Short abstract
Noncoding RNAs transcribed from control elements in DNA provide an anti-silencing mechanism by targeting chromatin-modifying enzymes to these genes.
Abstract
Expression of the genetic information encoded in our genomes is usually regulated by proteins interacting with the DNA. In some cases, however, noncoding RNAs transcribed from DNA control elements cooperate with histone-modifying enzymes to regulate gene expression, as has recently been shown for noncoding RNA originating from Polycomb- and Trithorax-group response elements.
The large majority of DNA sequences of most eukaryotic genomes do not possess any coding capacity. Nevertheless, many of these sequences are transcribed, generating a vast amount of noncoding RNA [1,2]. We are still far from fully understanding the role of this seemingly nonproductive large-scale transcription. In some situations, however, the process of intergenic transcription and the noncoding RNAs generated have been linked to the epigenetic control of gene expression. Intergenic transcripts appear to influence the way chromatin - the complex of DNA and the histones that package it - controls gene expression [3].
A prominent example of this type of epigenetic gene regulation is the mechanism of cellular memory, well studied in the fruit fly Drosophila melanogaster. Cellular memory is the inheritance of a cell's gene-expression program through cell division, and this memory is a prerequisite for the maintenance of cell fates throughout development. In Drosophila, cis-regulatory DNA element called PRE/TREs consisting of Polycomb group response elements and Trithorax group response elements are central components of the mechanism for establishing cellular memory. Poly-comb group (PcG) and Trithorax group (TrxG) proteins are targeted to these elements, promoting the formation of repressive (via PcG) or transcriptionally competent (via TrxG) chromatin structures [4]. In tissues where a PRE/TRE-controlled gene is active, transcription through the PRE and TRE sequences can be observed [5,6]. This suggests that the transcription acts as an anti-silencing mechanism, preventing the PcG proteins from repressing their target genes [7]. A recent study by Sanchez-Elsner et al. [8] now provides a first insight into the molecular basis of this anti-silencing mechanism. The authors show that the noncoding RNAs originating from a PRE/TRE stay associated with the element and anchor a histone-modifying enzyme, thus directly controlling the establishment of an epigenetically activated chromatin structure.
Sanchez-Elsner et al. [8] use the regulation of the Drosophila homeotic gene Ultrabithorax (Ubx) as a model to investigate the function of noncoding RNAs generated by transcription through PRE/TREs. Ubx specifies the identities of cells that give rise to the haltere and the third leg of the adult fly. Once established in appropriate segments during embryogenesis, the maintenance of Ubx expression depends on the continuous presence of functional TrxG proteins, in particular the histone methyltransferases ASH1 (Absent, Small and Homeotic discs) and TRX (Trithorax) [9]. The targeting of these proteins to Ubx depends on two PRE/TREs, one of which (bithorax, bx) is located within the third intron of Ubx, while the bithoraxoid (bxd) PRE/TRE lies 22 kb upstream of the Ubx promoter [10,11]. The bxd PRE/TRE is in close proximity to one of two characterized enhancers that drive Ubx expression in imaginal discs [10], the larval clusters of cells from which adult external structures will form during metamorphosis. Using the technique of rapid amplification of cDNA ends, Sanchez-Elsner et al. [8] identified three capped and polyadenylated transcripts originating from the bxd PRE/TRE in sense orientation with respect to its target gene Ubx. These noncoding RNAs, named tre1 (949 nucleotides), tre2 (1,109 nucleotides), and tre3 (350 nucleotides), are transcribed in a pattern overlapping the expression of Ubx, which is highly active in third-leg and haltere imaginal discs, but absent, for example, in wing disc tissue. The histone methyltransferase ASH1 follows the distribution of noncoding tre1, tre2 and tre3 transcripts, associating with the bxd PRE/TRE only in third-leg and haltere discs. This results in the enrichment of ASH1-dependent histone modifications such as histone H3 lysine 4 methylation specifically in these tissues.
As ASH1 has been reported to bind single-stranded nucleic acids in vitro [12], Sanchez-Elsner et al. [8] went one step further and demonstrated that the targeting of ASH1 to the bxd PRE/TRE directly depends on its interaction with the noncoding tre1, tre2 and tre3 transcripts via its SET protein domain, which has been identified as having histone methyl-transferase activity. ASH1 turned out to possess a surprising specificity in that it binds in vitro sense tre1, tre2 and tre3 RNAs, but not the antisense counterparts. The interaction was further confirmed by the finding that these RNAs also co-precipitate with chromatin-bound ASH1, and that the association of ASH1 with the bxd PRE/TRE is abolished by removing single-stranded RNAs (ssRNAs) through RNAse digestion. The authors therefore propose that the specificity of ASH1 targeting to a PRE/TRE might rely on the formation of a complex consisting of ASH1, stretches of ssDNA exposed during ongoing transcription, and the noncoding RNAs generated by this process (Figure 1).
Figure 1.
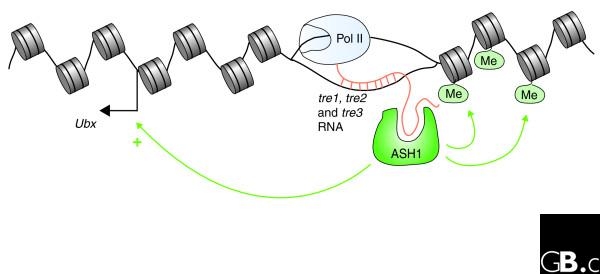
RNA polymerase II (Pol II) transcription through the bxd PRE/TRE leads to the generation of noncoding tre1, tre2 and tre3 RNAs. These RNAs remain tethered to their site of origin, presumably via base pairing with the unwound single-stranded DNA. The histone methyltransferase ASH1 specifically interacts with tre1, tre2 and tre3 RNAs and thus becomes targeted to the PRE/TRE. Through its SET domain, ASH1 catalyzes the trimethylation (Me) of lysines 4 and 9 on histone H3, and of lysine 20 on histone H4. This leads to the activation of the chromatin structure and, as a consequence, the stimulation of Ubx expression.
This targeting mechanism is reminiscent of the fly's strategy for compensating for the differences in X-chromosome gene dosage in males and females. In males, the single X chromosome becomes coated with the chromatin-modifying dosage compensation complex (DCC), consisting of six proteins and one of two redundant male-specific noncoding RNAs, roX1 or roX2 [13]. Assembly of the DCC presumably takes place on nascent roX1 or 2 RNAs while they are still tethered to the DNA template on the X chromosome. From these initial nucleation sites, the DCC spreads along the remainder of the X chromosome via more than 30 additional 'entry sites' [14]. The outcome is the chromosome-wide modification of chromatin structure by the histone acetyltransferases present in the complex, inducing a twofold upregulation of transcription [15]. Although the initial recruitment of the DCC and ASH1 seem to rely on very similar mechanisms, the association of ASH1 with the chromatin has to remain fairly restricted to PRE/TREs, as extensive spreading of ASH1 into neighboring regulatory regions would have deleterious consequences. Indeed, over the past few years several other mechanisms for targeting chromatin protein complexes via RNA moieties have become evident, such as the tethering of RNA-induced transcriptional silencing (RITS) complexes to yeast heterochromatin [13,16].
How is the action of tre1, tre2 and tre3 RNAs confined to the bxd PRE/TRE? Sanchez-Elsner et al. [8] propose that these RNAs remain associated with the chromatin at their site of production, as they are only present in the chromatin-bound fraction of ASH1 and not in the soluble pool. The production and tethering of tre1, tre2 and tre3 RNAs to the chromatin is, however, independent of ASH1, as it is not inhibited by its absence. This is an intriguing result, because it also implies that the intergenic promoters are regulated by a different set of factors than the target gene promoters, which require the continuous presence of functional ASH1 protein [9].
The most surprising finding from the work of Sanchez-Elsner et al. [8] is the observation that sense tre1, tre2 and tre3 transcripts expressed ectopically from a transgene can associate with the endogenous bxd PRE/TRE locus in trans. This results in the recruitment of ASH1 and, as a consequence, in the expression of Ubx in tissues in which this gene is normally kept silent. By contrast, antisense transcription of tre1, tre2 and tre3 RNAs does not have this activating effect, but as the authors mention, attenuates Ubx expression in its normal domains. Activating trans-effects for other PRE/TREs have not been observed so far. The PRE/TRE Frontoabdominal-7 (Fab-7) controls the homeotic gene Abdominal-B (AbdB); ectopic expression of Fab-7 from a transgene in all tissues throughout development does not affect the epigenetic state of the endogenous Fab-7 site, as the expression of the AbdB target gene remains normal [7]. In this case, however, large transcripts encompassing the entire PRE/TRE were produced, suggesting that the precise size and structure of the bxd transcripts used by Sanchez-Elsner et al. [8] were crucial for tethering ASH1 to the element.
The results presented by Sanchez-Elsner et al. [8] provide the first direct link between transcription through a PRE/TRE and the activation of chromatin via the interaction of noncoding RNAs with the histone methyltransferase ASH1 placing activating marks on histones. Once a PRE/TRE has been switched into the epigenetically activated mode, how is this information transmitted from mother to daughter cells during proliferation? Covalently modified histones presumably play a pivotal role in this process: during DNA replication, they may become distributed among daughter strands in the same semiconservative way as newly synthesized DNA [17]. In addition, modifications such as methylated lysine 4 of histone H3 (which is also set by ASH1 [18]) have been shown to survive mitosis, and might thus function to signal the activated state into the following cell cycle [19]. In the context of the bxd PRE/TRE, the recruitment of ASH1 to the chromatin depends crucially on the presence of tre1, tre2 and tre3 RNAs. This raises the intriguing possibility that noncoding RNAs, tethered to other target loci, might also serve as epigenetic 'bookmarks' to ensure the re-targeting of ASH1 to a PRE/TRE, thus ensuring the full reestablishment of epigenetic activation once the cells have traversed critical stages of the cell cycle.
Acknowledgments
Acknowledgements
We thank C. Beisel and M. Tariq for discussions and critical reading of the manuscript. S.S. is funded by a PhD fellowship of the Boehringer Ingelheim Fonds. The research of R.P. is funded by grants from the Deutsche Forschungsgemeinschaft.
References
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Tritho-rax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Bienz M. Imaginal disc silencers from Ultrabithorax: evidence for Polycomb response elements. Mech Dev. 1994;48:255–266. doi: 10.1016/0925-4773(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Krajewski WA, Nakamura T, Mazo A, Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen AA, Panning B. Epigenetic gene regulation by non-coding RNAs. Curr Opin Cell Biol. 2003;15:281–289. doi: 10.1016/S0955-0674(03)00041-3. [DOI] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/S1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]


