine triphosphate (Boehringer Mannheim). Frozen sections (4–6 μm thick) were cut and fixed in 3% buffered paraformaldehyde for 1h at room temperature(20–22°C). The sections were rinsed in 2 × SSC/0.25% acetic anhydride (Fisher Scientific, Springfield, NJ, USA) for 15 min at room temperature. After a rinsing step with 0.1mol/l tri-ethanolamine-HCl (pH8.0), prehybridization was performed. After the prehybridization, digoxigenin-labelled antisense or sense probe (for control) was applied to the tissue specimens in 15 l prehybridization buffer. The slides were sealed with nail polish and hybridized for 12 h in a humid chamber at 50°C. Then slides were washed at room temperature with SSC and sodium chloride-Tris-EDTA (STE) buffer. After digestion for 1 h at 37°C with 20 g/ml RNAse A (Boehringer Mannheim), slides were rinsed with 2 × SSC, 50% formamide for 5 min, followed by 1 × SSC, 0.1% sodium dodecyl sulphate for 10 min, and 0.5 × SSC, 0.1% sodium dodecyl sulphate for 15 min at 50°C. The slides were washed and developed using antidigoxigenin-5 nm gold-labelled antibody complex (Goldmark Biologicals, Phillipsburg, NJ, USA) according to a modification to the protocol of Komminoth et al [9].
Immunogold-silver immunohistochemistry for CENP-E
RA and OA synovial fibroblasts cultured in chamber wells and snap frozen sections (4–6 μm thick) were fixed for 5 min in acetone and covered with a 4% milk, 2% normal horse serum Tris buffer for 30 min at room temperature (20–22°C) to block nonspecific binding. Then, slides were washed and incubated with monoclonal mouse anti-CENP-E-antibody [10,11], or antifibroblast antibodies (Dianova, Hamburg, Germany). The slides were washed and incubated with a biotinylated goat antimouse antibody (Dianova) diluted 1 : 600 in Tris buffer. After washing, sections were incubated with peroxidase-conjugated streptavidin (Dianova), diluted 1 : 600 in Tris buffer. After a rinsing step, 6nm gold-labelled goat antihorseradish peroxidase (Dianove), diluted 1 : 40 in Tris buffer was applied. Signal detection by the immunogold-silver technique was performed as described above.
Immunohistochemical double labelling (alkaline phosphatase anti-alkaline phosphatase method)
Double labelling was performed using the alkaline phosphatase anti-alkaline phosphatase method with mono-clonal antibodies against human fibroblasts (Dianova). Sections were covered with a 4% milk, 2% normal horse serum buffer for 30 min at room temperature to block nonspecific binding, washed in Tris-NaCl (pH7.5) and incubated for 45 min at room temperature (20–22°C) with the primary antibodies diluted 1 : 50 to 1 : 100 in Tris-NaCl (pH 7.5). Colour development was performed as previously published [3].
Negative controls were performed in each of the techniques described above by omitting the primary antibody, incubation with isotype-matched controls or using the sense probe in the in situ hybridization assay.
Glucocorticoid stimulation
Three of the RA fibroblast populations showing upregulation of CENP-E (RA08, RA09 and RA21) and three populations showing no upregulation (RA10, RA17 and RA22) were adjusted to 0.5 × 106 cells and incubated with physiological concentrations of 10-7 and 10-9 mmol/l prednisolone for 6 h. Thereafter, CENP-E messenger (m)RNA was measured using reverse transcription PCR as outlined above.
Serum starvation
Three of the RA fibroblast populations showing upregulation of CENP-E (RA08, RA09 and RA21) and three populations showing no upregulation (RA10, RA17 and RA22) were adjusted to 0.5 × 106 cells and were cultured for 72 h without fetal calf serum. Thereafter, CENP-E mRNA was measured using reverse transcription PCR as outlined above.
Growth rate
Cells (105) of RA fibroblast populations showing upregulation of CENP-E (RA08, RA09 and RA21) and showing no upregulation (RA10, RA17 and RA22) were cultured in quintuplicate in 24 well plates. To evaluate the growth rate, cell counts were performed on days 2, 3, 4 and 5, and increase in cell numbers from day to day was evaluated using a Neubauer counting chamber and measured as percentage per day.
Results
RNA arbitrarily primed polymerase chain reaction
Total RNA, prepared from synovial fibroblasts from 10 patients with RA and four patients with OA, was used for RAP-PCR. In total, approximately 150 RNAs were amplified using RAP-PCR per primer pair, of which most were expressed both by RA and OA synovial fibroblasts. Approximately 2% of the RNAs showed a polymorphism within the RA group. Five RAP-products were differentially expressed between RA and nonRA synovial fibroblasts, and one PCR product was suitable for further analysis. The autoradiography of the original RAP-PCR gel is shown in Figure 1, and the arrowheads indicate the complementary DNA fragment that we isolated for further characterization.
Figure 1.
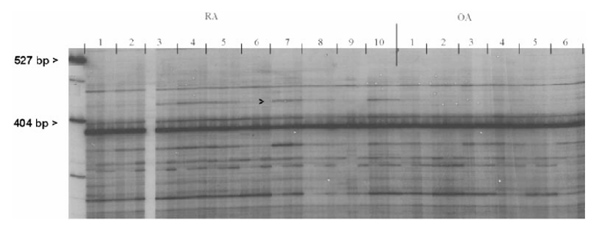
Original fingerprint [RNA arbitrarily primed (RAP)-polymerase chain reaction (PCR)] of RNA of synovial fibroblasts derived from 10 different patients with rheumatoid arthritis and six patients with osteoarthritis. Each RAP-PCR was performed in triplicate using three different RNA concentrations. Left two lanes: DNA marker. Triplicate lanes 1–10 show the RAP-PCR of the synovial fibroblasts of the 10 rheumatoid arthritis patients, and lanes 11–16 show the RAP-PCR of those of the osteoarthritis patients. The arrow indicates the differentially displayed gene fragment seen in the rheumatoid arthritis patients (434 base pairs), which was used for subsequent analysis.
Cloning and sequencing of differentially amplified RNA arbitrarily primed product
The indicated complementary DNA fragment of 434 base pairs from RA mRNA corresponded to nucleotides 6615-7048 in the human centromere kinesin-like protein CENP-E mRNA (GenBank®accession No. emb/Z15005) The RAP-PCR product was located entirely within the open reading frame. The isolated sequence shared greater than 99% nucleic acid (P = 2.9e-169) identity with the human centromere kinesin-like protein CENP-E. Two base changes at positions 6624 (A to C) and 6739 (A to G) did not result in alteration of the amino acid sequence, and therefore 100% amino acid identity could be confirmed. The amplification of 10 clones of the cloned RAP product revealed the presence of CENP-E mRNA in every fibroblast culture examined.
We confirmed the predicted upregulation of CENP-E in RA using quantitative reverse transcription PCR. When analyzed by phosphor imaging densitometry, significant upregulation of CENP-E mRNA production in RA synovial fibroblasts as compared with OA synovial fibroblasts (173.000± 46.000 PAU) was observed in eight out of 12 synovial fibroblasts populations of patients with RA. CENP-E upregulation among the fibroblast populations ranged from approximately 50% (271.000 PAU, culture RA04) to fivefold (961.000PAU, culture RA21) upregulation (Fig. 2a). In six additional synovial fibroblast populations, the RA fibroblasts also showed an upregulation of CENP-E expression as compared with psoriatic arthritis fibroblasts (Fig. 2b). Of interest, neither therapy with disease-modifying antirheumatic drugs such as methotrexate, gold, resochine or cyclosporine A, nor therapy with oral steroids showed an influence on CENP-E expression in the RA fibroblasts. Of the eight RA fibroblast populations from RA patients receiving disease-modifying antirheumatic drugs, five showed an CENP-E upregulation; and of the eight fibroblast populations from RA patients receiving steroids, four showed a CENP-E upregulation. Furthermore, no difference in upregulation was seen in the RA synovial fibroblasts obtained from the intraosseus invasion areas (RA3K and RA9K), indicating a general upregulation of CENP-E in all synovial tissue compartments.
Figure 2.
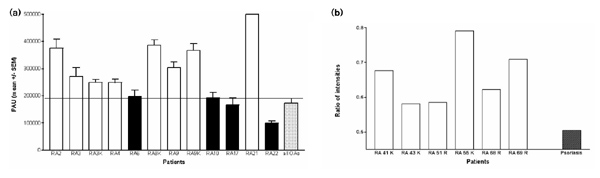
Quantitative reverse transcription polymerase chain reaction of the CENP-E gene segment followed by phosphor imaging densitometry evaluation. The polymerase chain reaction MIMIC® technique was used. (a) Significant upregulation in CENP-E in eight out of 12 rheumatoid arthritis synovial fibroblast cultures (white bars) as compared with all osteoarthritis fibroblast cultures (dotted bar; total n = 4); whereas four rheumatoid arthritis fibroblast populations did not show an upregulation in CENP-E upregulation (black bars). Values are shown as phosphor imager arbitrary units (PAU, mean ± standard error of the mean). Each MIMIC® was performed in quadruplicate. P < 0.05 was considered statistically significant. (b) Significant upregulation in six additional rheumatoid arthritis synovial fibroblast cultures (white bars) as compared with psoriatic arthritis fibroblasts (generous gift from E Märker-Herrmann, University of Mainz, Germany). Ratio of intensities are shown (when compared to standard 18S RNA).
Confirmation of CENP-E protein synthesis
Synthesis of CENP-E was not only confirmed on the mRNA level as outlined above, but also on the protein level by immunohistochemistry using monoclonal antibodies against human CENP-E. As illustrated in Figure 3, cultured rheumatoid synovial fibroblasts expressed intensive amounts of CENP-E (Fig. 3a), whereas in OA fibroblasts CENP-E production was below detection level (Fig. 3b). Figure 3c shows the positive control using antifibroblast antibodies.
Figure 3.
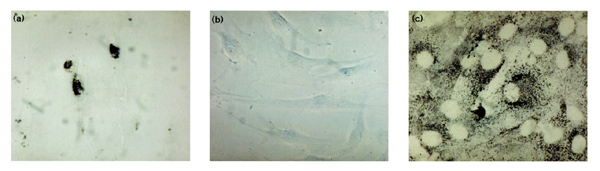
Immunocytochemistry of cultured rheumatoid arthritis fibroblasts using monoclonal antibodies against human CENP-E protein [10,11]. Note the strong expression of CENP-E protein rheumatoid fibroblasts (a) in comparison with the absence of CENP-E protein in osteoarthritis fibroblasts (b). (c) The positive control was performed with antifibroblast antibodies. Immunogold-silver staining, original magnifications × 800 (a, b) and × 1000 (c).
Expression of CENP-E in rheumatoid synovium
Numerous synovial cells of the patients with RA showed a positive in situ signal for the isolated CENP-E gene segment, confirming CENP-E mRNA production in rheumatoid synovium, whereas in OA synovial tissue CENP-E mRNA could not be detected. CENP-E expressing cells were found throughout the synovium, both in the lining layer as well as in the sublining. Because numerous CENP-E expressing cells showed an atypical fibroblast phenotype, we performed double-labelling using monoclonal fibroblast antibodies. The majority of CENP-E protein-expressing cells could be double-labelled with fibroblast antibodies, indicating that the CENP-E production seen in the cultured cells is not an effect caused by culture conditions. Figures 4a and 4b show synovial fibroblasts intensively expressing mRNA for the CENP-E gene segment isolated by RAP-PCR.
Figure 4.
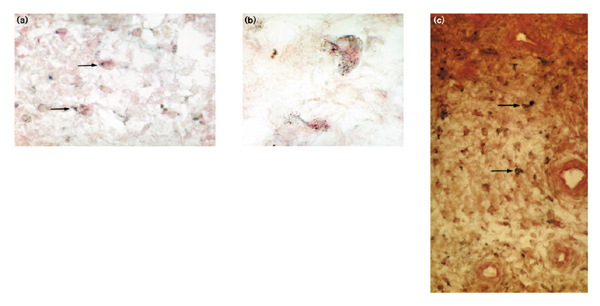
Demonstration of CENP-E messenger RNA and protein in rheumatoid synovium by in situ hybridization and immunohistochemistry. (a, b) A digoxigenin-labelled RNA probe transcribed from the amplified RNA arbitrarily primed-PCR gene product is used on rheumatoid arthritis snap-frozen sections. Double-labelling for CENP-E messenger RNA (black staining), and alkaline phosphatase antialkaline phosphatase counterstaining using anti-fibroblast antibodies (red staining) shows CENP-E expression in numerous fibroblasts througout the synovium (a, arrows). (b) The intensive CENP-E messenger RNA expression in two synovial fibroblasts. (c) Shows numerous fibroblasts expressing CENP-E protein [black staining (arrows), counterstaining with fast red]. Original magnifications × 300 (a, c) and × 600 (b).
To evaluate whether CENP-E mRNA production led to protein production detectable by immunohistochemistry, monoclonal antibodies directed against human CENP-E [10,11] were used. The most intensive CENP-E protein expression was found within highly inflamed areas in the sublining, and, to a lesser extent, in the lining layer. Figure 4c shows a synovial section with numerous synovial fibroblasts intensively expressing CENP-E protein.
Functional assays
As outlined above, CENP-E expression was independent from medication. This was further confirmed by analysis of the effect of prednisolone on CENP-E expression, which revealed no alteration in CENP-E mRNA after exposure to different (physiological) concentrations of prednisolone (Fig. 5). Serum starvation was also not able to suppress CENP-E mRNA completely. As shown in Figure 5, although in lesser amounts, CENP-E mRNA can still be detected in all RA synovial fibroblast populations, even after numerous days of serum starvation. Determination of the growth of CENP-E high and low expressors showed that the growth rate was slightly increased in the CENP-E high expressors, but by statistical criteria independent from the amount of CENP-E expression (Table 2). Also, the doubling time of CENP-E high and low expressors did not differ (data not shown).
Figure 5.
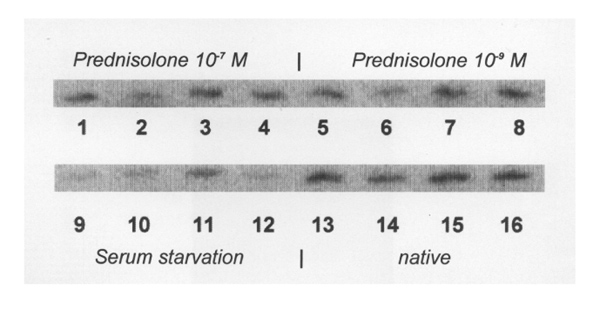
CENP-E regulation. The figure shows CENP-E messenger RNA in CENP-E high (right lanes 3/4, 7/8, 11/12 and 15/16) and CENP-E low (left lanes 1/2, 5/6, 9/10 and 13/14) expressing rheumatoid arthritis synovial fibroblast populations after different prednisolone exposure (lanes 1–8) and serum starvation (lanes 9–12). As compared with unstimulated fibroblasts (lanes 13–16) no significant alteration in CENP-E messenger RNA expression, either in CENP-E high- or in CENP-E low-expressing fibroblast populations, can be observed after prednisolone exposure. Serum starvation was also not able to suppress CENP-E messenger RNA completely (lanes 9–12).
Table 2.
Growth rate of CENP-E-expressing rheumatoid synovial fibroblasts
| Growth rate | |
| (increase factor of cells per 24 h, | |
| mean ± standard derivation) | |
| CENP-E high | 1.14 ± 0.19 |
| CENP-E low | 1.03 ± 0.15 |
Determination of the growth rate of CENP-E high and low expressors reveals that the growth rate is slightly higher, but not significantly different with regard to the amount of CENP-E expression.
Discussion
Since its introduction in 1992 [12], numerous variants of the differential display method [13] and continuous improvements including RAP-PCR [14] have proved to have both efficiency and reliability in examination of differentially regulated genes. Although the majority of experimental approaches have addressed malignant diseases [13,15,16], the results of this study reveal that, along with differential subtraction [17] and the original differential display [18], RAP-PCR is a particularly suitable method to identify differentially expressed genes in rheumatoid synovial fibroblasts.
The mRNA, which has been found to be upregulated in rheumatoid synovial fibroblasts, codes for a kinesin-like motor protein named CENP-E, which was first characterized in 1991 [10]. It is a member of a family of centromere-associated proteins, of which six (CENP-A to CENP-F) are currently known. The complete CENP-E gene spans a length of 8371 bases, the open reading frame encodes 2663 amino acids, and the respective protein has a molecular weight of 312 kDa and is a member of the human centromere-kinetochore complex [11]. CENP-E itself is a kinetochore motor, which accumulates transiently at kinetochores in the G2 phase of the cell cycle before mitosis takes place, appears to modulate chromosome movement and spindle elongation, and is degraded at the end of mitosis [11,19,20,21,22]. Consistently, inhibition of the CENP-E protein by specific antibodies causes cell cycle arrest at metaphase [11]. In addition, further research showed that kinesin-like proteins, including CENP-E, specifically regulate microtubule formation and the consecutive movement of chromosomes during mitosis, and therefore are crucial for cell division [23].
Interestingly, the presence or upregulation of CENP-E has never been associated with RA, although antibodies to the CENP family are frequently found in numerous rheumatic diseases [24], and CENP-E has recently been proposed to be an autoantigen in the limited form of systemic sclerosis [25].
The three-dimensional structure of CENP-E includes a coiled-coil domain. This has important functions that have links to known pathways in RA pathophysiology. Coiled-coil domains can also be found in jun and fos oncogene products [26,27], which are frequently upregulated in RA synovial fibroblasts [28]. They are also involved in DNA binding and transactivation processes, resembling the situation in activating protein-1 (Jun/Fos)-dependent DNA-binding in rheumatoid synovium [29]. Most interestingly, these coiled-coil motifs are crucial for the assembly of viral proteins [30,31], and the upregulation of CENP-E might reflect the influence of infectious agents in RA synovium [1,32]. In addition, CENP-E may also be involved in activation of rheumatoid synovial fibroblasts, because presence of CENP-E has been found to correlate with the active state of centomeres in translocations [33]. These pathways may include a distinct dysregulation of the cell cycle (eg rare activation steps in the formation of neocentromeres [34,35]), because rheumatoid synovial fibroblasts, although they have a transformed appearance, do not reveal an increased rate of proliferation in vitro or in vivo [36,37].
We also performed experiments showing that serum starvation decreased, but not completely inhibited CENP-E mRNA expression, showing that CENP-E is related to but does not completely depend on proliferation of these cells. In addition, we determined the growth rate of CENP-E high and low expressors, showing that it was independent on the amount of CENP-E expression, supporting the statement that upregulation of CENP-E reflects an activated RA fibroblast phenotype.
In summary, the results of this study support the hypothesis that CENP-E, presumably independently from medication, may not only be upregulated, but also be involved in RA pathophysiology.
Acknowledgments
Acknowledgments
This study was supported by grants of the German Research Society (DFG # Mu 1383/1-1, and Ku 1024/6-1), and the Olga Mayenfisch Stiftung, Zürich. The authors wish to thank Birgit Riepl and Christopher Benzing for excellent technical assistance, and Professor E Märker-Herrmann for providing the psoriatic arthritis synovial fibroblasts.
References
- Gay S, Gay RE, Koopman WJ. Molecular and cellular mechanisms of joint destruction in rheumatoid arthritis: two cellular mechanisms explain joint destruction? Ann Rheum Dis. 1993;52:S39–S47. doi: 10.1136/ard.52.suppl_1.s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Ladner U, Gay RE, Gay S. Molecular biology of cartilage and bone destruction. Curr Opin Rheumatol. 1998;10:212–219. doi: 10.1097/00002281-199805000-00010. [DOI] [PubMed] [Google Scholar]
- Müller-Ladner U, Kriegsmann J, Franklin B, et al. Synovial fibroblasts of patients with rheumatoid arthritis attach to and invade normal human cartilage when engrafted into SCID mice. Am J Pathol. 1996;149:1607–1615. [PMC free article] [PubMed] [Google Scholar]
- Welsh J, McClelland M. Arbitrarily primed PCR. Trends Genet. 1996;5:108–114. doi: 10.1016/s0168-9525(00)89058-7. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display. Materials and methods. Methods in Molecular Biology, vol 87, Totowa: Humana Press. 1997.
- Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Mathieu-Daudé F, Cheng R, Welsh J, McClelland M. Screening of differentially amplified cDNA products from RNA arbitrarily primed PCR fingerprints using single strand conformation polymorphism (SSCP) gels. . Nucleic Acids Res. 1996;24:1504–1507. doi: 10.1093/nar/24.8.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. Molecular Cloning Protocols and Applications, New York: Raven Press, 1997.
- Komminoth P, Merk FB, Leav I, Wolfe HJ, Roth J. Comparison of 35S-and digoxigenin-labeled RNA and oligonucletide probes for in situ hybridization. Histochemistry. 1992;98:217–228. doi: 10.1007/BF00271035. [DOI] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, et al. CENP-E, a novel centromere associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liang PL, Averboukh L, Keyomarsi K, Sager R, Pardee AB. Differential display and cloning of messenger RNAs from human breast cancer versus mammary epithelial cells. Cancer Res. 1992;52:6966–6968. [PubMed] [Google Scholar]
- McClelland M, Mathieu-Daudé F, Welsh J. RNA fingerprinting and differential display using arbitrarily primed PCR. Trends Genet. 1995;11:242–245. doi: 10.1016/s0168-9525(00)89058-7. [DOI] [PubMed] [Google Scholar]
- Shinoura N, Shamraj OI, Hugenholz H, et al. Identification and partial sequence of a cDNA that is differentially expressed in human brain tumors. Cancer Lett. 1995;89:215–220. doi: 10.1016/0304-3835(95)03690-x. [DOI] [PubMed] [Google Scholar]
- Yeatman TJ, Mao W. Identification of a differentially expressed message associated with colon cancer liver metastasis using an improved method of differential display. Nucleic Acids Res. 1995;19:4007–4008. doi: 10.1093/nar/23.19.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Selby J, Häupl T, Winchester R. Use of a differential subtraction method to identify genes that characterize the phenotype of cultured rheumatoid arthritis synoviocytes. Arthritis Rheum. 1998;41:1356–1364. doi: 10.1002/1529-0131(199808)41:8<1356::AID-ART4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Mangasser-Stephan K, Dooley S, Welter C, Mutschler W, Hanselmann RG. Identification of human semaphorin E gene expression in rheumatoid synovial cells by mRNA differential display. Biochem Biophys Res Commun. 1997;234:153–156. doi: 10.1006/bbrc.1997.6607. [DOI] [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Wilson L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. . Cell Motil Cytoskeleton. 1996;35:121–133. doi: 10.1002/(SICI)1097-0169(1996)35:2<121::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Schaar BT, Yen TJ, Wilson L. Mitotic HeLa cells contain a CENP-E-associated minus end-directed microtubule motor. . EMBO J. 1995;14:918–926. doi: 10.1002/j.1460-2075.1995.tb07073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Coulson RM, Yen TJ, Cleveland DW. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J Cell Biol. 1994;125:1303–1312. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinteochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. . J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mühlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic disease. Semin Arthritis Rheum. 1995;24:323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- Rattner JB, Rees J, Arnett FC, et al. The centromere kinesin-like protein CENP-E. An autoantigen in systemic sclerosis. Arthritis Rheum. 1996;39:1355–1361. doi: 10.1002/art.1780390813. [DOI] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. . Science. 1998;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, Adashi E, Graves BJ, McKnight SL. Isolation of a recombinant copy of the gene encoding C/EBP. . Genes Dev. 1998;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Müller-Ladner U, Kriegsmann J, Gay RE, Gay S. Oncogenes in rheumatoid arthritis. Rheum Dis Clin N Am. 1995;21:675–690. [PubMed] [Google Scholar]
- Asahara H, Fujisawa K, Kobata T, et al. Direct evidence of high DNA-binding activity of transcription factor AP-1 in the synovium of RA. Arthritis Rheum. 1997;40:911–917. doi: 10.1002/art.1780400520. [DOI] [PubMed] [Google Scholar]
- Tucker S, Srinivas R, Copmans R. Molecular domains involved in oligomerization of the Friend leukemia virus envelope protein. Virology. 1991;16:2431–2444. doi: 10.1016/0042-6822(91)90542-j. [DOI] [PubMed] [Google Scholar]
- Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- Müller-Ladner U, Gay RE, Gay S. Retroviral sequences in rheumatoid arthritis synovium. Intern Rev Immunol. 1998;17:273–290. doi: 10.3109/08830189809054406. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary componenets of functional centromeres. Hum Mol Genet. 1995;4:2189–2997. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Centromeres take flight: alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- Mohr W, Beneke K, Mohing W. Proliferation of synovial lining cells and fibroblasts. Ann Rheum Dis. 1975;34:219–224. doi: 10.1136/ard.34.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicher WK, Heer AH, Trabandt A, et al. Overexpression of zinc-finger transcription factor Z-225/egr-1 in synoviocytes from RA patients. J Immunol. 1994;152:5940–5948. [PubMed] [Google Scholar]


