Abstract
In Epstein–Barr virus (EBV)-associated tumors in nonimmunocompromised patients, EBV gene expression is highly restricted. EBV-encoded nuclear antigen (EBNA)-1 is expressed, whereas the immunogenic and proliferative EBNAs are not. This pattern of EBNA expression is generated by usage of the BamHI-Q promoter (Qp). We have determined that the JAK/STAT pathway positively regulates Qp activity. In transient-transfection assays, a Qp–CAT reporter was activated by cotransfected JAK-1 and by treatment of cells with the cytokine IL-6. The ability of Qp to bind signal transducer and activator of transcription (STAT) proteins was directly demonstrated by electrophoretic mobility-shift assay, and mutation of potential STAT-binding sites reduced Qp responsiveness to Janus kinase (JAK)-1. Consistent with a role for STATs in Qp function, Qp using Burkitt’s lymphoma Rael cells and cultured nasopharyngeal carcinoma (NPC) cells contained nuclear STAT protein. We investigated whether the inability to maintain EBV-positive NPC cell lines in culture was related to Qp activity. Passaging of the NPC cell line HK666 led to activation of expression of BZLF1, which encodes Zta and loss of Qp function. Transient expression assays linked Zta expression to the down-regulation of Qp. Cotransfection of Zta reduced Qp activity in reporter assays. This negative regulation required Zta DNA-binding activity. We provide evidence that Zta up-regulation of p53 leads to p53-mediated interference with JAK/STAT activation of Qp. The data imply that JAK/STAT signaling has a role in EBV-associated malignancies.
Epstein–Barr virus (EBV) is a ubiquitous herpesvirus that is carried by the majority of the population as a latent, persistent infection. Primary exposure to EBV may result in infectious mononucleosis, and EBV is also associated with both B cell and epithelial malignancies (1). Different forms of EBV latency are recognized, and these are defined by the extent of latent viral gene expression (2). During primary exposure, EBV infection of B cells gives rise to a population of cells that express the full spectrum of EBV-encoded nuclear antigens (EBNAs) and latency membrane proteins (LMPs) as well as the BamHI-A rightward transcripts and the polymerase III transcribed, noncoding EBERs. This expression pattern, which has been termed latency III (3), is also seen in lymphoblastoid cell lines in culture. Among the genes expressed in latency III are those for the growth-proliferative and highly immunogenic EBNA-2- and EBNA-3- family proteins (4). The question of how a lifelong latent infection could persist in the face of an active cytotoxic T cell response has recently been clarified by the recognition that in vivo latency in healthy EBV-seropositive individuals takes place in resting B cells with a memory B cell phenotype (5, 6). In these cells, EBV gene expression is extremely limited. The only viral transcripts consistently detected are those for LMP-2A and the BamHI-A rightward transcripts (7–9). EBV-associated tumors demonstrate a third pattern of latency-gene expression (latency I/II) in which only EBNA-1 and the BamHI-A rightward transcripts are expressed (latency I) or there is variable expression of the latency membrane proteins LMP-1, LMP-2A, and LMP-2B in additon to EBNA-1 and the BamHI-A rightward transcripts (latency II) (1).
The differing pattern of expression of the EBNAs in the various forms of latency is mediated by the use of alternative promoters. The BamHI-W promoter, which is constitutively active in B cells, is used on initial infection to drive expression of the EBNAs (10), the individual EBNA transcripts being generated through differential splicing. EBNA-1 and EBNA-2 then activate the BamHI-C promoter (Cp), which drives the latency III EBNA expression pattern seen in infectious mononucleosis and in lymphoblastoid cell lines. In EBV-associated tumors, the latency I/II pattern is the consequence of a switch from the Cp to the TATA-less BamHI-Q promoter (Qp) (11, 12). Methylation of the Cp is a major factor in loss of Cp activity (13–15). Several factors have been identified as playing a role in Qp regulation. Qp is negatively regulated by the downstream EBNA-1 binding sites and positively regulated by E2F-family proteins, which can displace EBNA-1 (16, 17). Interferon response factors (IRFs) also regulate Qp activity. IRF-2 and IRF-7 negatively regulate Qp (18–20), whereas IRF-1 and IRF-2 may provide positive regulation (21). The active use of Qp in EBV-associated tumors and its repression during in vivo latency in peripheral blood B cells points to Qp regulation as an important factor in EBV pathogenesis. In the present study, we demonstrate positive regulation of Qp by cellular Janus kinases (JAKs) and signal transducers and activators of transcription (STATs) (reviewed in ref. 22). The overlap between a potential STAT-binding site and the IRF-binding site in Qp suggests that STATs and IRFs may regulate Qp in a reciprocal manner.
Qp is used to express EBNA-1, which is essential for the maintenance of the EBV genome in dividing cells (23). The realization that the JAK/STAT pathway regulated Qp activity led us to evaluate the possibility that loss of Qp function might be contributing to the inability to maintain EBV genome-positive cells in cultures established from epithelial tumors such as nasopharyngeal carcinoma. We found that passaging of the NPC tumor cell line HK666 (24) led to loss of Qp activity concurrent with induction of expression of the lytic transactivator Zta. Evidence is presented that the mechanism of Qp down-regulation by Zta involves p53-mediated interference with the JAK/STAT pathway.
MATERIALS AND METHODS
Cell Lines and Plasmids.
Rael B cells and the nasopharyngeal carcinoma cell line HK666 (24) were cultured in RPMI medium 1640 plus 10% fetal calf serum. HeLa–Zta was a gift from E. Flemington (Dana-Farber Cancer Institute, Boston, MA). Hep3B and HeLa–Zta cells (25) were grown in DMEM plus 10% fetal calf serum. Zta expression was induced in HeLa–Zta by removal of tetracycline (1%) from the culture medium.
Qp-chloramphenicol acetyltransferase (CAT) and Qp-CAT mutants were constructed in the vector pCAT-BASIC (Promega). Qp-CAT contains Qp DNA sequences from −100 to +36. QpIRFmAT was generated by PCR amplification of the −100-to-+36 region from the plasmid FQUmAT-CAT (21) which was provided by S. Speck (Washington University School of Medicine, St. Louis, MO). Qp STATm1 and STATm2 contain the mutations shown in Fig. 1A introduced into the Qp-CAT background. STAT–CAT contains three copies of the SIF-binding element from the c-Fos promoter (26) in the vector pBL20 (27). STAT-1 was cloned in the vector pRK-5 and JAK-1 in pSRα (28). OriLyt-CAT (pDH123) is regulated by the BHLF-1 promoter and contains 1,000 bp of the EBV lytic origin (29). The Zta and Zta activation domain-deletion mutants have been described (30). The Zta DNA-binding mutant Zdbm1 (178EEL) was obtained from E. Flemington (Dana–Farber Cancer Institute) (31). The Zta dimerization domain mutant Zdmm is the Z214R/218R mutant (32) recloned into an SG5-based (Stratagene) vector as pDH331. The following plasmids were obtained from G. Hayward (Johns Hopkins University): p53-CAT contains three copies of the p53-binding site ligated into a modified pA10-CAT vector; the HHV-8 K8 expression vector, pCJC581, contains a K8 cDNA cloned into the modified SG5 vector pJH272. Expression plasmids for wild-type p53 and the p53 mutants G245 and R248 were obtained from K. Kinzler (Johns Hopkins University).
Figure 1.
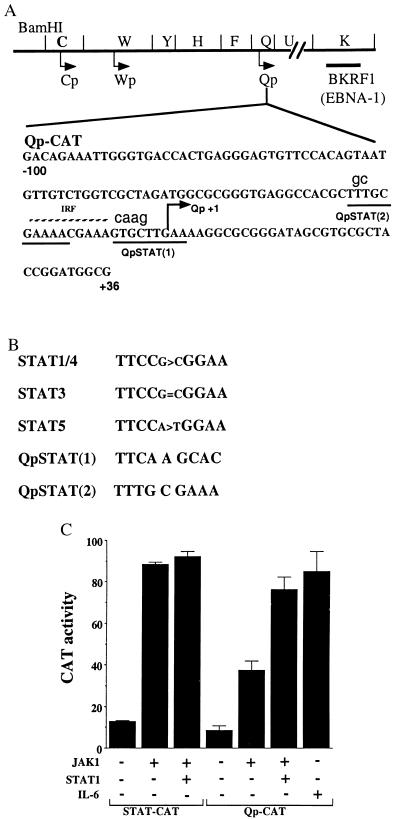
Qp contains potential STAT-binding sites. (A) The relative locations on the EBV genome of the Qp, Cp, and BamHI-W promoter (Wp) latency promoters and the ORF (BKRF1) encoding EBNA-1 are illustrated. The DNA sequence from −100 to +36 that was included in the Qp-CAT reporter is shown. The position of the IRF-binding site (broken line), two potential STAT-binding sites (underlined), and RNA start site (arrow) are indicated. The nucleotides in lowercase show the mutations introduced into QpSTATm1–CAT and QpSTATm2–CAT. (B) Comparison of STAT DNA-binding sequences with the potential Qp STAT sequences (38). (C) Activation of Qp-CAT by JAK-1. HeLa cells were transfected with a control STAT–CAT reporter or Qp-CAT reporter. As indicated, cells were also cotransfected with JAK-1 or JAK-1 plus STAT-1 or treated with IL-6. The results shown are an average of two experiments with the standard deviation indicated.
DNA Transfection and CAT Assays.
DNA transfections and CAT assays were performed as described (33). HeLa cells were transfected by using a calcium phosphate procedure, 1 or 2 μg of reporter DNA, and 1 or 2 μg of effector DNA and were harvested 30–40 hr after transfection. The transfection protocol was modified for Hep3B cells. The transfected cells were grown for only 6–8 hr in 3% CO2 at 35°C before the medium was replaced and the cells were transferred to growth at 37°C and 5% CO2. For IL-6 treatment, transfected cells were incubated in fresh medium at 37°C in 5% CO2 for 2–3 hr, after which IL-6 (100 ng/ml; R&D Systems) was added and growth continued for a further 14–16 hr. CAT activity was quantitated by using an Instant Imager (Packard).
Immunofluorescence and Immunohistochemistry.
HK666 and Rael cells were fixed in 1% paraformaldehyde and stained in an indirect immunofluorescence assay as described (33) by using anti-STAT-4 polyclonal antibody (1:50; Santa Cruz Biotechnology). Slides were viewed by using a Leitz fluorescence microscope, and the images were captured by using the program image pro plus (Media Cybernetics, Silver Spring, MD). Immunohistochemistry for Zta was performed on HK666 cells by using reagents from Dako with standard procedures. Anti-BZLF1 mAb (1:100) was the primary antibody, and biotin-conjugated anti-mouse Ig was the secondary antibody. Positive cells were visualized by using StreptABComplex/horseradish peroxidase.
Reverse Transcription (RT)–PCR.
Polyadenylated RNA was isolated from HK666 cells and amplified by RT-PCR, and the products were detected by Southern blotting with described protocols (8). PCR products were generated by using 30 cycles of amplification and the following primer pairs: Qp–EBNA-1, 5′-GCGGGATAGCGTGCGCTA-3′ and 5′-CTTCTGGTCCAGATGTGTCTC-3′; BZLF1, 5′-AGCAGACATTGGTGTTCCAC-3′ and 5′-ACATCTGCTTCAACAGGAGG-3′. The oligonucleotide probes used for hybridization were: Qp–EBNA-1, 5′-ATGCCCTGAGACTACTCTCT-3′ and BZLF1, 5′-GCGCAGCCTGTCATTTTCAG-3′.
Electrophoretic Mobility-Shift Assay.
The sequences of the sense strand of the double-stranded DNA probes and competitor oligonucleotides used are: Qp probe, 5′-CTAGACGCTTTGCGAAAACGAAAGTGCTTGAAAAGGCGGATC-3′; QpSTATm, 5′-CTAGACGCgcTGCGAAAACGAAAGTGCccGAAAAGGCGGATC-3′; QpIRFmAT, 5′-GACGCTTTGCGAAAAatAAAGTGCTTGAAAAGGCC-3′; STAT-4, 5′-GATCGAGCCTGATTTCCCCGAAATGATGAGCGATC-3′; Flag, 5′-GATCTGACTACAAGGACGACGATGACAAGTAGGATC-3′. Mutations are shown in lower case. The assays were performed as described (34) with protein–DNA complexes being analyzed on nondenaturing 4% polyacrylamide gels. In the competition assays, 100-fold excess of unlabeled double-stranded oligonucleotide competitor was incubated with the 32P-end-labeled probe and purified STAT-4 protein (0.2 μg) for 30 min at room temperature, followed by gel electrophoretic analysis. Anti-STAT-4 polyclonal antibody (2 μg; Santa Cruz Biotechnology) and control anti-Flag antibody (2 μg; Sigma) were added to the STAT-4/DNA mixture after binding, and the incubation was continued at room temperature for an additional hour.
RESULTS
Up-Regulation of Qp by JAK/STAT Signaling.
The latency Qp contains an upstream binding site for IRFs (18–21) and downstream binding sites for E2F1 (17, 35) and for the EBV EBNA-1 protein (16, 36). An inspection of the Qp DNA sequence revealed that Qp also contains two potential STAT-binding sites that flank and partially overlap with the IRF-binding site (Fig. 1 A and B). To ascertain whether Qp was responsive to STAT activation, cotransfection assays were performed in HeLa cells. Assay conditions were first established by using a STAT–CAT reporter containing three STAT-binding sites. Cotransfection of the STAT–CAT reporter with JAK-1, which phosphorylates STATs and facilitates their dimerization and relocalization from the cytoplasm to the cell nucleus, resulted in an 8-fold stimulation of reporter expression (Fig. 1C). Cotransfection of an expression plasmid for STAT-1 marginally increased the response over that seen with JAK-1 alone.
Cotransfection with JAK-1 increased expression of Qp-CAT 4-fold over basal levels (Fig. 1C). Cotransfection of JAK-1 plus STAT-1 resulted in a 2-fold-increased response over that seen with JAK-1 alone. The cytokine IL-6 activates three Janus kinases (JAK-1, JAK-2, and TYK-2), and treatment of HeLa cells with IL-6 induces STAT-3 DNA-binding activity (37). IL-6 treatment resulted in strong activation of Qp-CAT expression (Fig. 1C).
Demonstration of STAT Binding to Qp.
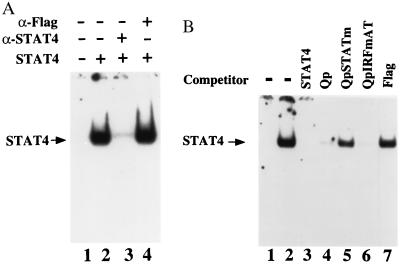
The Qp sequences do not conform to any preferred STAT-binding sites (Fig. 1B). STAT-family proteins recognize similar DNA sequences. STAT binding to Qp was tested by using purified, activated STAT-4 protein isolated from cultures infected with a STAT-4-expressing baculovirus vector (28). In random binding-site selection assays, STAT-4 shows an identical binding-site preference to STAT-1 and STAT-3 and a similar preference to STAT-5 (38). In the electrophoretic mobility-shift assay, Qp probe bound to activated STAT-4 (Fig. 2A, lane 2), and the STAT-4–DNA complex was disrupted by incubation with anti-STAT-4 rabbit antibody (lane 3) but not by incubation with a nonspecific antibody (anti-Flag antibody, lane 4). The specificity of the Qp–STAT-4 interaction was further established by the use of competitor oligonucleotides (Fig. 2B). The STAT-4–DNA complex was not detected when the incubation contained competitor STAT-4 concensus oligonucleotide (lane 3), or Qp oligonucleotide (lane 4), or a mutated Qp oligonucleotide competitor that lacked IRF-binding ability (QpIRFmAT; lane 6). STAT-4 binding was not affected by competition with an irrelevant oligonucleotide (Flag, lane 7) or by competition with the Qp oligonucleotide in which both STAT sequences were mutated (QpSTATm; lane 5). These results indicate that the region of Qp previously described to bind IRFs also contains a functional STAT-binding site or sites.
Figure 2.
Qp binds STAT-4. (A) Electrophoretic mobility-shift assay showing binding of purified STAT-4 to Qp probe (lane 2). The DNA–protein complex was disrupted by anti-STAT-4 antibody (lane 3) but not by control antibody (lane 4). (B) Electrophoretic mobility-shift assay illustrating the specificity of STAT-4 binding. STAT-4 binding was competed by unlabeled STAT-4 (lane 3), Qp (lane 4), and QpIRFmAT (lane 6) oligonucleotides but not by QpSTAT mutant (lane 5) or control oligonucleotides (lane 7). Lane 1 contains Qp probe alone.
Mutation of the Potential STAT-Binding Sites in Qp Affects Basal Activity and Activation by JAK-1.
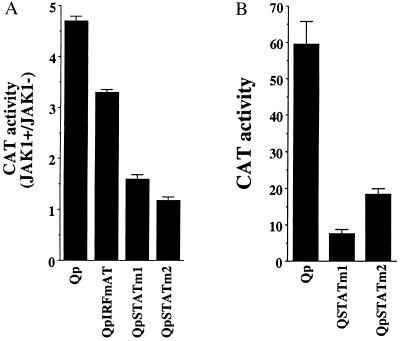
To confirm that STATs contributed to the JAK response of Qp, mutations were introduced into the two potential STAT sites (Fig. 1A). These mutations were placed in sequences that were nonoverlapping with the IRF-binding site. A Qp-CAT reporter carrying a previously described mutation that eliminates IRF binding (QP-IRFmAT; ref. 21) still had 3-fold greater expression when cotransfected with JAK-1 (Fig. 3A). The mutations introduced into the potential STAT-binding sites had a more deleterious effect on JAK responsiveness. Mutation of the promoter-proximal STAT-binding site (STATm1) resulted in a 1.5-fold-increase in expression in the presence of JAK-1, and mutation of the second potential STAT-binding site (STATm2) abolished responsiveness.
Figure 3.
Effect of mutations in the potential STAT-binding sites on JAK activation (A) and basal activity (B) of Qp-CAT in transfected HeLa cells. The results shown are an average of two experiments with the standard deviation indicated.
The effect of mutations in the STAT sites on basal Qp activity was also examined in HeLa cells with the Qp-CAT reporter (Fig. 3B). Mutation of either potential STAT site significantly diminished basal Qp activity. The promoter-proximal STAT site mutated in STATm1 overlaps with a mapped initiation site for Qp messages. It is possible that the m1 mutation may be interfering with the formation of an RNA initiation complex and that the apparent reduction in responsiveness to JAK-1 on the part of this Qp construction is a reflection of impaired RNA initiation. This caveat does not apply to the STATm2 mutation. The inability of the m2 mutant to respond to cotransfection with JAK-1 is consistent with JAK activation of Qp having a component that requires STAT binding. The presence of STATs in the nucleus to act as transcription factors is normally dependent on their activation, which occurs predominantly through JAK-mediated phosphorylation. However, certain cell lines have been found to contain constitutively activated STATs, and the reduction in Qp basal activity seen with the Qp STATm2 mutant may reflect some constitutive STAT activation in HeLa cells. The IRF mutation also reduced basal activity, as previously reported (21).
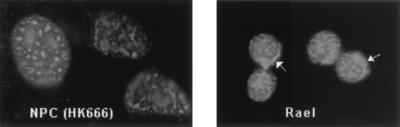
The intracellular localization of one of the STATs, STAT-4, was examined in an EBV-positive cell line that uses Qp to express EBNA-1. In the latently infected Rael B cell line, an indirect immunofluorescence assay revealed the presence of both nuclear and cytoplasmic STAT-4 (Fig. 4). STAT-4 expression is reported to have a restricted tissue distribution, but interestingly, nuclear STAT-4 staining was also detected by immunofluorescence in the EBV-positive nasopharyngeal carcinoma cell line HK666 (Fig. 4). Taken together, these results support a role for STATs in the positive regulation of Qp, and because EBV-associated tumors in nonimmunocompromised individuals invariably have a pattern of Qp usage, the data further imply that JAK/STAT signaling may be an important factor in establishment of these tumors.
Figure 4.
Constitutively activated STAT-4 is present in EBV+ Rael B cells and HK666 nasopharyngeal carcinoma cells. Indirect immunofluorescence assay showing the presence of nuclear STAT-4 protein in HK666 cells and nuclear plus cytoplasmic STAT-4 in Rael. Arrowheads indicate cytoplasmic staining in Rael.
Passaging of NPC Cells Leads to Induction of Zta Expression and Loss of Qp Function.
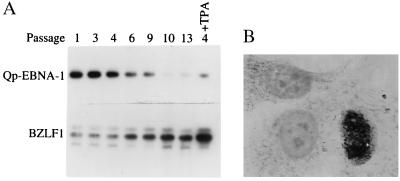
Epithelial cell lines established from EBV-positive NPC inevitably lose their EBV genomes on passaging. We wondered whether regulation of Qp activity might be a contributing factor. We first used RT-PCR to examine EBV promoter activity in the recently established NPC cell line HK666 (24). Qp activity was tested by using a primer pair spanning the EBNA-1 Q and K exons, and lytic promoter activity was tested by using a primer pair spanning exon 1 and exon 3 of the BZLF-1 gene that encodes the Zta lytic transactivator. Passaging of HK666 cells led to a decrease in Qp-driven EBNA-1 transcripts and a reciprocal increase in transcription of BZLF-1 (Fig. 5A). (Note that the passage numbers represent passaging at Johns Hopkins and not passage number from the time of establishment of the cell line.) Treatment of HK666 cells with n-butyrate and 12-O-tetradecanoylphorbol 13-acetate (passage 4) exacerbated the differential expression by decreasing Qp-driven EBNA-1 expression and increasing BZLF-1 expression (Fig. 5A). To determine the extent of EBV lytic activation in HK666 cells, an immunohistochemical assay was performed. Zta-expressing cells were detected by immunoperoxidase staining (Fig. 5B). Thus, passaging of HK666 cells led to loss of Qp activity and induction of Zta mRNA and protein expression.
Figure 5.
Passaging of HK666 leads to induction of BZLF-1 and loss of Qp expression. (A) Expression of Qp-initiated EBNA-1 transcripts and BZLF-1 transcripts in HK666 cells was examined by RT-PCR. Southern blots of the DNA products were probed for BKRF-1 (EBNA-1) (Upper) and BZLF-1 (Lower). Cells at passage 4 were also tested after treatment with 12-O-tetradecanoylphorbol 13-acetate (TPA) (20 ng/ml) and n-butyrate (3 mM) for 48 hr (+TPA). (B) Detection of Zta protein in HK666 cells by immunoperoxidase staining.
Zta Down-Regulates Qp.
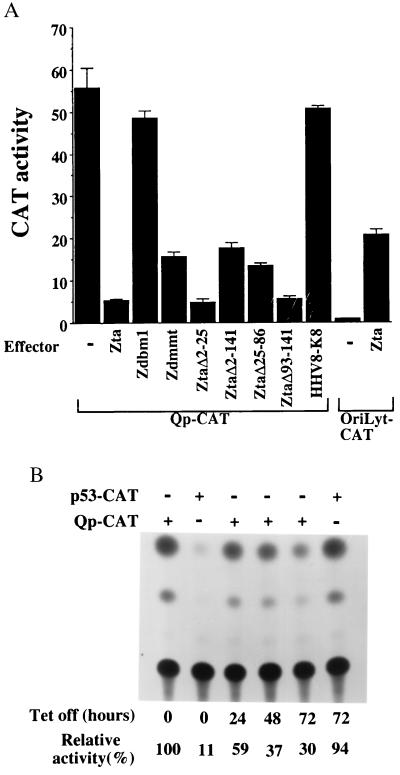
Zta down-regulates the EBV latency Cp and LMP-1 promoters (39). Because the loss of Qp activity on passaging of HK666 cells correlated with the activation of BZLF-1 expression, we examined whether Zta might also have a role in down-regulation of Qp. In transient-expression assays in HeLa cells, cotransfection of Zta dramatically reduced Qp-CAT expression (Fig. 6A). Zta mutants carrying deletions within the activation domain (ZΔ2–25; ZΔ25–86 and ZΔ93–141; ref. 30) retained the ability to significantly repress Qp activity, as did a Zta mutant in which the entire activation domain is deleted (ZΔ2–141). A Zta variant carrying a mutation in the dimerization domain (Zdmmt; ref. 32) also retained the ability to repress Qp-CAT, albeit with reduced efficiency. On the other hand, a Zta variant carrying a mutation in the DNA-binding domain (Zdbm1; ref. 31) did not repress Qp-CAT activity. These results imply that the Zta DNA-binding domain is critical for Zta-mediated down-regulation of Qp. As a control for the specificity of the negative regulation, the cells were also transfected with an oriLyt-CAT reporter. In this reporter, CAT expression is driven by the EBV BHLF-1 oriLyt promoter and enhancer, which contain multiple binding sites for Zta (29). As expected, this reporter was activated by cotransfection with Zta (Fig. 6A). In a separate control, cells were cotransfected with an expression plasmid for K8, an HHV-8 homolog of EBV Zta. K8 did not have a significant effect on Qp-CAT expression.
Figure 6.
Zta down-regulates Qp-CAT expression. (A) Qp-CAT was cotransfected into HeLa cells with Zta or Zta variants carrying mutations in the DNA-binding domain (Zdbm1) or the dimerization domain (Zdmmt) or deletions in the activation domain (ZtaΔ2–25, ZtaΔ2–141, ZtaΔ25–86, ZtaΔ93–141). The HHV8-K8 effector and oriLyt-CAT target served as controls for the specificity of the Zta-mediated repression of Qp-CAT. The results shown are an average of two experiments, with the standard deviation indicated. (B) Induction of Zta in the HeLa-Zta cell line by removal of tetracycline from the medium led to reduced expression of transfected Qp-CAT. The p53-CAT reporter served as a positive control for Zta expression. CAT activity is expressed relative to that of Qp-CAT in the presence of tetracycline (set at 100%).
CAT-reporter assays were also performed by using a cell line that expresses Zta in a tetracycline-regulated manner (25). Release of tetracycline control induces Zta expression, and ≈50% of the cells expressed Zta at 72 hr after induction. Induction of Zta in this setting also led to decreased expression of the transfected Qp-CAT reporter (Fig. 6B). Zta has been shown to induce accumulation of p53 (40). A CAT reporter carrying three p53-binding sites (p53-CAT) was included as a control in this experiment. p53-CAT had limited expression in the presence of tetracycline, and expression was activated 72 hr after removal of the drug, consistent with Zta expression in the culture at this time.
Negative Regulation of Qp Involves p53 Interference with JAK/STAT Signaling.
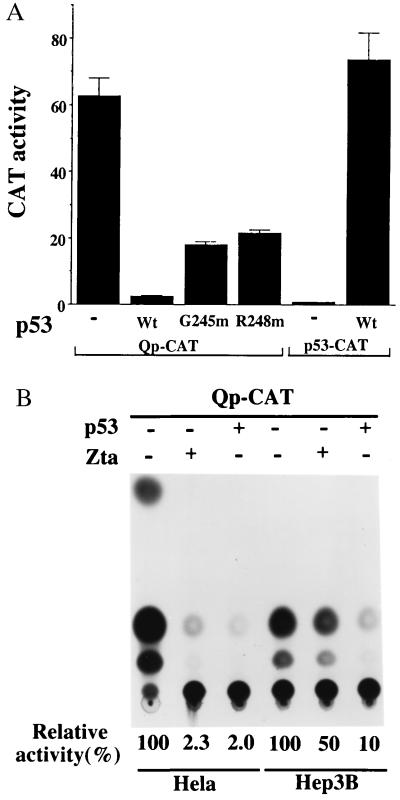
Zta-mediated growth arrest did not require the Zta-activation domain (40, 41). Zta-mediated repression of Qp was also independent of Zta transcriptional activation function (Fig. 6A). This similarity in functional requirements led us to examine whether Zta suppression of Qp activity might be related to the Zta-induced accumulation of p53. Cotransfection of the Qp-CAT reporter with a p53 expression vector resulted in down-regulation of CAT activity similar to that mediated by Zta (Fig. 7A). Two p53 variants that have mutations in the DNA-binding domain and have altered DNA-binding specificity (G245 m and R248 m) (42) were less effective than wild-type p53 at inhibiting Qp-CAT (Fig. 7A). p53-CAT was included as a control in this experiment. In contrast to Qp-CAT, p53-CAT was efficiently activated by cotransfection of wild-type p53. To further establish that p53 was an essential intermediate in Zta-mediated down-regulation of Qp, the ability of Zta to affect Qp activity was compared in HeLa cells and Hep3B cells, which carry a deletion in the p53 gene (Fig. 7B). As already demonstrated, Qp activity was severely inhibited in HeLa cells by cotransfection of either p53 or Zta expression vectors. In contrast, p53 and Zta had discordant effects in Hep3B cells. p53 retained the ability to down-regulate Qp in Hep3B cells, but Zta cotransfection had a minimal effect in the p53-null cells. These observations are consistent with p53 acting downstream of Zta in repression of Qp.
Figure 7.
p53 is a downstream effector of Qp repression. (A) Qp-CAT was down-regulated by coexpression of p53. Qp-CAT was cotransfected into HeLa cells with either wild-type (Wt) or mutant (G245 m, R248 m) p53. p53-CAT formed a positive control for p53 function. The results shown are an average of two experiments, with the standard deviation indicated. (B) Zta is an ineffective repressor of Qp-CAT in p53− cells. The effect of cotransfection of Zta or p53 on Qp-CAT activity was compared in transfected HeLa and p53− Hep3B cells. Qp-CAT activity in the absence of cotransfected Zta and p53 is set at 100%.
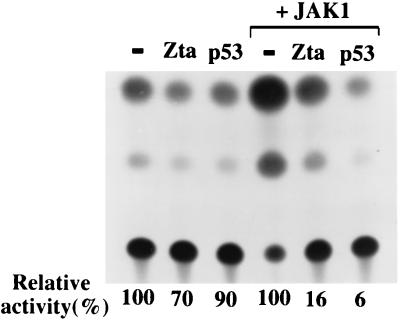
To understand how p53 might inhibit Qp activity, we returned to our initial observation that Qp is positively regulated by the JAK/STAT pathway. The effect of Zta and p53 on STAT-mediated activation was examined in transfection assays by using a STAT–CAT reporter (Fig. 8). Cotransfection of Zta or p53 with STAT–CAT resulted in a marginal repression of CAT activity in HeLa cells. However, JAK-1 activation of the STAT–CAT reporter was inhibited by both Zta and p53. Western blot analysis showed that expression of JAK-1 itself was not affected by coexpression with Zta or p53 (data not shown). In summary, the experiments suggest that Zta, through stabilization of p53, can interfere with JAK/STAT activation of Qp. Qp regulates EBNA-1 synthesis in NPC and in the majority of EBV-associated tumors, and EBNA-1 is required for maintenance of the EBV genome as an episome. The inability to retain the EBV genome in cell lines established from EBV-positive epithelial tumors may therefore be related to induction of Zta expression on passaging and subsequent negative regulation of the JAK/STAT pathway.
Figure 8.
Zta and p53 interfere with JAK/STAT activation of a STAT–CAT reporter. The effect of cotransfected Zta and p53 on STAT–CAT activity was compared in HeLa cells in the presence or absence of cotransfected JAK-1. The activity of STAT–CAT was set at 100% in each case.
DISCUSSION
The patterns of EBV gene expression associated with the different forms of EBV latency derive from the regulated activity of the latency Cp and Qp promoters. The latency Cp used in primary infection drives expression of the full spectrum of EBNAs. Qp is not active in group III latency. Down-regulation of Qp may be mediated through binding of EBNA-1 (expressed from Cp) to the Qp locus EBNA-1 binding sites along with negative regulation by promoter-bound IRF-7 and IRF-2 (16, 18–20).
EBV has evolved a strategy for long-term latent infection in peripheral B cells in the face of immune surveillance by establishing a form of in vivo latency in resting B cells (5, 6). In these cells, both Cp and Qp are repressed. Methylation of Cp is a key factor in Cp repression in peripheral blood B cells (43), but Qp is hypomethylated in all situations in which it has been examined (15, 44, 45). Qp is a TATA-less promoter, and the quiescence of Qp in this setting may reflect the lack of positive regulatory factors. We have shown that STATs positively regulate Qp. Activated STATs have not been detected in peripheral blood B cells (46), but STATs are activated in B cells after cross-linking of the surface IgM antigen receptor and engagement of the CD40 receptor as well as in response to cytokines (47). Although Qp is not active in latently infected, resting B cells, Qp activity has been detected in some studies of peripheral blood B cells (9, 48). Maintenance of the episomal EBV genome in dividing cells depends on the EBV origin binding protein, EBNA-1. There is no need for EBNA-1 expression in resting B cells, but if these cells were periodically stimulated into transient proliferation, then EBNA-1 would be required to maintain the infection. Regulation of Qp by E2F family proteins has been described, and this would provide one mechanism to link Qp driven expression of EBNA-1 to the cell cycle (35). Our observations suggest a second mechanism. If resting B cells are activated through costimulation with antigen and CD40 ligand, then this mitogenic signal would also result in STAT tyrosine phosphorylation and concomitant up-regulation of Qp. EBNA-1 contains a repeat region that interferes with processing and MHC presentation, and hence transient expression of EBNA-1 would not result in immune clearance of the cells (49).
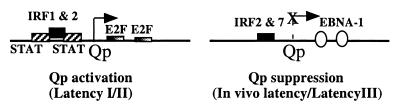
The setting in which Qp-directed EBNA-1 synthesis is consistently detected is in EBV-associated tumors. Tumor cells are actively cycling cells, and positive displacement of EBNA-1 from the downstream Qp locus by E2F proteins may be a factor in Qp expression in this setting (Fig. 9). It has been noted (50) that STAT-1 and STAT-3 are constitutively activated in the EBV-positive B cell lines Namalwa, Akata, and Daudi. We also detected activated, nuclear STAT-4 in the EBV-positive cell line Rael and in the NPC cell line HK666. Our experiments indicate that STATs positively regulate Qp, and the presence of nuclear STATs in EBV associated tumor derived cell lines is consistent with such a role. We do not yet know whether both potential Qp STAT-binding sites are functional or which STAT-family members are the biologically relevant activators. Different STATs may contribute in epithelial versus B cell tumors. IRF-1 and IRF-2 can also stimulate Qp (Fig. 9). However, the overlap between the Qp IRF-binding site and one of the STAT-binding sites described in our work makes it possible that binding of the IRF and STAT factors may be mutually exclusive.
Figure 9.
Summary of Qp regulation in the different forms of EBV latency.
Cell lines established from nasopharyngeal carcinoma and gastric carcinoma tumors do not retain the EBV genome on passaging. Our data suggest that induction of expression of Zta in culture contributes to down-regulation of Qp activity. Because Qp drives EBNA-1 synthesis, this would result in loss of EBNA-1 and subsequent loss of episomal EBV DNA, which depends on EBNA-1 for replication. In an analogous situation, induction of Zta expression in cultured B cell lines has been shown to lead to loss of EBV genomes and the outgrowth of cells that either contained a reduced number of EBV episomal genomes or retained only integrated genomes (51, 52). Transcripts for BZLF-1 have been detected by RT-PCR in NPC and Burkitt’s lymphoma tissues (53, 54). Induction of the complete cycle of viral replication would result in cell death. However, Zta itself is not cytotoxic (40). A more transient induction is apparently compatible with long-term cell survival, and a proportion of the cell population can grow out while carrying the effects of Zta expression and consequent repression of Qp activity, in the form of loss of EBV genomes. This scenario has several consequences. It suggests that the incorporation of steps to prevent Zta induction and to stimulate the JAK/STAT pathway should be beneficial for the maintenance in culture of EBV-positive epithelial tumor cells. More generally, the association of EBV with malignant diseases such as Burkitt’s lymphoma has been clouded by the isolation of EBV-negative tumors. Our data reinforce the point raised by others (51) that the lack of EBV genomes in cells within a tumor that is otherwise consistently associated with EBV may reflect transient Zta induction and subsequent loss of EBV genomes from the cells.
Zta induces accumulation of p53 (40) and our results are consistent with Zta repression of Qp being effected through p53. Zta-mediated down-regulation of Qp-CAT was ineffective in p53-null cells, and the requirements for Zta repression of Qp mirrored the requirements for p53 induction in that the integrity of the Zta DNA-binding domain was essential for both functions, whereas the Zta activation domain was dispensable in each case. The only EBV-positive NPC-derived cell line that has been maintained long term is NPC-KT, which was generated by fusing NPC cells with the AdAH cell line (55). Interestingly, both AdAH and the resultant NPC-KT cells express the human papillomavirus E6 protein and hence have low levels of p53 as a consequence of p53 destabilization by E6 (40). That the induction of p53 should interfere with JAK/STAT activation of Qp is also consistent with known properties of p53. p53 has been found to mask STAT DNA-binding activity and functional signaling through a mechanism that requires a functional p53 DNA-binding domain (56). In our experiments, two p53 variants carrying mutations in the DNA-binding domain were less effective at down-regulating Qp than the wild-type protein. Wild-type p53 masking reduces STAT-3 and STAT-5 DNA-binding activity but does not affect STAT-1 function (56, 57). Because the loss of Qp activity in the passaged NPC cell line appears to be effected through Zta-mediated stabilization of p53, one implication is that STAT-3 and/or STAT-5 may be biologically relevant STATs for Qp activation. STAT-3 is one of the STATs activated in B lymphocytes by antigen receptor engagement (46). Regulation of Qp by the JAK/STAT pathway fits well with the known usage of Qp in the different forms of in vivo latency and has implications for EBV-associated tumorigenesis.
Acknowledgments
We thank T. Hoey for purified STAT-4; E. Flemington, S. Speck, D. Nathans, K. Kinzler, P. Lieberman, and G. Hayward for their generous gifts of plasmids; E. Flemington for the Tet-regulated Zta cell line; J. Nicholas for Hep-3B cells; Q. Tao for assistance with immunohistochemistry, and Feng Chang for manuscript preparation. This work was supported by Grant RO1 CA30356 from the National Institutes of Health to S.D.H. and Grant P01 CA15396 to R.F.A.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- RT
reverse transcription
- CAT
chloramphenicol acetyltransferase
- EBNA
EBV-encoded nuclear antigen
- STAT
signal transducer and activator of transcription
- NPC
nasopharyngeal carcinoma
- LMP
latency membrane proteins
- JAK
Janus kinase
- Qp
BamHI-Q promoter
- Cp
BamHI-C promoter
- IRF
interferon response factors
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Rickinson A B, Kieff E. In: Field’s Virology. Field B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2397–2446. [Google Scholar]
- 2.Kieff E. In: Field’s Virology. Field B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2343–2396. [Google Scholar]
- 3.Rowe M, Lear A L, Croom-Carter D, Davies A H, Rickinson A B. J Virol. 1992;66:122–131. doi: 10.1128/jvi.66.1.122-131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rickinson A B, Moss D J. Annu Rev Immunol. 1997;15:405–431. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 5.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 7.Qu L, Rowe D T. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Smith P, Ambinder R F, Hayward S D. Blood. 1999;93:3026–3032. [PubMed] [Google Scholar]
- 9.Tierney R J, Steven N, Young L S, Rickinson A B. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlager S, Speck S H, Woisetschlager M. J Virol. 1996;70:3561–3570. doi: 10.1128/jvi.70.6.3561-3570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nonkwelo C, Skinner J, Bell A, Rickinson A, Sample J. J Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer B C, Strominger J L, Speck S H. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minarovits J, Li-Fu H, Minarovits-Kormuta S, Klein G, Ernberg I. Virology. 1994;200:661–667. doi: 10.1006/viro.1994.1229. [DOI] [PubMed] [Google Scholar]
- 14.Robertson K D, Swinnen L J, Zong J C, Gulley M L, Ambinder R F. Blood. 1996;88:3129–3136. [PubMed] [Google Scholar]
- 15.Schaefer B C, Strominger J L, Speck S H. Mol Cell Biol. 1997;17:364–377. doi: 10.1128/mcb.17.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sample J, Henson E B D, Sample C. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung N S, Wilson J, Davenport M, Sista N D, Pagano J S. Mol Cell Biol. 1994;14:7144–7152. doi: 10.1128/mcb.14.11.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nonkwelo C, Ruf I K, Sample J. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Pagano J S. Mol Cell Biol. 1999;19:3216–3223. doi: 10.1128/mcb.19.4.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Pagano J S. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer B C, Paulson E, Strominger J I, Speck S H. Mol Cell Biol. 1997;17:873–886. doi: 10.1128/mcb.17.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leonard W J, O’Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 23.Lee M A, Diamond M E, Yates J L. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui A B, Cheung S T, Fong Y, Lo K W, Huang D P. Cancer Genet Cytogenet. 1998;101:83–88. doi: 10.1016/s0165-4608(97)00231-8. [DOI] [PubMed] [Google Scholar]
- 25.Cayrol C, Flemington E K. J Virol. 1995;69:4206–4212. doi: 10.1128/jvi.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner B J, Hayes T E, Hoban C J, Cochran B H. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer T S, Sanders L K, Nathans D. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Sun Y-L, Hoey T. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman P M, Hardwick J M, Hayward S D. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flemington E K, Lytle J P, Carol C, Borras A M, Speck S H. Mol Cell Biol. 1994;14:3041–3052. doi: 10.1128/mcb.14.5.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flemington E, Speck S H. Proc Natl Acad Sci USA. 1990;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling P D, Ryon J J, Hayward S D. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling P D, Hsieh J J-D, Ruf I K, Rawlins D R, Hayward S D. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davenport M G, Pagano J S. J Virol. 1999;73:3154–3161. doi: 10.1128/jvi.73.4.3154-3161.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawlins D R, Milman G, Hayward S D, Hayward G S. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 37.Song M M, Shuai K. J Biol Chem. 1998;273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 38.Ihle J N. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 39.Kenney S, Kamine J, Holley-Guthrie E, Lin J C, Mar E C, Pagano J. J Virol. 1989;63:1729–1736. doi: 10.1128/jvi.63.4.1729-1736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cayrol C, Flemington E K. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 41.Cayrol C, Flemington E K. J Biol Chem. 1996;271:31799–31802. doi: 10.1074/jbc.271.50.31799. [DOI] [PubMed] [Google Scholar]
- 42.Ko L J, Prives C. Genes Devel. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 43.Robertson K D, Ambinder R F. Blood. 1997;90:4480–4484. [PubMed] [Google Scholar]
- 44.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. Blood. 1998;91:1373–1381. [PubMed] [Google Scholar]
- 45.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. J Virol. 1998;72:7075–7083. doi: 10.1128/jvi.72.9.7075-7083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karras J G, Wang Z, Huo L, Howard R G, Frank D A, Rothstein T L. J Exp Med. 1997;185:1035–1042. doi: 10.1084/jem.185.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karras J G, Wang Z, Huo L, Frank D A, Rothstein T L. J Immunol. 1997;159:4350–4355. [PubMed] [Google Scholar]
- 48.Chen F, Zou J-Z, di Renzo L, Winberg G, Hu L-F, Klein E, Klein G, Ernberg I. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci M G. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber-Nordt R M, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart R, Mertelsmann R, Finke J. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 51.Srinivas S K, Sample J T, Sixbey J W. J Infect Dis. 1998;177:1705–1709. doi: 10.1086/517427. [DOI] [PubMed] [Google Scholar]
- 52.Takada K, Ji Z, Fujiwara S, Shimizu N, Tanabe-Tochikura A. J Virol. 1992;66:5590–5593. doi: 10.1128/jvi.66.9.5590-5593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J X, Chen H L, Zong Y S, Chan K H, Nicholas J, Middeldorp J M, Sham J S T, Griffin B E, Ng M H. J Med Virol. 1998;55:227–233. doi: 10.1002/(sici)1096-9071(199807)55:3<227::aid-jmv8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 54.Labrecque L G, Xue S A, Kazembe P, Phillips J, Lampert I, Wedderburn N, Griffin B E. Int J Cancer. 1999;81:6–11. doi: 10.1002/(sici)1097-0215(19990331)81:1<6::aid-ijc2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 55.Takimoto T, Kamide M, Umeda R. Arch Otorhinolaryngol. 1984;239:87–92. doi: 10.1007/BF00454266. [DOI] [PubMed] [Google Scholar]
- 56.Fritsche M, Mundt M, Merkle C, Hajne R, Groner B. Mol Cell Endocrinol. 1998;143:143–154. doi: 10.1016/s0303-7207(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 57.Rayanade R J, Patel K, Ndubuisi M, Sharma S, Omura S, Etlinger J D, Pine R, Sehgal P B. J Biol Chem. 1997;272:4659–4662. doi: 10.1074/jbc.272.8.4659. [DOI] [PubMed] [Google Scholar]