Short abstract
A literature search shows that biologists do not follow the official nomenclature guidelines for a substantial fraction of gene names.
Abstract
Current usage of gene nomenclature is ambiguous and impairs the efficient handling of scientific information. Therefore it is important to propose guidelines to deal with this problem. This study attempts to evaluate the success of HUGO nomenclature for human genes. The results indicate that HUGO guidelines are not supported by the scientific community.
Ambiguous gene names impose a serious hurdle for the analysis of a wide range of high-throughput data, such as microarray experiments or protein-interaction maps. This sort of ambiguity also limits the efficiency of genome analysis and annotation and slows the implementation of automatic text-mining systems for using bibliographic information [1,2]. While systems for automatic gene name recognition in other domains (such as in business or news reports) perform very well, the best systems in the biological field perform just slightly better than 80% [3].
Genes are commonly named using functional terms, such as 'insulin' or 'tumor necrosis factor', or symbols consisting of abbreviations such as INS for insulin or TNF for tumor necrosis factor. Functional names are usually unique, in the sense that a given name refers only to one gene family, even if not always to a single gene of the family. Ambiguity exists because often more than one functional name is used to refer to the same gene (synonymy), and also many functional names are descriptive of some phenotype of the gene (such as 'deafness' or 'wingless'), a practice that creates many complications [4]. The use of symbols should alleviate some of the problems created by the use of functional names, but in practice seems to produce even more ambiguities. In addition to extended synonymy (with many symbols describing the same gene), a given symbol can also be used to describe different genes (homonymy). Moreover, many other meanings can match the abbreviation used for the gene name (acronyms). Text-mining systems are severely limited by these factors, as ambiguities decrease the precision in the retrieval of correct articles, and synonyms limit the number of total retrieved articles.
These limitations potentially impair the effective application of text mining and natural language processing (NLP) techniques in genomics. For instance, the comparison of microarray data from different sources requires the exact mapping of the names used by different authors. This task can be greatly complicated by ambiguous names such as 'PAP', which can refer to five different human genes, and will therefore be impossible to classify in the absence of additional information. In this type of situation, valuable experimental information could be lost because of nomenclature problems that could be solved by the use of standard names.
Standard nomenclatures, strictly following naming guidelines, are the most obvious solution to the problem. Indeed, considerable community effort has gone into the creation of these standards for gene symbols in organisms such as yeast, mouse, fly, and, of course, human. An illustrative example is the valuable effort of HUGO nomenclature for human genes [5,6]. A single official symbol is proposed for every gene, and the aliases (alternative symbols, synonyms) for each gene are also listed. The obvious concern is the extent to which scientists follow these nomenclature rules. Other instances of standard nomenclatures, such as enzymatic codes (EC numbers), have been loosely followed.
We carried out a study to assess the relative success of HUGO guidelines by measuring the progress in the usage of official gene symbols in recent years. We analyzed PubMed abstracts for the period 1994-2004, collecting information regarding the mention of human gene symbols and the frequency with which official symbols were mentioned in comparison with their aliases. It is painfully obvious that the community has not widely adopted the HUGO guidelines. It is equally obvious that there is no clear tendency that this situating is improving, as the proportion of official symbols that are used predominantly has only increased slightly, from 35% in 1994 to 44% in 2004 (Figure 1). Accordingly, a small decrease in the cases where the official name was not mentioned at all is observed (from 23% in 1994 to 14% in 2004). Despite this minor progress, it is still true that aliases are used more often than official symbols, and as many as 14% of genes are never mentioned using the recommended official symbols.
Figure 1.
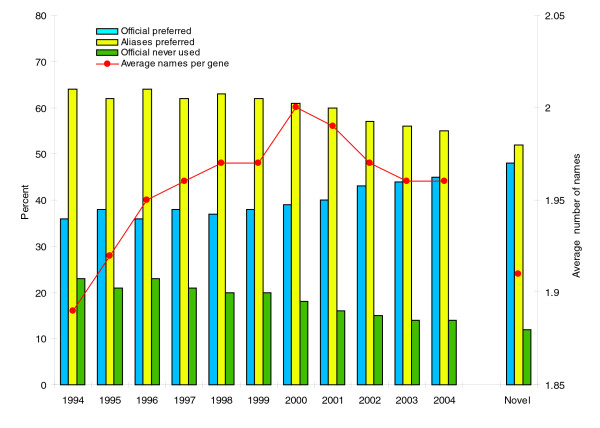
Usage of HUGO nomenclature in the past ten years. We analyzed PubMed abstracts for the period 1994-2004, collecting information about the human genes mentioned on the abstracts, and noting how such mention was made (official symbol or other aliases). Names were detected using Text Detective (BioAlma SL), a gene name recognition software that is able to recognize human gene names in texts with high recall and precision, distinguishing real instances of the gene from other uses and meanings of the same name [13]. Text Detective combines gene name recognition with standardization of citations, using HUGO nomenclature in the case of human genes. Additional results (the yeast results discussed in the text) were obtained using the Information Interlinked Over Proteins (iHOP) system [14] in order to discard possible biases due to the name-recognition software used. The percentage of genes that are cited predominantly by their official name is used as a measure of the support for official names. Blue bars show the percentage of genes for which the official name is favored (the official name is mentioned more often than aliases). Yellow bars show the inverse, the percentage of genes for which aliases are favored. Green bars show the percentage of cases in which the official name is never used, and all mentions correspond to aliases. Also, the average number of names per gene is shown, computed as the total number of names used divided by the total number of genes. The last column, labeled 'Novel', takes into account only those genes whose first mention in the literature occurred in the year 2000 or later.
A positive observation is that this small increment is in part due to new genes that are named preferentially according to the official standards. The genes mentioned for the first time after the year 2000 have a higher proportion of official symbols and a smaller number of synonyms (Figure 1); however, it can still be argued that it is only a question of time for these genes to acquire new synonyms. Furthermore, highly referenced genes are cited notably more often by unofficial gene names. For example, in 2004, only 38% of genes cited in more than 50 articles were named predominantly by following HUGO, whereas scarcely cited genes more often followed the standards (54% in 2004).
The tendency to improve the situation by replacing aliases in favor of HUGO official symbols is, unfortunately, weak. The changes in name usage, either from official to aliases or from aliases to official, are not very frequent, and the nomenclature of most genes remains rather stable with time. These findings seem to confirm the intuition that researchers remain attached to their favorite names.
This trend is not species-dependent. For example, in yeast, where there is also a proposed standard nomenclature [7], there is not a tendency to replace aliases with official names (the usage of official names has remained approximately the same in recent years as in the past), even if in this community official names are used more often (85% of the genes are preferentially cited using official names).
Many of the occasional transitions are in fact produced after the publication of a prominent paper describing an important discovery regarding a gene, which usually produces a chain of subsequent studies that tend to use the new name. For instance, in the mid-1990s the gene for intestinal trefoil factor 3 was cited predominantly under the alias ITF. But since 1998, the official name TFF3 has been preferred, apparently influenced by a paper describing the regulation that the gene exerts on the expression of catenin and cadherin, with important consequences for epithelial cell adhesion, migration, and survival [8], which gave rise to the use of the symbol TFF3 for that gene. Therefore, it would appear that important scientific papers influence nomenclature usage even more than does the adoption of standards (Figure 2a).
Figure 2.
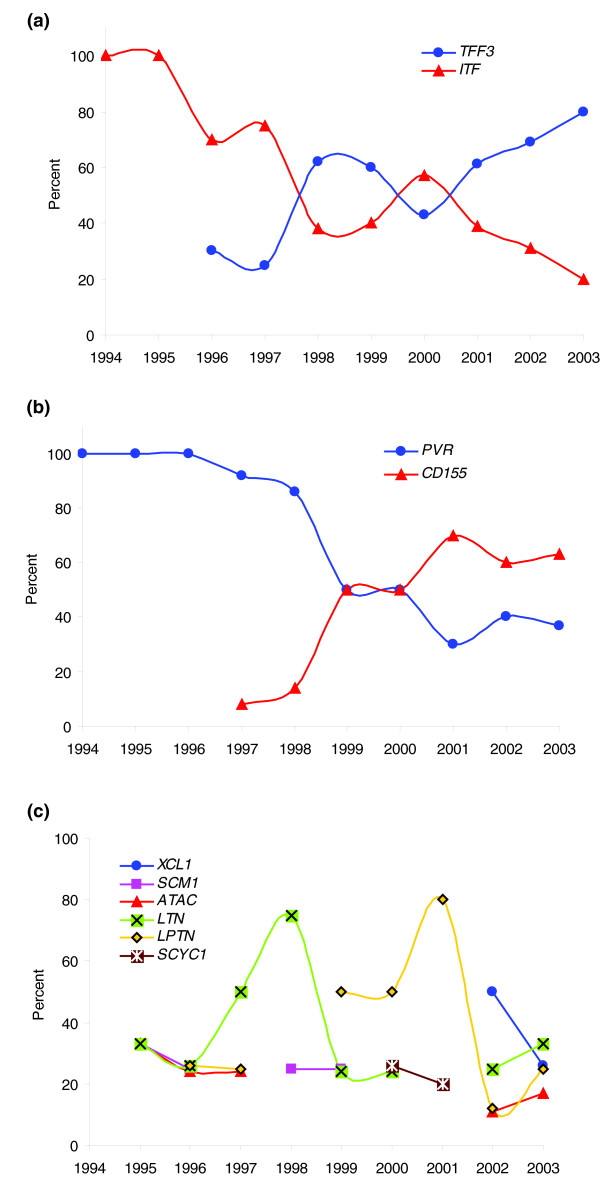
Plot of the evolution of the usage of different names. The plots show, for each year, the percentage usage of each of the names. (a) Intestinal trefoil factor 3 (official name, TFF3); (b) poliovirus receptor (official name, PVR); (c) lymphotactin (official name, XCL1).
A similar case is illustrated in Figure 2b for the gene encoding the poliovirus receptor. In the mid-1990s, the only symbol used was PVR (which is today the official name for the gene). The alternative name CD155 for the protein appeared for the first time in 1997, but gained greater acceptance after the publication in the late nineties of several articles describing structural aspects of the CD155 protein [9] that are critical to the interaction with the virus (CD nomenclature for cell-surface proteins follows a long established standard nomenclature). These articles named the gene as CD155, and this has been the preferred name since then. In this case, HUGO nomenclature apparently did not take this fact into account, since the establishment of PVR as the official gene name took place in 2003.
Finally, Figure 2c shows an interesting case of the persistence of several different names for one gene, that for the chemokine lymphotactin. The cloning of this gene was reported almost simultaneously by three independent groups in Japan, Germany and the USA in 1995 [10-12]. The three groups named the gene differently (SCM1, ATAC and LTN, respectively). These names have all been used since then, as well as LPTN and, lately, the official name XCL1. It is interesting to notice that the three groups reporting the discovery kept using their own names for the gene, at least until very recently, a trend that can be observed also in the previous examples.
The problem of linking names in texts with the molecules they refer to can only be solved by a concerted community effort to explicitly mention the official names and/or the corresponding database accession numbers (such as these of UniProt or Refseq for proteins, and GenBank for genes). The use of accession numbers has the advantage of providing a unique and unambiguous reference that is also a direct link to the real biological object. But it does have some drawbacks. Citing accession numbers instead of gene or protein names would seriously affect the clarity and readability of the text. From this point of view, names and accession numbers must coexist. This could be done, for instance, by citing only names in the main text, and including accession numbers for the protein or gene names used in the text in a separate section. Also, our experience is that mapping between different databases is not exempt from problems. For instance, a single nucleotide sequence often has several different entries, corresponding to splice variants, polymorphisms or regions of the genome. Also, for these references to be really useful, they would have to cover all the mentions of genes including anaphoric (the use of a linguistic unit, such as the pronoun 'it' to refer to a previous mention of the name) and other forms of implicit mentions, and to take into account the difference between individual genes and proteins and general protein names referring to, for instance, protein familes (that is, 'tubulin beta1 protein' can be assigned to a well defined molecule, but 'tubulin' cannot, since it can refer to several different molecules). It would be important to develop adequate tools to facilitate the introduction of names and identifiers at the time of writing papers, and to enable the posterior recovery by both humans and software tools.
The task of tagging genes and proteins in papers with the corresponding official names and/or database entries will require the collaboration of authors, journals and grant agencies, and could be facilitated by the development of adequate text-mining methods.
Acknowledgments
Acknowledgements
J.T. developed the gene name recognition system Text Detective as part of his work at BioAlma SL (Tres Cantos, Madrid, Spain). This work was partly supported by research grants ENFIN LSGH-CT-2005-518254 (VI Framework Programme, European Comission), ESPAÑOL BIO2004-00875 (Spanish Ministry of Education and Science), and Fundación BBVA.
References
- Petsko GA. What's in a name? Genome Biol. 2002;3:comment1005.1–1005.2. doi: 10.1186/gb-2002-3-4-comment1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman S. Tough mining: the challenges of searching the scientific literature. PLoS Biol. 2003;1:e48. doi: 10.1371/journal.pbio.0000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh A, Morgan A, Colosimo M, Hirschman L. BioCreAtIvE task 1A: gene mention finding evaluation. BMC Bioinformatics. 2005;6(Suppl 1):S2. doi: 10.1186/1471-2105-6-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu H, Friedman C. Gene name ambiguity of eukaryotic nomenclatures. Bioinformatics. 2005;21:248–256. doi: 10.1093/bioinformatics/bth496. [DOI] [PubMed] [Google Scholar]
- Wain HM, Lush M, Ducluzeau F, Povey S. Genew: the human nomenclature database. Nucleic Acids Res. 2002;30:169–171. doi: 10.1093/nar/30.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGO Gene Nomenclature Committee http://www.gene.ucl.ac.uk/nomenclature
- Saccharomyces Genome Database (SGD) http://www.yeastgenome.org
- Efstathiou JA, Noda M, Rowan A, Dixon C, Chinery R, Jawhari A, Hattori T, Wright NA, Bodmer WF, Pignatelli M. Intestinal trefoil factor controls the expression of the adenomatous polyposis coli-catenin and the E-cadherin-catenin complexes in human colon carcinoma cells. Proc Natl Acad Sci USA. 1998;95:3122–3127. doi: 10.1073/pnas.95.6.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol. 1999;73:958–964. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Imai T, Kakizaki M, Nishimura M, Yoshie O. Molecular cloning of a novel C or gamma type chemokine, SCM-1. FEBS Lett. 1995;360:155–159. doi: 10.1016/0014-5793(95)00093-O. [DOI] [PubMed] [Google Scholar]
- Muller S, Dorner B, Korthauer U, Mages HW, D'Apuzzo M, Senger G, Kroczek RA. Cloning of ATAC, an activation-induced, chemokine-related molecule exclusively expressed in CD8+ T lymphocytes. Eur J Immunol. 1995;25:1744–1748. doi: 10.1002/eji.1830250638. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Kelner GS, Kleyensteuber S, Schall TJ, Weiss MC, Yssel H, Schneider PV, Cocks BG, Bacon KB, Zlotnik A. Molecular cloning and functional characterization of human lymphotactin. J Immunol. 1995;155:203–209. [PubMed] [Google Scholar]
- Tamames J. Text Detective: a rule-based system for gene annotation in biomedical texts. BMC Bioinformatics. 2005;6(Suppl 1):S10. doi: 10.1186/1471-2105-6-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]


