Abstract
Gene expression profiling is a relatively new technology for the study of breast cancers, but within the past few years there has been a rapid rise in interest in its potential to improve the clinical management of breast cancer. This technology has contributed to our knowledge of the molecular pathology of breast tumours and shows promise as a tool to predict response to therapy and outcome, such as risk of metastasis. Microarray technology is continually developing and it is becoming apparent that, despite the various platforms available, robust conclusions can still be drawn that apply across the different array types. Gene expression profiling is beginning to appear in the breast cancer clinic but it is not yet fully evaluated. This review explores the questions that must be addressed before this technology can become an everyday clinical tool.
Introduction
Ongoing development of a wide spectrum of therapies for breast cancer has increased the complexity of breast cancer management in recent years. Clinicians must make decisions regarding the best treatment for an individual woman, but the disease parameters currently used to decide upon treatment are relatively unsophisticated. Furthermore, although these parameters may predict proportions of the population in which an outcome may occur fairly accurately, they cannot identify in which women the outcome will occur. This leads to over-treatment of many patients and inadequate treatment of others. High-throughput genomic technologies have been used to delineate tumour subtypes and response to therapy, and to identify patient characteristics that may influence tumour behaviour. One of the keys to improving breast cancer management is the targeting of treatment to those who will truly benefit, thereby avoiding iatrogenic morbidity in those who will not. The use of gene expression profiling is unfortunately not straightforward, because differences between the platforms used in studies have led to inconsistencies in results and considerable scepticism. However, as is discussed below, these problems are not insurmountable and gene expression profiling shows considerable promise, but large clinical studies are now needed.
Historical perspective
Clinically measurable characteristics (for instance, tumour size, spread to lymph nodes, distant metastases and histological appearance [tumour grade]) and patient characteristics (such as age and menopausal status) give an approximate guide to tumour behaviour and formed the basis of the earliest prognostic indices. Advances in immunohistochemistry added oestrogen receptor (ER)-α and progesterone receptor status, and most recently growth factor receptor status (epidermal growth factor receptor and ERBB2 over-expression), Ki-67 and others. Some or all of these factors are included in indices such as The International Consensus Guidelines of St Gallen [1] and the Nottingham Prognostic Index [2], which are widely used to guide treatment. It has been obvious for years that breast cancer is a heterogeneous disease, and more recently it has become apparent that dissection of the molecular basis of malignancies classifies tumours into increasingly large numbers of subtypes that require individualized treatment.
The advent of genomics and proteomics could be regarded as a natural progression from the use of only a few markers to the combination of large numbers of markers – 'tumour signatures' – that can define tumour subtypes and predict outcome.
Gene expression approaches to classification, prognostication and determining response to treatments
The primary focus of work using gene expression microarrays in the breast cancer field has been the molecular subclassification of breast cancer put forward by Perou and Sorlie and their coworkers [3-6] and the prognosis profiles proposed by the Amsterdam [7,8] and Rotterdam [9] groups (Table 1). The luminal and basal-like subtypes have been repeatedly identified and validated by gene expression analysis as the most distinctive of three or more molecular subtypes of breast cancer. Tumours that are positive for ERBB2 are predominantly identified as a distinct subtype of breast cancer by the 'intrinsic' subtype classification [3-6] and among the 'molecular apocrine' tumours [10]. However, ERBB2-positive tumours are also found within other classes, which presumably reflects heterogeneous expression of ER and their distinctive therapeutic and clinical attributes. The molecular subtypes identified are associated with significantly different clinical outcomes [4,11] and are likely to respond best to different treatment approaches.
Table 1.
Gene expression studies of breast carcinomas aiming to improve clinical management
| Gene expression profiles and classifiers | Tissue/population included | Samples (n) | Platform | Number of differentially expressed cDNAs/oligos/probe sets (total) | Ref. |
| Overall outcome/metastasis | PBC (node -ve, age <55 years) | 117 | oligo Agilent Hu25k | 70 (24,479) | [7] |
| PBC (node ±ve, age <53 years) | 295 | oligo Agilent Hu25k | 70 (24,479) | [8] | |
| PBC (node -ve, any age) | 287 | Affymetrix U133A | 76 (22,283) | [9] | |
| PBC (node -ve, ER+ only) | 668 | qRT-PCR | 21 genes | [32,33] | |
| PBC (population and validation cohorts) | 448 | Affymetrix U133 set | 64 (44,792) | [37] | |
| Tumour classification | PBC (+benign tissues) | 65 | cDNA (Stanford) | 476 (8102) | [3] |
| PBC (+benign tissues) | 78 | cDNA (Stanford) | 476 (8102) | [4] | |
| PBC (+benign tissues) | 122 | cDNA (Stanford) | 476 (8102) | [5] | |
| PBC (node ±ve) | 20 | Agilent, Applied Biosystems, cDNA (Stanford) | [6] | ||
| PBC + metastases | 105 | Agilent oligo (1Av1, 1Av2) | 1300 (>17,000) | [11] | |
| PBC (node ±) | 99 | cDNA (NCI) | 706 (7650) | [38] | |
| ER status | PBC (2–5 cm, node -ve) | 47 | cDNA | 100 (6728) | [39] |
| PBC (1.5–5 cm, node ±ve, ER/PR +ve or -ve) | 49 | Affymetrix HuGeneFL | 100 (5600) | [40] | |
| PRC (stage I-II, node -ve, ER and PR +ve) | 26 | SAGE | 520 (>50,000) | [41] | |
| Nodal status | PBC (ER ±ve, node ±ve) | 49 | Affymetrix U95A | 100 (12,626) | [42] |
| Apocrine | PBC (large operable/advanced inoperable/inflammatory) | 49 | Affymetrix U133A | 520 (22,283) | [10] |
| Wound Response | PBC (node ±ve, age <55 years) | 295 | Oligo Agilent Hu25k | 442 (24,479) | [16] |
| Proliferation | PBC (node ±ve, age <55 years) | 311 | Oligo Agilent Hu25k | 50 (24,479) | [17] |
| Grade | PBC (node ±ve, ER ±ve) | 189 | Affymetrix U133A | 128 (22,283) | [12] |
| Grade/progression | Normal breast, ADH, DCIS, IDC (LCM) | 61 | cDNA (Research Genetics) | 200 (1940) | [43] |
| Hereditary breast cancer | PBC (sporadic, BRCA1, BRCA2) | 22 | DNA clones | 176 (6512) | [18] |
| Fibroblasts from normal breast (BRCA1, normal controls) | 28 | cDNA (IMAGE) | 47 (5603) | [44] | |
| Tamoxifen resistant | PBC (responsive and nonresponsive to tamoxifen) | 112 | cDNA (NKI) | 81 (19,200) | [21] |
| ER response | MCF-7 cells and luminal breast tumours | 65 | cDNA (Agilent) | 822 (>17,000) | [13] |
| Neoadjuvant chemotherapy response | PBC (docetaxel) | 24 | Affymetrix U95A | 92 (12,626) | [20] |
| PBC (doxorubicin/cyclophosphamide) | 40 | Affymetrix U133A | 253 (22,283) | [19] | |
| PBC (doxorubicin) | 36 | Affymetrix U133A plus2.0 | 38 (54,678) | [14] | |
| PBC (gemcitabine/epirubicin/docetaxel) | 100 | Custom made oligo | 512 (21,329) | [45] | |
| PBC (doxorubicin/cyclophosphamide) | 16 | cDNA (NCI) | 137 (7650) | [46] |
Whilst we have endeavoured to highlight the major studies in this field, we regret any offence to authors of other important work not included in this table. ADH, atypical ductal hyperplasia; DCIS, ductal carcinoma in situ; ER, oestrogen receptor; IDC, invasive ductal carcinoma; LCM, laser capture microdissection; PBC, primary breast carcinoma; PR, progesterone receptor; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Traditional classifications of tumours may provide clear-cut treatment options in high and low risk cases, but often tumours fall into an 'intermediate' group; it is in these borderline cases that improvements are most urgently required. In these cases the 'safe' option is to over-treat, benefiting a relatively small minority of cases and exposing the rest to side effects unnecessarily. Conversely, a more conservative approach may avoid unwarranted treatment and additionally reduce costs, but some women that would benefit may go untreated. Studies that examine links between gene expression and known prognostic factors such as grade [12] and ER status [13] may be beneficial for this intermediate group. Tumour grade is an excellent example; grade 3 tumours are at much higher risk of recurrence than are grade 1 tumours, but 30–60% of tumours are classified as histological grade 2. Sotiriou and coworkers [12] recently used gene expression profiling to reclassify grade 2 tumours into two groups resembling grade 1 and grade 3 tumours with low and high risk of recurrence, respectively, thereby better characterizing these tumours conventionally considered to be 'intermediate risk'.
Expression profiling has uncovered signatures associated with distinct phenotypes (Table 1), such as profiles exhibited by hypoxic tumours or inflammatory breast cancers [14]. A recent study [15] showed that medullary breast cancer is a subgroup of basal breast cancers. Novel aspects of tumour behaviour have been identified by the expression of specific functional sets of genes such as 'wound response' genes [16]. A 'proliferation signature' has been shown to identify particular patient groups who have an extremely poor outcome in a subpopulation of breast cancer patients [17]. Gene expression studies have been used to distinguish between sporadic tumours and tumours from women with BRCA1 and BRCA2 germline mutations [18]. Tumours from BRCA1 mutation carriers have been found to be frequently of the basal subtype, whereas BRCA2 tumours fall mainly within the luminal A category [5]. Observations such as these may further our understanding of the pathogenesis of tumours in mutation carriers.
Response to treatment has also been studied using expression microarrays. A gene expression profile from pretreatment tumour biopsies has been found to predict response to combination doxorubicin/cyclophosphamide treatment [19]. The genes comprising this profile differed from those in the same group's previously reported profile for predicting response to docetaxel therapy [20]. Similarly, response to tamoxifen can be predicted by expression profiling [21]. These exciting results suggest that predictive profiles could potentially be found for any given chemotherapy or endocrine regimen.
Influences of genetic background on outcome
The finding that tumours from BRCA1 mutation carriers are predominantly of the basal subtype clearly demonstrates that genetic background can predispose to a particular tumour subtype and outcome. Furthermore, a study showed that oncogene-induced mouse mammary tumors from 31 inbred strains exhibit differing rates of metastasis [22]. The study also reported distinct gene expression profiles according to genetic background. This suggests that the tendency for metastatic disease to develop may have a germline component. The existence of a common 11-gene 'death from cancer' signature from both epithelial and nonepithelial malignancies [23] may lend support to this hypothesis. The implication of a germline polymorphism as opposed to a somatic mutation for assessment of metastasis risk is that risk assessment can be done using any tissue at the time of diagnosis or even before diagnosis of a primary tumor [24]. The existence of inherited metastasis risk factors (or prospective metastatic biomarkers) has potentially highly significant implications for our models of metastasis, clinical prognosis and the development of tailored treatment. However, the specific genes and mechanisms responsible for this heritable effect on metastasis remain to be fully identified, and more evidence is required to establish their influence on human disease.
Cost/benefits
Although the actual cost of performing gene expression analysis is likely to be higher than those of determining conventional clinical and histopathological markers, the potential savings in terms of avoiding over-treatment, based on existing guidelines, could be substantial.
Cost/benefit analyses of using a new 70-gene commercially available assay as an alternative to clinical guidelines to identify high-risk patients for adjuvant chemotherapy were recently described [25]. The study found that the use of gene expression profiling to guide treatment would result in a cost saving of US$2882 per woman treated, but in a greater number of women developing recurrent disease and a smaller gain in quality-adjusted life years (QALYs), compared with existing methods. As with any test that defines groups within a population, a gene expression threshold must be chosen to discriminate between high and low risk women. Adjustments to the level at which this test threshold was set had a greater effect on the number of QALYs obtained by gene expression profiling than did adjustments of the estimates of chemotherapy-associated risks and benefits. The discrimination threshold of the gene expression profile could not be adjusted to a level that would detect high risk individuals sensitively enough to match the performance of the conventional guidelines in terms of QALYs gained. This study highlights the extreme importance of setting the discrimination level correctly and concluded that further refinement and testing are needed before routine clinical use of the 70-gene assay can be recommended.
Reliability of microarray data
The genes that make up a gene expression signature are by their nature dependent on the patient and tumour characteristics, array platform, normalization method and statistical thresholds for gene selection or the classification algorithm employed. Using a particular dataset to generate a predictive profile has its own inherent bias based on its attributes. By simply changing the members of 'training' and 'test' sets, Ein-Dor and coworkers [26] were able to identify more than 1000 genes that are related to survival, even when using the same dataset and methods as van't Veer and colleagues [7,8], and found that many different but equally predictive lists of 70 genes could have been produced from the same analysis. Rather than indicating that the data are meaningless, this suggests that the predictive value is real but highly context dependent.
The comparability of different microarray platforms is a major issue. The lack of overlap (three genes) between the 70-gene signature of the Amsterdam group (cDNA arrays) [7,8] and the 76-gene signature of the Rotterdam group (Affymetrix oligonucleotide arrays) [9] has been claimed as evidence that genomic approaches are unreliable. A multiple random validation strategy has been suggested to combat this problem [27]. However, although validation is essential, a predictive signature determined from a highly selected group of samples cannot possibly be expected to replicate the exact findings when applied to a different group of samples using a different gene expression platform. The differences between signatures in different studies are due to varying inclusion criteria (age, lymph node status, diameter of tumour, adjuvant treatment, among other factors), the platform (cDNA or oligonucleotide arrays, or reverse transcription polymerase chain reaction) and data analysis methods used in each study.
Nevertheless, Sorlie and coworkers [6] recently showed that even with different array platforms the breast cancer subtypes are distinguishable at the unsupervised level. Despite some variation in the most differentially expressed genes identified by each array platform, there was a highly significant overlap at the pathway level. This confirms that distinct molecular mechanisms underlie the clinically relevant subtypes of breast cancer, and that perturbations in these mechanisms can be detected reliably by different platforms. Similarly, Hu and colleagues [11] recently described and validated a refined 'intrinsic' classification signature that is conserved across microarray platforms and also uncovered a possible 'new' subtype characterized by the high expression of interferon-regulated genes.
Validation of new findings with independent data is essential for us to believe that the results are indeed 'real', and this has been encouraged and facilitated by making it a prerequisite for publication that gene expression datasets are publicly accessible according to the MIAME (Minimum Information About a Microarray Experiment) guidelines [28], allowing complete datasets to be downloaded. Public access has permitted studies in which gene expression data from several studies are re-analyzed. For example, meta-analyses of multiple experiments using different platforms have resulted in new predictive signatures that perform as well or better than the platform specific signature [29,30] (Figure 1). These approaches remove the inherent bias of a single microarray platform and are able to concentrate on genes that are consistently differentially expressed, regardless of the technology used. However, they may be limited by the number of common genes represented.
Figure 1.
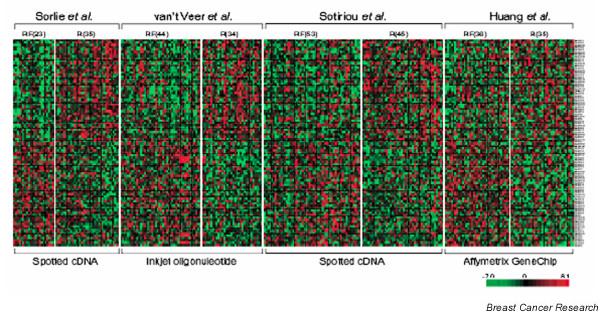
Combining datasets. Combining data from multiple gene expression studies of human breast tumours reveals significant overlap, despite inherent differences in the technology used. One example, shown here, is a 90-gene meta-signature reported by Shen and coworkers [30], which achieved equal or better prognostic performance compared with the individual signatures derived from four studies of breast cancer recurrence using different microarray platforms. Primary tumours were taken at diagnosis from patients who later had recurrent (R) or recurrence-free (RF) disease. Individual heat maps show increased expression (red) and decreased expression (green) of the raw data from four separate experiments using different platforms. With permission from Shen and coworkers [30].
Improvements in technology and analysis
Advances in gene expression technology are continually being made, with increasing numbers of more clearly defined transcripts represented on each new generation of array. Improvements in RNA extraction, quantification and quality assessment (such as the RNA integrity number [31]) will increase reliability of expression profiling. Improved labelling and amplification protocols enable profiling to be performed on smaller amounts of tissue.
Laser capture microdissection has been used to ensure that RNA is isolated from tumour rather than normal tissue. Alternatively, small amounts of RNA have been isolated from formalin fixed, paraffin embedded blocks rather than fresh frozen tissue, and can be used in assays such as the multigene reverse transcription polymerase chain reaction classifier commercially known as OncotypeDX [32,33]. These are important considerations because taking fresh tissue for gene expression analysis from a tiny tumour can compromise histopathology reporting. These improved methods will be required to circumvent the logistical difficulties, costs and time associated with collecting fresh tissue straight from a tumour at the time of surgery. Multicentre studies will be facilitated by techniques for extraction of intact RNA from preserved tissues, because these will obviate the need to collect and transport frozen material.
Although much of the additional information from expression profiling of primary tumours is consistent with that for existing markers, there have been some conflicting results; for example, some women found to be ERBB2 positive by immunohistochemistry were classified in the luminal cluster and not the ErbB2 group [4]. It may be difficult to choose the most appropriate treatment when different assays conflict, and of course existing technologies should not be discounted. Fluorescent in situ hybridization is still considered the most reliable and consistent method to determine ERBB2 status [34].
Trials: what is required
Large trials are now needed to evaluate fully the use of genomic signatures. Recruitment has begun for the MINDACT (Microarray for Node-negative Disease Avoids Chemotherapy) trial, which will be run under the auspices of the TRANSBIG consortium and coordinated by the EORTC (European Organisation for Research and Treatment of Cancer). It will study women with node-negative disease who are aged 18–75 years. One of its primary aims is to establish whether women with a good prognosis gene expression signature (assessed using Mammaprint, a commercially available form of the 70-gene profile reported by van't Veer and coworkers [7,8]) but poor prognosis clinicopathological status (as assessed using the Adjuvant! Online software [35]) can be spared chemotherapy without reducing metastasis-free survival. One of the secondary objectives is to identify and validate expression profiles that predict response to chemotherapy and endocrine therapy. Another large trial currently underway is being run by the US Intergroup PACCT (Program for the Assessment of Clinical Cancer Tests) to assess the OncotypeDX signature in 10,046 women aged 18–75 years with operable, node negative, ER positive and/or progesterone receptor positive breast cancer. In this study, patients with an intermediate risk OncotypeDX score will be randomized to combination chemotherapy and endocrine therapy (the usual treatment) or endocrine therapy alone. These two studies are seen as the best way to establish whether the genomic signatures will lead to the desired goal of better targeting treatment to those who will benefit and so yielding improvements in breast cancer mortality and morbidity.
With individualization of therapy, it is inevitable that women in a study will be undergoing many different treatment regimens. Consequently, it may become more difficult to obtain sufficiently high numbers of women with similar regimens to compare with each other to evaluate new tests with adequate statistical power. Many of the profiles have been developed in well defined groups of women (Table 1), and clearly it is important that validation takes place in equivalent groups of women undergoing the same treatment, because prognostic differences evident in one group may not be apparent in a differently treated group. It is likely that new profiles will be developed in the years ahead, and the need for large numbers of patients will necessitate large multinational trials.
New drugs are rapidly being developed. In order for gene expression profiling to be useful, the initiation of trials evidently needs to keep pace with the introduction of new treatments that are shown to be best practice.
Conclusion
Fundamentally, there is a major leap from the observation that gene expression profiles can predict outcome to the use of these profiles in treatment decisions. In recent years great advances in our knowledge of the molecular biology of breast tumours have been achieved using genomic approaches. It is perhaps not surprising that genomic signatures tend to outperform the pre-existing clinical models when one considers the large numbers of additional factors that are being taken into account, and that precision of measurement of these factors is continuous (gene expression levels) rather than categorical (for example, tumour grade and lymph node status). Despite their promise, some caution is required with the new approaches. A study comparing the power of gene expression measurements with that of conventional prognostic markers found that transcriptional profiling approaches did not perform noticeably better than indices constructed from the clinical variables [36].
The challenge that lies ahead is to convert the increased information potentially available from gene expression profiling of breast tumours into useful tools that can optimize clinical decisions and tailor treatment regimens to an individual patient, ultimately improving outcome and reducing overall costs. This will take place through advances in sample processing, microarray technology and statistical analysis techniques to obtain consistency. Finally, investment in trials of this genomic technology is needed now if savings in costs of treatment, particularly with the rise in availability of expensive drugs, are to be made in the long term.
Abbreviations
ER = oestrogen receptor; QALY = quality-adjusted life year.
Competing interests
The authors declare that they have no competing interests.
Note
This article is part of a review series on High-throughput genomic technology in research and clinical management of breast cancer, edited by Yudi Pawitan and Per Hall.
Other articles in the series can be found online at http://breast-cancer-research.com/articles/review-series.asp?series=bcr_Genomic
Contributor Information
Andrew H Sims, Email: Andrew.H.Sims@manchester.ac.uk.
Kai Ren Ong, Email: Kai-Ren.Ong@manchester.ac.uk.
Robert B Clarke, Email: Robert.Clarke@manchester.ac.uk.
Anthony Howell, Email: anthony.howell@christie-tr.nwest.nhs.uk.
References
- Goldhirsch A, Glick JH, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol. 2005;16:1569–1583. doi: 10.1093/annonc/mdi326. [DOI] [PubMed] [Google Scholar]
- Haybittle JL, Blamey RW, Elston CW, Johnson J, Doyle PJ, Campbell FC, Nicholson RI, Griffiths K. A prognostic index in primary breast cancer. Br J Cancer. 1982;45:361–366. doi: 10.1038/bjc.1982.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, Borresen-Dale AL. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol. 2006;24:1656–1664. doi: 10.1200/JCO.2005.03.2755. [DOI] [PubMed] [Google Scholar]
- Dressman HK, Hans C, Bild A, Olson JA, Rosen E, Marcom PK, Liotcheva VB, Jones EL, Vujaskovic Z, Marks J, et al. Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin Cancer Res. 2006;12:819–826. doi: 10.1158/1078-0432.CCR-05-1447. [DOI] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Cervera N, Charafe-Jauffret E, Mamessier E, Adelaide J, Debono S, Houvenaeghel G, Maraninchi D, Viens P, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 2006;66:4636–4644. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, van't Veer L, Lamb J, He YD, Mao M, Fine BM, Bernards R, van de Vijver M, Deutsch P, Sachs A, et al. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005;65:4059–4066. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- Hedenfalk I, Ringner M, Ben-Dor A, Yakhini Z, Chen Y, Chebil G, Ach R, Loman N, Olsson H, Meltzer P, et al. Molecular classification of familial non-BRCA1/BRCA2 breast cancer. Proc Natl Acad Sci USA. 2003;100:2532–2537. doi: 10.1073/pnas.0533805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleator S, Tsimelzon A, Ashworth A, Dowsett M, Dexter T, Powles T, Hilsenbeck S, Wong H, Osborne CK, O'Connell P, et al. Gene expression patterns for doxorubicin (Adriamycin) and cyclophosphamide (Cytoxan) (AC) response and resistance. Breast Cancer Res Treat. 2006;95:229–233. doi: 10.1007/s10549-005-9009-7. [DOI] [PubMed] [Google Scholar]
- Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, Mohsin S, Osborne CK, Chamness GC, Allred DC, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- Jansen MP, Foekens JA, van Staveren IL, Dirkzwager-Kiel MM, Ritstier K, Look MP, Meijer-van Gelder ME, Sieuwerts AM, Portengen H, Dorssers LC, et al. Molecular classification of tamoxifen-resistant breast carcinomas by gene expression profiling. J Clin Oncol. 2005;23:732–740. doi: 10.1200/JCO.2005.05.145. [DOI] [PubMed] [Google Scholar]
- Qiu TH, Chandramouli GV, Hunter KW, Alkharouf NW, Green JE, Liu ET. Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res. 2004;64:5973–5981. doi: 10.1158/0008-5472.CAN-04-0242. [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KW, Crawford NP. Germ line polymorphism in metastatic progression. Cancer Res. 2006;66:1251–1254. doi: 10.1158/0008-5472.CAN-05-3705. [DOI] [PubMed] [Google Scholar]
- Oestreicher N, Ramsey SD, Linden HM, McCune JS, van't Veer LJ, Burke W, Veenstra DL. Gene expression profiling and breast cancer care: what are the potential benefits and policy implications? Genet Med. 2005;7:380–389. doi: 10.1097/01.gim.0000170776.31248.75. [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- Microarray Gene Expression Data Society: MGED Society http://www.mged.org
- Warnat P, Eils R, Brors B. Cross-platform analysis of cancer microarray data improves gene expression based classification of phenotypes. BMC Bioinformatics. 2005;6:265. doi: 10.1186/1471-2105-6-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Ghosh D, Chinnaiyan AM. Prognostic meta-signature of breast cancer developed by two-stage mixture modeling of microarray data. BMC Genomics. 2004;5:94. doi: 10.1186/1471-2164-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- Perez EA, Suman VJ, Davidson NE, Martino S, Kaufman PA, Lingle WL, Flynn PJ, Ingle JN, Visscher D, Jenkins RB. HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol. 2006;24:3032–3038. doi: 10.1200/JCO.2005.03.4744. [DOI] [PubMed] [Google Scholar]
- Adjuvant! Online. Decision making tools for health care professionals http://www.adjuvantonline.com
- Eden P, Ritz C, Rose C, Ferno M, Peterson C. "Good Old" clinical markers have similar power in breast cancer prognosis as microarray gene expression profilers. Eur J Cancer. 2004;40:1837–1841. doi: 10.1016/j.ejca.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K, Sahin A, Aldaz CM. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Williams RD, Cattini N, Copeland M, Giddings I, Wooster R, tePoele RH, Workman P, Gusterson B, Peacock J, et al. Gene expression profiling after radiation-induced DNA damage is strongly predictive of BRCA1 mutation carrier status. Clin Cancer Res. 2004;10:958–963. doi: 10.1158/1078-0432.CCR-1067-3. [DOI] [PubMed] [Google Scholar]
- Thuerigen O, Schneeweiss A, Toedt G, Warnat P, Hahn M, Kramer H, Brors B, Rudlowski C, Benner A, Schuetz F, et al. Gene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancer. J Clin Oncol. 2006;24:1839–1845. doi: 10.1200/JCO.2005.04.7019. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Powles TJ, Dowsett M, Jazaeri AA, Feldman AL, Assersohn L, Gadisetti C, Libutti SK, Liu ET. Gene expression profiles derived from fine needle aspiration correlate with response to systemic chemotherapy in breast cancer. Breast Cancer Res. 2002;4:R3. doi: 10.1186/bcr433. [DOI] [PMC free article] [PubMed] [Google Scholar]


