Abstract
The origin of the H5N1 influenza viruses that killed six of eighteen infected humans in 1997 and were highly pathogenic in chickens has not been resolved. These H5N1 viruses transmitted directly to humans from infected poultry. In the poultry markets in Hong Kong, both H5N1 and H9N2 influenza viruses were cocirculating, raising the possibility of genetic reassortment. Here we analyze the antigenic and genetic features of H9N2 influenza viruses with different epidemiological backgrounds. The results suggest that the H9N2 influenza viruses of domestic ducks have become established in the domestic poultry of Asia. Phylogenetic and antigenic analyses of the H9N2 viruses isolated from Hong Kong markets suggest three distinct sublineages. Among the chicken H9N2 viruses, six of the gene segments were apparently derived from an earlier chicken H9N2 virus isolated in China, whereas the PB1 and PB2 genes are closely related to those of the H5N1 viruses and a quail H9N2 virus—A/quail/Hong Kong/G1/97 (Qa/HK/G1/97)—suggesting that many of the 1997 chicken H9 isolates in the markets were reassortants. The similarity of the internal genes of Qa/HK/G1/97 virus to those of the H5N1 influenza viruses suggests that the quail virus may have been the internal gene donor. Our findings indicate that the human and poultry H5N1 influenza viruses in Hong Kong in 1997 were reassortants that obtained internal gene segments from Qa/HK/G1/97. However, we cannot be certain whether the replicate complex of H5N1 originated from Qa/HK/G1/97 or whether the reverse transfer occurred; the available evidence supports the former proposal.
The natural reservoirs of influenza A viruses are the aquatic birds of the world (1). In these hosts, the 15 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes cause no disease signs and are in evolutionary stasis. After transfer to new avian or mammalian hosts, the viruses evolve rapidly and cause mild respiratory and occasional severe disease (2). The avian H5N1 influenza virus that transmitted to poultry and humans in 1997 caused high mortality in both species (3–6) and is unusual in having a large proportion of amino acid substitutions in all gene products except in the surface genes (7). It was therefore proposed that this H5N1 influenza virus might be a reassortant. The present study supports this contention and indicates that the donor of the replication gene complex may be an H9N2 influenza virus.
Surveillance studies in poultry markets in Hong Kong in 1997 indicated that two subtypes of influenza viruses were cocirculating. H5N1 influenza viruses were isolated from approximately 20% of chickens tested, whereas H9N2 viruses were also isolated from approximately 5% (8), raising the possibility of genetic exchange between these viruses after coinfection of the same host. Both subtypes were also isolated from ducks and geese, indicating a wide distribution in avian hosts.
In domestic avian species in North America, H9N2 influenza viruses occur primarily in turkeys, occasionally in quail, and rarely if ever in chickens. The H9N2 virus subtype was first isolated from turkeys in 1966 (9), when the virus was associated with mild respiratory disease. In Asia, long-term surveillance in live poultry markets in Hong Kong from 1975 to 1985 detected H9N2 influenza viruses in apparently healthy ducks but not in chickens (10). Since the early 1990’s, H9N2 influenza viruses have become widespread in domestic chickens in Asia (11–13).
The present report characterizes the genomes of H9N2 viruses isolated in 1997 from the Hong Kong poultry markets. H9N2 viruses isolated from Eurasia and America were also included because of a limited number of publications on H9 influenza viruses (GenBank contains only a single H9 HA sequence). Results of the present study indicate multiple lineages of H9N2 viruses in Asia and at least three distinguishable subgroups in Hong Kong poultry. They also suggest that reassortment occurred between the H9N2 and H5N1 viruses, possibly involving the acquisition of internal genes from Qa/HK/G1/97 by the H5N1 viruses that infected humans in 1997.
MATERIALS AND METHODS
Viruses and Serological Assays.
The viruses used in this study are given in Fig. 1 and Table 1. Those collected in Hong Kong in December of 1997 were from live-poultry markets throughout the city. Fecal samples from chickens, ducks, pigeons, and quail were inoculated into embryonated chicken eggs as described (14–15). Hyperimmune rabbit antiserum to the prototype H9N2 strain, A/turkey/Wisconsin/66 (Ty/WI/66), and postinfection chicken sera (see below) were treated with receptor-destroying enzyme (16) and used in hemagglutination inhibition assays.
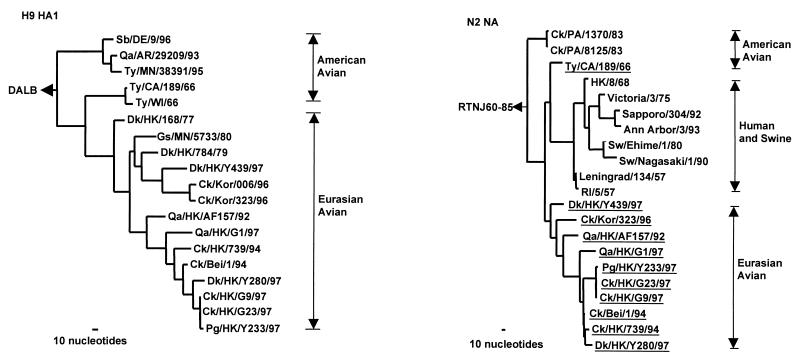
Figure 1.
Phylogenetic trees for the H9 HA1 and N2 NA genes of influenza A viruses. The nucleotide sequences of the HA1 and NA genes were analyzed with paup by using a maximum-parsimony algorithm. Nucleotides 43 to 1,047 (1,005 bp) of H9 HAs and nucleotides 21 to 1,393 (1,373 bp) of N2 NA were used for the phylogenetic analysis. The HA1 phylogenetic tree is rooted to duck/Alberta/60/76, DALB (H12N5). The N2 NA phylogenetic tree is rooted to A/ruddy turnstone/New Jersey/60/85, RTNJ60–85 (H4N9). The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Virus names and abbreviations are listed in Table 1, and the remaining sequences can be found in GenBank by using the following accession numbers: D90305, M12051, M11925, U42630, J02173, U42776, U43421, D00713, D29659, L37330, and M11205. All viruses underlined in the NA tree are H9N2 influenza viruses.
Table 1.
Crossreactivity between H9N2 influenza viruses in hemagglutination inhibition tests
| H9N2 viruses* | Antibody titers with antisera to influenza viruses
|
||||||
|---|---|---|---|---|---|---|---|
| Ty/WI/66 [R] | Ty/MN/38391/95 [C] | Sb/DE/9/96 [C] | Qa/HK/G1/97 [C] | Ck/HK/G9/97 [C] | Dk/HK/Y280/97 [C] | Ab/HK/M603/98 [C] | |
| Ty/CA/189/66 | 1280 | 1280 | 1280 | 20 | 40 | 20 | < |
| Ty/WI/66 | 320 | 640 | 640 | 20 | 20 | < | < |
| Gs/MN/5733/80 | 320 | 1280 | ≥2560 | 40 | 80 | 160 | < |
| Qa/AR/29209/93 | 320 | 640 | 640 | 20 | 40 | 20 | < |
| Ty/MN/38391/95 | 320 | 320 | 1280 | < | 40 | 20 | < |
| Sb/DE/9/96 | 160 | 640 | 640 | < | 20 | 20 | < |
| Ck/Kor/006/96 | 160 | 1280 | 320 | 20 | 40 | 20 | < |
| Ck/Kor/323/96 | 80 | 640 | 160 | < | 40 | 20 | < |
| Ck/Bei/1/94 | 80 | 320 | 80 | 40 | 320 | 320 | < |
| Qa/HK/G1/97 | 80 | 1280 | 80 | 1280 | 160 | 80 | < |
| Ck/HK/G9/97 | 640 | ≥2560 | 1280 | 320 | ≥2560 | >2560 | < |
| Ck/HK/G23/97 | 320 | ≥2560 | 640 | 320 | 2560 | ≥2560 | < |
| Pg/HK/Y233/97 | 640 | ≥2560 | 1280 | 1280 | ≥2560 | >2560 | < |
| Dk/HK/Y280/97 | 160 | 1280 | 80 | 80 | 1280 | 640 | < |
| Dk/HK/Y439/97 | 160 | 640 | 320 | 20 | 40 | 20 | < |
| Ab/HK/M603/98(H11N1) | < | < | < | < | < | < | 320 |
Virus abbreviation: animals: turkey, Ty; goose, Gs; quail, Qa; chicken, Ck; shorebird, SB; pigeon, Pg, aquatic bird, Ab. Place: Arkansas, AR; California, CA; Delaware, DE; Wisconsin, WI; Minnesota, MN; Beijing, Bei; Korea, Kor; Hong Kong, HK. [R] = hyperimmune rabbit antiserum, [C] = postinfection chicken antisera.
The H9N2 viruses collected in Hong Kong in 1992–94 were from chickens and quail with mild clinical disease that responded to antibiotic treatment, indicating that infection involved both influenza virus and bacteria. The H9N2 Beijing isolates were from chickens infected with virus that had produced 40% mortality in Guangdong Province in 1994 (11). Each of the viruses used for sequence analysis was cloned twice in eggs at limiting dilution. All viruses were handled in a biohazard level 3 facility approved by the United States Department of Agriculture.
Chicken Antisera.
Three-week-old specific pathogen-free chickens were inoculated intranasally and orally with 1.0 ml of allantoic fluid (egg infection dose, >106.5/ml). The chickens were bled 3 wk postinfection and boosted intravenously with 1.0 ml of infectious virus.
RNA Extraction and PCR.
Viral RNA was extracted from infected allantoic fluid (RNeasy Mini Kit, Qiagen, Chatsworth, CA). After reverse transcription, cDNA was amplified by PCR, as described previously (17). PCR products were purified with the QIAquick PCR Purification Kit (Qiagen).
Gene Sequencing and Analysis.
PCR products were sequenced by using synthetic oligonucleotides by the Center for Biotechnology at St. Jude Children’s Research Hospital. Reactions were performed with Rhodamine Dye-Terminator Cycle Sequencing Ready Reaction Kits with AmpliTaqDNA Polymerase FS (Perkin–Elmer/Applied Biosystems). Samples were electrophoresed and analyzed on model 377 DNA sequencers (Perkin–Elmer/Applied Biosystems).
All sequence data were edited and translated by using the wisconsin Sequence Analysis Package, Version 10.0 (GCG). Nucleotide and deduced amino acid sequences were aligned by the Feng–Doolittle progressive alignment method and manipulated with genedoc, version 2.3 (K. B. Nicholas at ketchup@cris.com). Phylogenetic analysis was performed with paup (Phylogenetic Analysis Using Parsimony, Version 4.0, D. Swofford, Illinois Natural History Survey, Champaign, IL). Trees with the shortest lengths were identified by implementing a heuristic search.
Nucleotide Sequence Accession Numbers.
The nucleotide sequences for all H9N2 influenza viruses used in this study are available from GenBank under accession numbers AF156373 through AF156485.
RESULTS
Influenza viruses of the H9N2 subtype were isolated from the fecal samples of apparently healthy chickens, ducks, geese, quail, and pigeons in Hong Kong poultry markets during December 1997. Additional H9N2 viruses were isolated from floor cages and other items in the markets. The isolates were characterized, both antigenically and genetically, to establish interrelationships.
Antigenic Analysis.
Postinfection chicken antisera to representative influenza viruses and a hyperimmune rabbit antiserum to Ty/WI/66 virus were used to determine the extent of antigenic diversity among H9N2 isolates from North America and Asia (Table 1). Rabbit antiserum to the prototype H9N2 virus reacted with all H9N2 viruses from North America and from Asia, but not with a heterologous H11N1 influenza virus-Aquatic bird/Hong Kong/M603/98 (Ab/HK/M603/98); the reactivity was lower with some of the H9N2 influenza viruses isolated in 1997 in Asia. Postinfection chicken antiserum to Ty/MN/95 crossreacted with H9N2 viruses from North America and Asia, whereas antiserum to Sb/DE/96 reacted with lower titers to some of the Asian H9N2 isolates. The most discriminating antisera were to the Asian H9N2 viruses; antiserum to Qa/HK/G1/97 reacted to low titers with American H9N2 isolates and demonstrated antigenic differences between the H9N2 viruses from Asia in 1994 and in 1997. Similarly, antisera to Ck/HK/G9/97 and to Dk/HK/Y280/97 showed antigenic differences between these homologous viruses and Qa/HK/G1/97 and earlier H9N2 isolates from Asia and America. Thus, antigenically distinguishable H9N2 influenza viruses were isolated from Asia and examples of these viruses were cocirculating in the live poultry markets in Hong Kong in 1997.
Sequence Analysis of the Hemagglutinin and Neuraminidase of H9N2 Viruses.
Comparison of the extent of homology between the HAs of representative H9N2 viruses from Hong Kong live poultry markets shows 91% nucleotide sequence homology between Ck/HK/G9/97 and Qa/HK/G1/97 and 84% homology between Ck/HK/G9/97 and Dk/HK/Y439/97 (Table 2).
Table 2.
Homology, %* of HA and NA genes between different H9N2 influenza viruses isolated in Hong Kong markets in 1997
| Qa/HK/G1/97 | Ck/HK/G9/97 | Dk/HK/Y439/97 | |
|---|---|---|---|
| Gene | NA | NA, HA | HA |
| Qa/HK/G1/97 | — | 91 | 84 |
| Ck/HK/G9/97 | 92 | — | 84 |
| Dk/HK/Y439/97 | 88 | 88 | — |
Homologies were calculated based on the nucleotide sequences of HA gene (43 to 1,572) and NA gene (21 to 1,393).
Phylogenetic relationships among the major surface glycoproteins of H9 influenza viruses are shown in Fig. 1. Two distinct lineages were apparent from the HA1 sequences of the H9N2 viruses. One of them included H9N2 viruses isolated in America, whereas the other comprised viruses isolated in both America and Eurasia. The Eurasian sublineage could be further divided into two branches: one that contained only viruses isolated in China, and one that comprised the viruses found in both China and Korea. Each branch has a duck or quail H9N2 virus close to the root. The H9N2 influenza viruses from the poultry markets in Hong Kong in 1997 occurred in each branch of the Eurasian lineage. Thus, Dk/HK/Y439/97 is grouped with H9N2 isolates from chickens in Korea; Qa/HK/G1/97 is grouped with earlier H9N2 isolates from quail in Hong Kong in 1992 and chicken/Hong Kong/G9/97 is grouped with isolates from ducks, pigeons, and chickens in Hong Kong in 1997. Hence, the phylogenetic analyses of HA indicate that different lineages of H9 influenza viruses cocirculated in Hong Kong poultry markets in 1997.
The American H9 HAs also form two subgroups (Fig. 1), one comprising recent isolates exclusively and the other consisting of earlier American isolates joined with Eurasian viruses. This apparent discrepancy could be interpreted in either of two ways: the American viruses were the precursors of the Eurasian sublineage or, more likely, we lack sufficient sequence data to establish the real phylogenetic relationship.
Comparison of the extent of homology between the NAs of representative H9N2 viruses shows 92% nucleotide sequence homology between Ck/HK/G9/97 and Qa/HK/G1/97, and 88% homology between Ck/HK/G9/97 and Dk/HK/Y439/97 (Table 2). Phylogenetic analysis of N2 NAs of the H9N2 influenza viruses also revealed separation into American and Eurasian lineages (Fig. 1). The latter contains human/swine and avian sublineages. The phylogenetic analysis of the Hong Kong N2 NAs in the Eurasian lineage shows no clear subdivisions.
The N2 NAs of the H9N2 viruses from poultry markets were widely dispersed in the Eurasian avian lineage. The N2 of Dk/HK/Y439/97 was located near the root of the Eurasian lineage; Qa/HK/G1/97 follows an earlier quail isolate from Hong Kong in 1992, and the isolates from 1994 were located near the terminus of the tree together with H9N2 isolates from chickens and pigeons from the poultry markets. Thus, multiple distinguishable N2 NAs were cocirculating in the live poultry markets in Hong Kong in 1997.
Analysis of the Internal Genes of H9N2 Influenza Viruses.
Surveillance in the poultry markets in Hong Kong in 1997 revealed that the H9 influenza viruses were cocirculating with the H5N1 influenza viruses, raising the possibility of genetic exchange between these viruses. We therefore determined the cross-homologies of H9 and H5 internal genes and constructed phylogenetic trees.
Table 3 shows the results of cross homology between the internal genes of the H9 isolates from Hong Kong poultry markets and those of HK/156/97, a representative human H5N1 virus. Of interest, the homologies for the PB2 and PB1 genes of some H9 and H5 viruses ranged from 97% to 98%, whereas those for others ranged from 85% to 90%. The Qa/HK/G1/97 virus showed greater than 98% cross-homologies, regardless of the internal genes tested. Taken together, the data suggest that the internal genes of some H9 influenza viruses are highly related to those of H5N1 Hong Kong influenza viruses.
Table 3.
Homology, %* between the human H5 (HK/156/97) and avian H9 influenza viruses isolated in Hong Kong in 1997
| Viral gene | % homology
|
|||||
|---|---|---|---|---|---|---|
| Qa/HK/G1/97 | Ck/HK/G9/97 | Ck/HK/G23/97 | Pg/HK/Y233/97 | Dk/HK/Y280/97 | Dk/HK/Y439/97 | |
| PB2 | 98 | 97 | 97 | 97 | 85 | 87 |
| PB1 | 98 | 98 | 98 | 98 | 89 | 90 |
| PA | 98 | 88 | 88 | 89 | 89 | 89 |
| NP | 98 | 90 | 90 | 90 | 89 | 94 |
| M | 98 | 96 | 96 | 96 | 96 | 92 |
| NS | 98 | 94 | 93 | 94 | 94 | 91 |
Homologies were calculated based on the nucleotide sequences of NP (45 to 1,499), M (6 to 989), NS (42 to 876), PB1 (25 to 2,242), PB2 (22 to 2,262), and PA (4 to 2,130).
The genes encoding the nonstructural (NS), matrix (M), nucleoprotein (NP), and the three polymerase proteins (PB2, PB1 and PA) were phylogenetically analyzed. To conserve space, we present only the terminal branches of the Eurasian avian lineage of NS and PB1 phylogenetic trees (Fig. 2), and to further conserve space, generalized phylogenies of the PB2, PA, NP, and M genes are presented (Fig. 3). The NS phylogenetic tree shows that most of the H9 NS genes from China form a subgroup within the Eurasian avian lineage that has a sister-group relationship with the NS genes of H5N1 influenza viruses isolated in Hong Kong in 1997. Like the HA gene, the NS gene of most current H9 influenza viruses is derived from an early chicken H9 virus identified in Beijing in 1994. It is noteworthy that the NS gene of Qa/HK/G1/97 is joined directly to the H5 virus group and that it maintains an out-group relationship with those viruses. The NS genes of Dk/HK/Y439/97 and two Korean chicken H9 viruses form another subgroup together with Dk/Nanchang/1944/93 within the Eurasian avian lineage. Similar relationships were observed among the M, NP, PA generalized phylogenies (Fig. 3). Thus, three distinct subgroups of H9N2 influenza viruses were cocirculating in the poultry markets in Hong Kong.
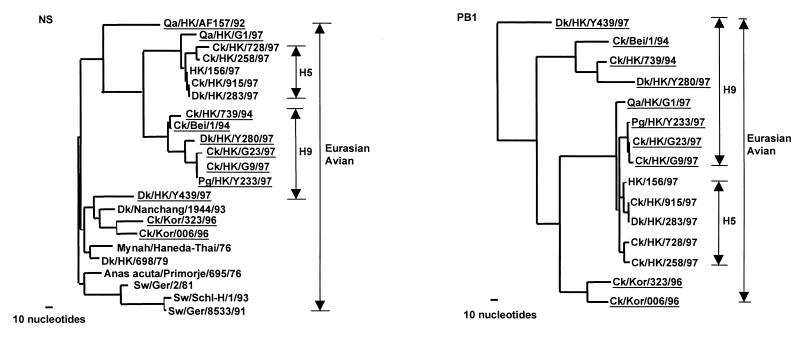
Figure 2.
Phylogenetic trees for the NS and PB1 of avian influenza viruses from Eurasia. Phylogenetic relationships were determined with the paup program. The nucleotide tree of the NS gene is rooted to A/Equine/Prague/1/56 (H7N7). The PB1 tree is rooted to B/Lee/40. Nucleotides 39 to 842 (804 bp) of NS gene and nucleotides 25 to 2,237 (2,213 bp) of PB1 gene were used for the phylogenetic analysis. Only the terminal branches of the Eurasian avian lineage of the trees are shown. Virus names and abbreviations are listed in Table 1 or can be found in ref. 7 or GenBank by using the following accession numbers: AF036360, U49492, U49493, M17070, M60800, M55484, Z46440, Z26865, and AF036362. All viruses underlined are H9N2 influenza viruses.
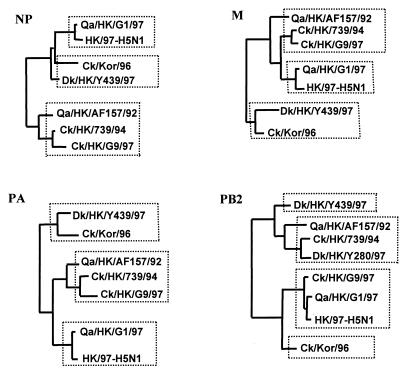
Figure 3.
Generalized phylogenies in diagrammatic form of NP, M, PB2, and PA genes of H9N2 influenza viruses. The nucleotide sequences of NP (1,381 bp), M (935 bp), PB2 (2,177 bp), and PA (2,125 bp) were used in the phylogenetic analysis. Only the major terminal branches of the Eurasian avian lineages of the trees are shown. Abbreviations are listed in Table 1.
The PB1 gene tree for H9 influenza viruses comprises four different subgroups within the Eurasian avian lineage (Fig. 2). Dk/HK/Y439/97 maintains an out-group relationship with most of the avian viruses tested, suggesting that its natural reservoir status has been preserved. The early H9 chicken and Dk/HK/Y280/97 separated into the second subgroup. The chicken and quail viruses isolated from Hong Kong poultry markets in 1997 form the third subgroup, which includes the H5 Hong Kong viruses and Qa/HK/G1/97. It is noteworthy that Qa/HK/G1/97 still keeps an out-group relation with all H5 influenza viruses. It is also noted that the common node of this subgroup is far from the preceding node (with more than 100 nucleotide differences), suggesting that the viruses in this virus subgroup of H9 and H5 influenza viruses may acquire their PB1 genes recently from a common precursor. The PB1 of chicken Korean H9 viruses clustered into another sublineage, the fourth subgroup, in the Eurasian avian lineage. A similar phylogenetic relation was also found in PB2 genes (Fig. 3). Thus, the phylogenetic analyses revealed that the chicken H9 influenza viruses from the poultry markets in Hong Kong in 1997 may be reassortant viruses, which acquired their PB1 and PB2 genes from H5-like or Qa/HK/G1/97-like influenza viruses.
Generalized phylogenies showing the main diagrammatic form of the NP, M, PA, and PB2 genes of H9N2 viruses are shown in Fig. 3. These diagrams show that the phylogenetic topology of NP, M, and PA genes of the H9N2 viruses that cocirculated in Hong Kong live poultry markets are similar to that of the NS gene (Fig. 2) and contain three groups. In contrast, the phylogenetic topology of PB2 gene is similar to that of the PB1 gene and has four groups. In all cases, the NP, M, PA, and PB2 genes of H5N1 viruses are linked with Qa/HK/G1/97 virus, indicating that they either share a common precursor or transmitted the gene complex from one to the other.
DISCUSSION
Influenza viruses of the H9 subtype have until recently received little attention with a single H9 sequence available in GenBank. The present studies indicate that, like other influenza hemagglutinin subtypes studied, the H9 influenza viruses can be separated geographically into lineages in Eurasia and in the Americas. Antigenic and phylogenetic analysis of the H9 hemagglutinin of the Eurasian lineage reveals at least two separate sublineages, whereas analysis of the genes encoding the internal proteins reveals at least three distinct lineages.
The available epidemiological data indicate that H9N2 influenza viruses were not detectable in chickens in southern China from 1975 to 1985, for extensive surveillance in live poultry markets in Hong Kong during this period detected the H9 subtype only in ducks (10). The first reported isolates of H9N2 in Asia from domestic poultry were in 1992 and 1994 (11), and by 1997 reports from Korea and Europe indicated widespread H9N2 influenza virus activity (12, 13, 18). The present studies indicate that the H9N2 influenza viruses from Asia may have established a stable lineage in chickens and other domestic poultry.
Homology analysis suggests that one of the H9N2 influenza viruses, Qa/HK/G1/97, is highly related to H5N1 influenza viruses in all of its six internal genes. Moreover, in each of the internal gene trees, Qa/HK/G1/97 is joined with the H5 virus subgroups and maintains an almost direct ancestor relationship in the topology. A key question is whether Qa/HK/G1/97 (H9N2) donated the six internal genes to the H5N1 viruses or whether the exchange was in the opposite direction. If the latter alternative holds, Qa/HK/G1/97 would have clustered directly within the H5N1 virus group. Because it shows a consistent out-group relationship in all phylogenetic trees, we postulate that the H5N1 Hong Kong influenza virus acquired its internal genes from Qa/HK/G1/97 or one of its precursors.
The presence of H9N2 influenza viruses in domestic chickens throughout the 1990’s in Asia and the absence of H5N1 until 1997 favor the possibility that the internal gene segments of the H5N1 virus originated from the H9N2 virus. It could be argued that the H5N1 influenza virus could have been circulating in a nonpathogenic form and that insufficient sampling was being done to detect these viruses. However, the earlier nonpathogenic H5 influenza viruses from Hong Kong in 1975 possessed distinctly different internal genes from the 1997 highly pathogenic H5N1 virus, arguing against this possibility (7).
A possible source of the H5 hemagglutinin gene in the H5N1 viruses in humans and poultry was from a virus related to A/Goose/Guangdong/1/96 (H5N1) (19). Influenza viruses of the H5N1 subtype have been isolated from geese from southern China in 1999, and preliminary characterization indicates that the HA is >99% homologous with the human HK/156/97 (H5N1) virus, but all the other genes including the NA are distinctly different (unpublished results). Thus the H5N1 viruses that infected humans in Hong Kong in 1997 were probably reassortants that obtained their H5 gene from a goose H5N1 virus and their internal gene complex from a Qa/HK/G1/97-like virus. The origin of the N1 neuraminidase is still unresolved.
Analysis of 14 H9N2 influenza viruses from the live poultry markets in Hong Kong in 1997 revealed only one Qa/HK/G1/97-like virus. Thus, Qa/HK/G1/97-like influenza viruses were less prevalent than H9N2 viruses from other lineages. The placement of the Qa/HK/G1/97 virus on a separate ancestral branch of the phylogenetic tree must be viewed as preliminary and additional H9N2 influenza viruses from Asia must be examined to support this proposal.
Acknowledgments
These studies were supported by Public Health Research Grant AI29680 and AI95357 from the National Institute of Allergy and Infectious Diseases, Cancer Center Support CORE Grant CA-21765, and the American Lebanese Syrian Associated Charities, the World Health Organization, the Department of Health (DOH), the Hong Kong Government, the Committee on Research and Conference Grants, and the Office of the Vice Chancellor, the University of Hong Kong. We thank Melissa Norwood and Dr. Lijuan Zhang for technical support for these studies and Alice Herren for preparation of this manuscript. We also thank John Gilbert for scientific editing.
ABBREVIATIONS
- HA
hemagglutinin
- NP
nucleoprotein
- NA
neuraminidase
- NS
nonstructural
- M
matrix
- Ty
turkey
- Gs
goose
- Qa
quail
- Ck
chicken
- Sb
shorebird
- Pg
pigeon, Ab, aquatic bird
- Bei
Beijing
- Kor
Korea
- HK
Hong Kong
Footnotes
References
- 1.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludwig S, Stitz L, Planz O, Van H, Fitch W M, Scholtissek C. Virology. 1995;212:555–561. doi: 10.1006/viro.1995.1513. [DOI] [PubMed] [Google Scholar]
- 3.de Jong J C, Claas E C J, Osterhaus A D M E, Webster R G, Lim W L. Nature (London) 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claas E C J, Osterhaus A D M E, Van Beck R, de Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 5.Subbarao K, Klimov A, Katz J, Regenery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 6.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. J Virol. 1998;72(8):6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou N N, Shortridge K F, Claas E C J, Krauss S L, Webster R G. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shortridge, K. F. (1999) Vaccine, in press. [DOI] [PubMed]
- 9.Homme P J, Easterday B C. Avian Dis. 1970;14(2):285–290. [PubMed] [Google Scholar]
- 10.Shortridge K F. Semin Respir Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- 11.Sun Y. In: Proceedings of the Fourth International Symposium on Avian Influenza. Swayne D E, Slemons R D, editors. United States Animal Health Association; 1997. (Rose Printing Company, Tallahassee, FL), pp. 47–49. [Google Scholar]
- 12.Guan Y. Ph.D. thesis. University of Hong Kong; 1997. pp. 105–116. [Google Scholar]
- 13.Mo I P, Song C S, Kim K S, Rhee J C. In: Proceedings of the Fourth International Symposium on Avian Influenza. Swayne D E, Slemons R D, editors. United States Animal Health Association; 1997. (Rose Printing Company, Tallahassee, FL), pp. 379–383. [Google Scholar]
- 14.Shortridge K F, Butterfield W K, Webster R G, Campbell C H. Bull W H O. 1977;55:15–20. [PMC free article] [PubMed] [Google Scholar]
- 15.Shortridge K F, Zhou N N, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti K G, et al. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- 16.Palmer D F, Coleman M, Dowdle W R, Schild G G. Advanced Laboratory Techniques for Influenza Diagnosis. Education, and Welfare: U. S. Department of Health; 1975. , Immunology Series No. 6, pp. 51–52, [Google Scholar]
- 17.Shu L L, Lin Y P, Wright S M, Shortridge K F, Webster R G. Virology. 1994;202:825–833. doi: 10.1006/viro.1994.1404. [DOI] [PubMed] [Google Scholar]
- 18.Alexander D J. In: Proceedings of the Fourth International Symposium on Avian Influenza. Swayne D E, Slemons R D, editors. United States Animal Health Association; 1997. (Rose Printing Company, Tallahassee, FL), pp. 9–13. [Google Scholar]
- 19.Xu X, Cox N, Guo Y. Am. Soc. Virol. 17th Annu. Meeting. 1998. , Vancouver, British Columbia, Canada W29–5, 110 (abstr.). [Google Scholar]