Abstract
Previous studies in rodents and sheep show that maternal nutrient restriction during pregnancy alters fetal renal development. To date, no studies using fetal baboon RNA with human Affymetrix gene chips have been published. In the present study we have (1) evaluated the specificity of the Affymetrix human gene array ‘Laboratory on a Chip’ system for use with fetal baboon mRNA and (2) investigated the effects of moderate maternal global nutrient restriction (NR; 70% of ad libitum animals) from early (30 days gestation (dG)) to mid-gestation (90 dG; term = 184 dG) on the fetal baboon kidney. Morphometric and blood measurements were made on 12 non-pregnant baboons before they were bred. All baboons were fed ad libitum until 30 days pregnant, at which time six control baboons continued to feed ad libitum (control – C) while six received 70% of the C diet on a weight adjusted basis. Fetal kidneys were collected following caesarean section at 90 dG, with samples flash frozen and fixed for histological assessment. Fetal hip circumference was decreased in the NR group (68 ± 2 versus 75 ± 2 mm), while fetal body weight and all other measurements of fetal size were not different between C and NR at 90 dG. Maternal body weight was decreased in the NR group (12.16 ± 0.34 versus 13.73 ± 0.55 kg). Having established the specificity of the Affymetrix system for fetal baboon mRNA, gene expression profiling of fetal kidneys in the context of our maternal nutrient restriction protocol shows that NR resulted in a down-regulation of genes in pathways related to RNA, DNA and protein biosynthesis, metabolism and catabolism. In contrast, genes in cell signal transduction, communication and transport pathways were up-regulated in the NR group. These changes indicate that even a moderate level of maternal global NR impacts fetal renal gene pathways. Our histological assessment of renal structure indicates decreased tubule density within the cortex of NR kidneys compared with controls. The number of glomerular cross-sections per unit area were unaffected by NR, suggesting that tubule tortuosity and/or tubule length was decreased in the NR kidney. Taken together the changes indicate that NR results in accelerated fetal renal differentiation. The negative impact of poor maternal nutrition on the fetal kidney may therefore be in part due to shortening of critical phases of renal growth resulting in decreased functional capacity in later life. These findings may have important implications for postnatal renal function, thereby contributing to the observed increased predisposition to hypertension and renal disease in the offspring of nutrient restricted mothers.
A compelling set of animal research studies exist to demonstrate that maternal nutritional deficiencies, in global intake (Gilbert et al. 2005) as well as in altered intake of micro- and macronutrients (Galaverna et al. 1995; Langley-Evans, 1996; Welham et al. 2005), have adverse effects on fetal growth and development and offspring physiology. Growth of the fetal kidney has received considerable attention due to the vital role it plays in blood pressure and body fluid homeostasis during fetal life, in the transition to an independent postnatal existence and throughout postnatal life (Ingelfinger & Woods, 2002; Rasch et al. 2004).
The effects of differing degrees of restriction in maternal nutrient availability (from 30 to 90% restriction) on development of fetal organs including the kidney has been extensively studied in rodents and sheep (Langley-Evans et al. 1996; Ingelfinger & Woods, 2002). There are several developmental differences between commonly studied laboratory species (e.g. rats and mice), large animals such as sheep, and primates. For example, most rodents are polytocous and deliver immature young. Although ruminants deliver fewer and more mature offspring, ruminant species have a different placental structure from primates. Oxygen and glucose consumption of the ruminant placenta are higher on a weight adjusted basis than those of the human placenta (Hay, 1994).
The mounting evidence for critical effects of maternal nutrition on fetal development in non-primate species supports the need to examine the effects of suboptimal maternal nutrition in a non-human primate model. We have developed a non-human primate model, the baboon, through which we can provide key comparative primate data to guide extrapolation of rodent and ruminant data to human pregnancy. The baboon is the non-human primate species in which the greatest amount of experimental data is available on maternal and fetal function (Ducsay et al. 1991; Hennessy et al. 1994; Koenen et al. 2002; Antonow-Schlorke et al. 2003; Pepe et al. 2003). We have developed a group housing system, described in detail elsewhere (Schlabritz-Loutsevitch et al. 2004), which allows each animal to maintain its normal physical and social activity while at the same time enabling us to regulate each animal's food intake and monitor weight daily.
The purpose of this study was twofold, first to perform an initial evaluation of specificity of the Affymetrix human gene array U133A Plus 2.0 ‘Laboratory on a Chip’ system for use with baboon mRNA, and second to utilize this system to determine the impact of moderate global maternal nutrient restriction (NR) from 30 days of gestation (dG) to 90 dG (term 184 dG) on genes involved in key pathways in the fetal baboon kidney. Gene expression profiling in kidneys of fetuses of NR mothers showed down-regulation of genes in pathways related to RNA, DNA and protein biosynthesis, metabolism and catabolism, actin cytoskeleton assembly, and apoptosis compared with kidneys of fetuses from ad libitum fed (control, C) mothers. In contrast, genes involved in cell signal transduction, communication and transport pathways were up-regulated. Expression profiles of apoptosis-related genes were confirmed by quantitative reverse transcription real time PCR (QRT-PCR). These changes indicate that even moderate maternal global nutrient restriction affects various fetal renal metabolic pathways differently. Histological analysis of 90 dG kidney sections showed decreased structure per unit area in the NR versus the C kidney. Taken together the changes indicate that 30% maternal NR accelerates fetal renal differentiation and inhibits kidney structure development. The negative impact of poor maternal nutrition on the fetal kidney may therefore be in part due to shortening of critical phases of growth.
Methods
Animal care and maintenance
All procedures were approved by the Southwest Foundation for Biomedical Research (SFBR) Institutional Animal Care and Use Committee and conducted in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care. Six baboons from two independently housed groups each consisting of 16 females and one male were studied. Each group was housed in a cage that was 3.5 m high with a floor area of 37 m2. Details of housing structure and environmental enrichment provided have been published elsewhere (Schlabritz-Loutsevitch et al. 2004). Maternal morphometric measurements were made prior to pregnancy to ensure homogeneity of females in the two groups.
System for controlling and recording individual feeding
Once a day prior to feeding, all baboons were run into individual feeding cages. Baboons passed along a chute, over a weighing scale and into one of the individual feeding cages. Once in the individual cages they were fed either between 07.00 h and 09.00 h or between 11.00 h and 13.00 h as previously described (Schlabritz-Loutsevitch et al. 2004). Food was provided as Purina Monkey Diet 5038, standard biscuits. Water was continuously available in the feeding cage through individual lixits and at several locations in the group housing.
Formation of stable grouping for the nutrient restriction study
Each group of 16 females was initially housed with a vasectomized male to establish a stable social group (Schlabritz-Loutsevitch et al. 2004). All female baboons were observed twice a day for well-being and three times a week for turgescence (sex skin swelling) and signs of vaginal bleeding to enable timing of pregnancy (Hendrickx, 2001). At the end of a 30-day period of adaptation to the feeding system, a fertile male was introduced into each breeding cage. Pregnancy was dated initially by following the changes in the swelling of the sex skin and confirmed at 30 dG by ultrasonography.
Diet and food consumption
The Purina Monkey Diet 5038 fed is described by the vendor as ‘a complete life-cycle diet for all Old World Primates.’ The biscuit contains stabilized vitamin C as well as all other required vitamins. Its basic composition is crude protein not less than 15%, crude fat not less than 5%, crude fibre not more than 6%, ash not more than 5% and added minerals not more than 3% (Schlabritz-Loutsevitch et al. 2004). At the start of the feeding period, each baboon was given 60 biscuits in the feeding tray. At the end of the 2-h feeding period after the baboons had returned to the group cage, the biscuits remaining in the tray and on the floor of the cage and in the pan were counted. Food consumption of animals, their weights and health status were recorded daily. The weight of each baboon was obtained as she crossed the electronic scale system (GSE 665; GSE Scale Systems, MI, USA). A commercial software application designed to capture weight data was modified to permit the recording of 50 individual measurements over 3 s. If the standard deviation of the weight measurement was greater than 0.01 of the mean weight, the weight was automatically discarded and the weighing procedure begun again. All baboons were fed ad libitum until 30 dG when six control baboons continued to feed ad libitum and six were fed 70% of feed consumed by controls on a weight adjusted basis.
Caesarean sections were performed at 90 dG under isoflurane anaesthesia (2%, 2 l min−1) to obtain the fetus and placenta. Techniques used and postoperative maintenance have been previously described in detail (Schlabritz-Loutsevitch et al. 2004). Analgesia was provided with buprenorphine hydrochloride at 0.015 mg kg−1 day−1 during 3 postoperative days (Buprenex® Injectable, Reckitt Benckiser Health care (UK) Ltd, Hull, UK). Fetal morphometrics were obtained at the time of caesarean section. In addition, fetal kidneys were collected and cut in half longitudinally. One half was immediately snap frozen in liquid nitrogen and then stored at −80°C until used for RNA extractions. The other kidney half was fixed in formalin and embedded in paraffin for histological analyses.
RNA isolation from tissue
RNA was isolated from tissue using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, approximately 100 mg section of frozen kidney was cut from one pole of a longitudinally sliced kidney half. The tissue was homogenized in 1 ml Trizol Reagent using a Power General Homogenizer (Omni International, Wilmington, DE, USA). Genomic DNA in the sample was sheared by passing the homogenate three times through a 22-gauge needle attached to a 1 ml syringe. The homogenized samples were incubated for 5 min at 25°C. Two hundred microlitres of chloroform was added to each sample, and the samples were shaken vigorously by hand for 15 s and incubated at 25°C for 3 min. Samples were then centrifuged at 4°C and 12 000 g for 15 min. The aqueous phase containing RNA was transferred to a fresh tube and the RNA precipitated by addition of 0.5 ml of isopropyl alcohol. Samples were incubated for 10 min at 25°C and then centrifuged at 4°C and 12 000 g for 10 min. The RNA precipitate was washed with 1 ml of 75% ethanol and centrifuged at 4°C and 7500 g for 5 min. The RNA was resuspended in 100 μl DEPC-treated water and stored at −80°C.
Preparation of cRNA probe for gene chip interrogation
Total RNA samples were shipped on dry ice to Genome Explorations, Inc. (Memphis, TN, USA) for RNA quality check, cRNA synthesis, and determination of gene expression profiles for each RNA sample by interrogation of the Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA). RNA Quality was checked using an Agilent Bioanalyser 2100 ‘Laboratory on a Chip’ system. RNA concentrations were confirmed by quantification using a dual beam spectrophotometer on approximately 200 ng of each RNA sample. Complementary RNA was synthesized and biotin labelled at Genome Explorations, Inc. using the MessageAmp™ aRNA Kit (catalogue no. 1750, Ambion, Austin, TX, USA) according to the manufacturer's instructions. Total RNA was used for first and second strand cDNA synthesis followed by an in vitro transcription step to synthesize biotin-labelled cRNA. The cRNA was quality checked and then hybridized to the Human Genome U133 Plus 2.0 Array (Affymetrix).
Gene chip data collection and analysis
Gene expression was detected using GCOS software (Affymetrix). Sequence data available for baboon genes show sequence differences in 3′UTRs compared with human genes. Since the majority of Affymetrix's probe sets are based on 3′UTR sequences and baboon RNA was used to screen human gene chips, we evaluated gene expression from unfiltered complete data sets in addition to the standard Affymetrix filtered data set. Analyses, including data normalization, transformation, t tests, and gene expression profile overlaying Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, were carried out using GeneSifter software (GeneSifter.Net, VizX Laboratories, Seattle, WA, USA).
To assess the frequency of individual probe mismatches within a gene's probe set and determine the impact of mismatches on ‘gene quality’ called by the Affymetrix software, we evaluated individual probe hybridizations (11 probes per probe set) for 40 genes on the human Affymetrix gene chip that were probed with 12 different RNA pools (480 observations). For this analysis, kidney RNA was pooled from the same animals used for interrogation of Affymetrix arrays. RNA was also pooled for lung and placental tissues for the control and experimental groups and pooled for these tissues from 175 dG placentas and fetuses (authors' unpublished data).
Statistical analysis
Fetal and maternal morphometric data analysis was performed using a one-sided Student's t test. Array data were all-median normalized and log2 transformed using GeneSifter software (GeneSifter.Net). Statistical analyses of array data were performed by t test using GeneSifter software.
Rationale for array analysis methods
To assess the impact of Affymetrix's filtering software on cross-species (baboon/human) probe-to-target hybridizations, we assessed the frequency of individual probe mismatches within a gene's probe (set) for 40 genes on the human Affymetrix gene chip that were probed with 12 different RNA pools. We also determined the relationship between probe mismatches within a gene's probe set on ‘gene quality’ called by the Affymetrix software.
Affymetrix filtering software indicates an ‘absent call’ for any gene with a detection P-value > 0.059 and assigns that gene a quality score of 0; genes with detection P-values < 0.05 are called ‘present’ and assigned quality scores of 1. We found that high intensity genes with as many as five probe mismatches in an 11 probe set for each gene had detection P-values < 0.05 and were called ‘present’, whereas less intense genes (70–410) with three mismatches had detection P-values > 0.05 and were called ‘absent’. Moderately intense genes (410–580) were called ‘present’ with zero, one, two, or three probe mismatches but were called ‘absent’ with four mismatches (< 410) or with 5 mismatches (< 580).
In the absence of baboon sequence data for these 40 genes, it is not possible to determine if absent calls are due to probe sequence mismatches for baboon RNA to human probe set hybridizations or due to cross-hybridization from gene family members. For genes with probe set mismatches of one or two probes, it is more likely that these mismatches are due to species sequence differences, whereas probe set mismatches of three or more probes could be argued for either case.
Based on the correlation between gene expression intensity and allowable probe mismatches within a gene's probe set, we analysed the gene expression profiles using both unfiltered (no quality minimum required) and filtered gene lists (minimum quality for group averages = 0.5) with the rationale that low expression genes with large variations in expression will not be statistically significantly different between the two groups (P < 0.05). Although filtered versus unfiltered gene lists impacts inclusion of individual genes in the dataset, this did not alter KEGG pathways (see below) with significant z-scores (data not shown). The final data set presented and analysed in this study (n = 685) only includes genes with minimum quality scores of 0.5.
Pathway analysis
To perform pathway analyses, we first created a ‘custom baboon array’ using GeneSifter. z-Score calculations defining significant gene categories and pathways are based on the total number of genes on the array. Thus, to accurately calculate z-scores using GeneSifter software, the array of baboon genes for which expression was detected on the human gene chip had to be defined. To do so, we merged expression array data from five baboon tissues at three fetal time points and three adult baboon tissues. Any gene from any baboon RNA sample with a marginal or present call on the human genechip (Affymetrix U133A 2.0) was considered expressed and included in the ‘custom baboon array’. Using this method, 16 186 of the 22 227 genes on the genechip were detected using baboon RNA. Thus, these 16 186 genes comprise the ‘custom baboon array’ from which z-scores were calculated.
Array data for significantly differently expressed genes were overlaid onto Ontological pathways (http://www.geneontology.org/) (Ashburner et al. 2000) and KEGG pathways (http://www.genome.jp/kegg/) (Kanehisa et al. 2004) using GeneSifter software. The ontological and KEGG pathway analyses provide detailed data on individual genes in the context of that gene's role in described biological/biochemical pathways. Pathways were considered significantly altered from the control gene expression profiles if the z-score for that pathway was less than −2 or greater than 2. z-Scores were calculated in GeneSifter using the following formula:
where R = total number of genes meeting selection criteria, N = total number of genes measured, r = number of genes meeting selection criteria with the specified GO term, and n = total number of genes measured with the specific GO term (Doniger et al. 2003).
QRT-PCR quantification of target gene abundance
To measure mRNA levels of target genes, we used the Assays-on-Demand system (Applied Biosystems, Foster City, CA, USA). Although no baboon specific primers and probes are available, we have successfully used the Assays-on-Demand system for more than 40 different baboon genes. We quantified mRNA according to manufacturer's instructions (Applied Biosystems). In brief, total RNA (50 ng) was reverse transcribed in a 100-μl reaction using a High-Capacity cDNA Archive Kit (Applied Biosystems). Complimentary DNA synthesis was followed by real-time-PCR using gene specific primers provided by the manufacturer (CREBBP, Hs00231733_m1; BCL2L13, Hs00209787_m1; SGK, Hs00178612_m1; VEGF, Hs00900054_m1; RHOB, Hs00269660_s1) TaqMan Universal PCR master mix (Applied Biosystems) and the target cDNA. 18S rRNA was quantified as an endogenous control using the Applied Biosystems human Assay-on-Demand probe set. All samples were assayed in triplicate.
For relative quantification of gene expression, the comparative threshold cycle (Ct) method was employed (described in User Bulletin 2 for ABI PRISM® 7700 Sequence Detection Systems). The value obtained for Ct represents the PCR cycle at which an increase in reporter fluorescence above a background signal can first be detected (10 times the standard deviation of the baseline). Using this approach, the endogenous control Ct values were subtracted from the gene of interest Ct values to derive a ΔCt value. The relative expression of the gene of interest was then evaluated using the expression 2-ΔΔCt, where the value for ΔΔCt was obtained by subtracting the ΔCt of the calibrator from each ΔCt, using the mean of the negative control as the calibrator.
Kidney morphology
Formalin-fixed 90 dG kidneys from control and experimental animals were paraffin embedded and cut into 5 μm sections. Kidney sections were photographed at 20× magnification. Two 5 μm paraffin cross-sections (50 μm apart) from approximately halfway between one pole and the hilus were stained with H&E. Each section was assessed using a Nikon E600 microscope (Nikon Instruments Inc., Lewisville, TX, USA) and a SPOT RT Color CCD camera (Diagnostic Instruments Inc., Sterling Heights, MI, USA). Using 20× magnification, photographs were taken of adjacent, non-overlapping fields around the periphery of each section immediately below the nephrogenic zone. An average of 15 photographs per kidney were examined. Cross sections of convoluted tubules, ducts and glomeruli were counted and the area/field occupied by convoluted tubules was calculated using ImageJ (Abramoff et al. 2004) and its cell counter (De Vos, 2006) and multiple region of interest (Abramoff et al. 2004) plugins. Glomerulus cross-sections that extended beyond the periphery of the field were ignored. The area of the field occupied by tubules was expressed as a percentage of the total field. The area and cross-section counts from the two sections for each animal were averaged.
Results
Morphometric measurements of mother, fetus and fetal kidney
Morphometric measures were collected on the mothers before pregnancy (Table 1A). There were no differences between the two sets of animals that were used for the ad libitum and NR groups. Average maternal weight decreased 1.30 kg in the NR mothers at 90 dG compared with preconception average weight (P < 0.05). Measures were also collected on the 90 dG fetuses and placentas (Table 1B). Hip circumference was less in NR fetuses than control fetuses (P < 0.05).
Table 1.
Morphometric characteristics of (A) non-pregnant baboons before pregnancy in ad libitum (Ad lib) fed (n = 6) and nutrient restricted (NR, n = 6) at 90 days' gestation and (B) their fetuses and placenta at 90 days' gestation
| Unit | Ad lib (n = 6) | NR (n = 6) | |
|---|---|---|---|
| A. Maternal non-pregnant | |||
| Weight at physical | kg | 13.44 ± 0.708 | 13.46 ± 0.484† |
| Weight –4 weeks | kg | 13.39 ± 0.647 | 13.02 ± 0.241† |
| Height | cm | 87.63 ± 1.602 | 87.68 ± 0.764 |
| Biparietal distance | cm | 9.40 ± 0.732 | 9.72 ± 0.334 |
| Abdominal distance | cm | 9.88 ± 1.047 | 11.07 ± 0.392 |
| Sterno-pubis distance | cm | 38.05 ± 1.199 | 39.37 ± 1.314 |
| Chest circumference | cm | 54.62 ± 1.893 | 54.50 ± 1.065 |
| Waist circumference | cm | 42.96 ± 1.638 | 49.58 ± 2.031† |
| Hip circumference | cm | 53.00 ± 3.329 | 52.83 ± 1.364 |
| B. 90 dG | |||
| Maternal and placenta | |||
| Body weight last day | kg | 13.73 ± 0.552 | 12.16 ± 0.339* |
| Body weight last week | kg | 13.46 ± 0.540 | 12.10 ± 0.307 |
| Placenta | g | 73.08 ± 6.510 | 62.93 ± 1.485 |
| Placenta diameter | cm | 8.20 ± 0.339 | 7.25 ± 0.316 |
| Fetal membranes | g | 12.86 ± 3.842 | 7.52 ± 1.036 |
| Umbilical cord length | cm | 14.10 ± 0.775 | 11.83 ± 0.901 |
| Fetal | |||
| Weight | g | 102.13 ± 4.278 | 95.43 ± 3.257 |
| Length | cm | 17.66 ± 0.333 | 17.58 ± 0.436 |
| Weight/length | g cm−1 | 5.77 ± 0.142 | 5.43 ± 0.145 |
| BMI | kg m−2 | 3.266 ± 0.048 | 3.10 ± 0.122 |
| Biparietal distance | mm | 33.50 ± 0.529 | 34.17 ± 0.703 |
| Abdominal distance | mm | 22.83 ± 3.198 | 29.17 ± 1.621 |
| Femur length | mm | 35.00 ± 1.506 | 32.50 ± 1.118 |
| Chest circumference | mm | 89.16 ± 3.005 | 87.92 ± 2.275 |
| Waist circumference | mm | 77.33 ± 2.667 | 75.83 ± 2.713 |
| Hip circumference | mm | 75.00 ± 1.826 | 68.33 ± 1.667* |
| Sterno-pubis | mm | 66.66 ± 5.426 | 70.83 ± 0.833 |
| Total kidney | g | 0.85 ± 0.122 | 0.83 ± 0.127 |
Values are means ± s.e.m.
P < 0.05 compared to ad lib fed group:
P < 0.05 compared with 90 dG maternal weight.
Overview of gene expression differences between control and nutrient restricted 90 dG kidneys
A pair-wise comparison of gene expression was performed between control (n = 6) and maternal nutrient restricted (n = 6) 90 dG fetal kidney RNA samples. Data were all-median normalized, log2 transformed and analysed using the custom baboon array. Data were then analysed by comparing control and experimental group averages for fold differences in expression at 1.5×, 1.8× and 2.0×. A more rigorous approach using pair-wise comparisons was performed on the data using a t test to identify genes expressed significantly differently between groups. This dataset was then filtered using a minimum quality score of 0.5. A final sorting of genes was performed by increasing the minimum gene expression difference between groups to 2.0×.
Evaluation of gene expression based on gene product function and different selection criteria
Table 2 presents the overview of gene expression differences between C and NR fetal kidneys. Genes identified in Table 2, using increasingly rigorous selection criteria, were grouped by biological function from Gene Ontology terms (Ashburner et al. 2000). Table 3 presents the number of genes differentially expressed in each category listed in the table. z-Scores for each category are also presented with significant z-scores (= −2 or = 2) indicated with bolded font. The data presented in this table show that one category of gene products regardless of gene selection criteria, general signal transducer activity, is significantly altered when comparing 90 dG kidney RNA samples from fetuses in the C and NR groups. The cellular processes category was significantly altered when the expressed genes were sorted according to fold change between C and NR but not when sorted according to P-values. This suggests that although more genes than expected by chance differ between C and NR samples in this category, and that the differences are as much as 2-fold, a number of these differentially expressed genes have large variations in gene expression within each sample group. An interesting note is that genes categorized as important for transcriptional regulatory activity only show a significant z-score by using a 1.8-fold minimum expression difference or the most rigorous selection method of statistical significance and quality score.
Table 2.
Summary of expression-based gene selection
| Expressed genes | ≥Fold change | P ≤ | Quality |
|---|---|---|---|
| 22277 | — | — | 0 |
| 2793 | 1.5 | — | 0 |
| 1205 | 1.8 | — | 0 |
| 742 | 2.0 | — | 0 |
| 1088 | 1.1 | 0.05 | 0 |
| 685 | 1.1 | 0.05 | 0.5 |
| 19 | 2.0 | 0.05 | 0.5 |
‘Expressed genes’ indicates the number of genes detected based on fold change between mean control and mean nutrient restricted RNA samples. ‘Quality’ indicates the Affymetrix assessment of probe to target hybridization quality where 0.5 quality indicates that at least one-half of all samples (control plus experimental, n = 12) were called ‘present’. All data were all-median normalized and log2 transformed.
Table 3.
Summary of gene expression by biological function category
| 1.5× 2793 | 1.8× 1205 | 2.0× 742 | 1.1×, P < 0.5 1088 | 1.1×, P < 0.5, Q = 0.5685 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene ontogeny categories | Gene total | No. of genes | z-score | No. of genes | z-score | No. of genes | z-score | No. of genes | z-score | No. of genes | z-score |
| Antioxidant activity | 24 | 2 | −1.7 | 1 | −0.99 | 1 | 2.69 | 2 | −0.89 | 2 | 0.63 |
| Behaviour | 55 | 14 | 0.83 | 7 | 1.34 | 5 | 1.14 | 7 | 0.72 | 4 | 0.68 |
| Binding | 5561 | 1315 | 1.61 | 588 | 1.87 | 367 | 1.82 | 542 | −0.55 | 340 | −1.73 |
| Catalytic activity | 3129 | 687 | −2.15 | 290 | −1.74 | 184 | −1.04 | 301 | −0.93 | 204 | 0.51 |
| Cellular process | 4110 | 1013 | 3.18 | 472 | 2.72 | 301 | 2.58 | 424 | 1.9 | 255 | −0.56 |
| Chaperone activity | 1 | 0 | −0.38 | 0 | −0.26 | 0 | −0.21 | 0 | −0.6 | 0 | −0.54 |
| Chaperone regulator activity | 5 | 1 | −0.85 | 1 | 1.85 | 0 | −0.21 | 0 | −0.27 | 0 | −0.24 |
| Development | 1130 | 303 | 3.11 | 129 | 1.04 | 87 | 1.77 | 110 | −0.31 | 71 | −0.52 |
| Enzyme regulator activity | 403 | 110 | 1.65 | 51 | 1.38 | 34 | 1.39 | 48 | 1.85 | 30 | 1.16 |
| Motor activity | 103 | 25 | 0.31 | 8 | −1.04 | 5 | −0.87 | 6 | −1.15 | 5 | −0.69 |
| Nutrient reservoir activity | 1 | 0 | −0.38 | 0 | −0.26 | 0 | −1.85 | 0 | −0.27 | 0 | −0.24 |
| Obsolete biological process | 9 | 0 | −1.14 | 0 | −0.79 | 0 | −0.63 | 0 | −0.8 | 0 | −0.72 |
| Physiological process | 6513 | 1506 | 0.35 | 664 | −0.9 | 407 | −1.66 | 655 | 1.05 | 425 | 1.21 |
| Reg. of biological process | 348 | 83 | 1.9 | 37 | 1.62 | 24 | 1.24 | 44 | 2.15 | 28 | 1.71 |
| Signal transducer activity | 1367 | 369 | 4.36 | 176 | 3.01 | 116 | 2.9 | 118 | −3.01 | 64 | −3.42 |
| Structural molecule activity | 476 | 105 | −1.05 | 46 | −0.41 | 25 | −0.87 | 43 | −0.83 | 28 | −0.75 |
| Transcription regulator act. | 832 | 194 | −1.53 | 72 | −2.38 | 47 | −1.37 | 87 | 0.55 | 55 | 2.16 |
| Translation regulator act. | 79 | 9 | −2.37 | 4 | −1.85 | 1 | −0.74 | 5 | −0.97 | 3 | −0.88 |
| Transporter activity | 933 | 239 | 2.67 | 113 | 1.45 | 70 | 1.94 | 92 | 0.29 | 59 | 0.76 |
| Viral life cycle | 26 | 8 | 1.6 | 3 | 1.05 | 1 | −0.77 | 2 | −0.93 | 2 | 0.51 |
The top row of the table includes abbreviated gene sorting information from Table 2 and the second row indicates the number of differentially expressed genes from the indicated selection criteria. ‘Gene total’ indicates the total number of genes on the Affymetrix chip in the stated category; ‘No. of genes’ indicates the number of genes differentially expressed in that category. Categories in bold font show at least one sorted group with a significant z-score.
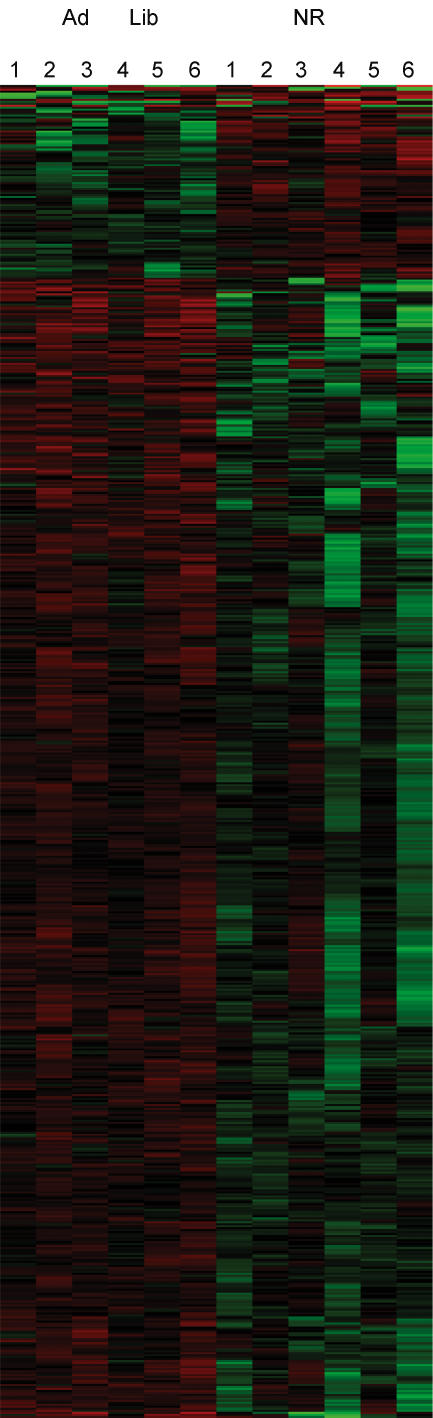
A general analysis of the genes (n = 685) selected based on P-values (P = 0.05) and probe set hybridization quality (Q = 0.5) show 96 genes significantly up- and 589 significantly down-regulated when comparing kidney RNA from C compared with NR fetuses. The graphical representation of this analysis, presented as a heat map in Fig. 1, provides an overall view of the variation within groups and between groups for this dataset. Array data from each kidney RNA sample are represented by one column in the heat map.
Figure 1. Heat Map of 685 significantly differently expressed genes comparing RNA samples from kidneys of 90 dG fetuses from control (n = 6) and nutrient restricted mothers (n = 6).
Control fetuses are represented in the 6 left columns (Ad Lib) and samples from fetuses of nutrient restricted mothers are represented in the 6 right columns (NR). Increased expression of experimental versus control samples is indicated in red and decreased expression of experimental versus control is represented in green.
Evaluation of gene expression according to ontological categorizations
We performed further analysis of the 685-gene set by examining the changes in gene expression between kidney samples from fetuses of C and NR mothers based on biological classifications established by the Gene Ontology Consortium (Ashburner et al. 2000). Gene products have been categorized based on the descriptive terms included in gene databases such as GenBank and LocusLink. Gene ontologies, organized in a hierarchical structure, allow categorization of each gene product based on current detailed information; this also allows placement of genes into multiple categories based on known gene product function or conserved domain-based prediction of gene product function. The detailed gene ontology report provides an overview of all genes expressed on the array grouped by gene ontology. An abbreviated form of the gene ontology report, which includes categories containing four or more genes, is provided in Tables 4 and 5. Table 4 shows those specific gene ontologies that are differentially up-regulated, and Table 5 those that are down-regulated. Since each gene may be included in multiple ontologies, the total number of genes listed in Tables 4 and 5 exceeds the total number of differentially expressed genes in the data set. The classification of differentially expressed genes suggests that groups of genes involved with electron, Golgi vesicle and nucleocytoplasmic transport, carbohydrate, sterol and steroid metabolism, metal ion binding, protein transport and targeting, protein tyrosine phosphatase and heterodimerization activity, regulation of cyclin-dependent protein kinase activity, and voltage-gated potassium channel activity are up-regulated in 90 dG fetal kidneys as a result of maternal nutrient restriction. This classification approach also indicates that groups of genes involved with DNA and RNA biosynthesis, metabolism, processing and packaging, and nucleic acid component biosynthesis and metabolism are down-regulated. Also down-regulated are groups of genes associated with protein biosynthesis and metabolism, cell growth, communication, organization, biogenesis, and surface receptor signal transduction, cytoskeleton organization and cytoskeleton biogenesis. Furthermore, genes relevant to protein complex assembly, are also down-regulated. Narrower categories down-regulated due to NR included ubiquitin-mediated proteolysis and cell cycle. Genes specific to cell cycle regulation and ubiquitin-mediated proteolysis are shown in Table 6.
Table 4.
Ontological categorization of up-regulated genes
| Up-regulated biological pathways | Diff exp genes | Up-reg | Down-reg | Tot on array | z-Score up |
|---|---|---|---|---|---|
| Electron transport | 19 | 6 | 13 | 236 | 2.61 |
| Golgi vesicle transport | 8 | 2 | 6 | 53 | 2.15 |
| Lyase activity | 8 | 3 | 5 | 102 | 2.08 |
| Main pathways of carbohydrate metabolism | 7 | 3 | 4 | 75 | 2.77 |
| Metal ion binding | 111 | 9 | 102 | 1927 | −2.52 |
| Nucleocytoplasmic transport | 11 | 3 | 8 | 88 | 2.43 |
| Protein heterodimerization activity | 2 | 2 | 0 | 27 | 3.46 |
| Protein targeting | 8 | 3 | 5 | 107 | 2.02 |
| Protein transport | 36 | 8 | 28 | 384 | 2.4 |
| Protein tyrosine phosphatase activity | 3 | 2 | 1 | 47 | 2.34 |
| Regulation of cyclin dependent protein kinase activity | 3 | 2 | 1 | 30 | 3.27 |
| Steroid metabolism | 10 | 3 | 7 | 104 | 2.08 |
| Sterol metabolism | 7 | 2 | 5 | 55 | 2.09 |
| Transition metal ion binding | 58 | 3 | 55 | 1042 | −2.37 |
| Voltage-gated potassium channel activity | 2 | 2 | 0 | 50 | 2.22 |
| Zinc ion binding | 42 | 0 | 42 | 790 | −2.9 |
‘Diff exp genes’ indicates the total number of genes differentially expressed on the array in that category; ‘Up-reg’ indicates the total number of up-regulated genes; ‘Down-reg’ indicates the total number of down-regulated genes; ‘Tot on array’ indicates the total number of genes on the array in that ontological category; and ‘z-Score up’ indicates the z-score for that category.
Table 5.
Ontological categorization of down-regulated genes
| Down regulated biological pathways | Diff exp genes | Up-reg | Down-reg | Tot on array | z-Score down |
|---|---|---|---|---|---|
| Actin binding | 21 | 3 | 18 | 160 | 3.27 |
| Actin polymerization and/or depolymerization | 4 | 0 | 4 | 21 | 2.74 |
| Amine biosynthesis | 7 | 0 | 7 | 50 | 2.66 |
| Amino acid derivative biosynthesis | 3 | 0 | 3 | 15 | 2.48 |
| Amino acid derivative metabolism | 5 | 0 | 5 | 35 | 2.3 |
| Biogenic amine biosynthesis | 3 | 0 | 3 | 12 | 2.98 |
| Biogenic amine metabolism | 5 | 0 | 5 | 29 | 2.79 |
| Carbon-carbon lyase activity | 5 | 1 | 4 | 26 | 2.24 |
| Carboxy-lyase activity | 4 | 0 | 4 | 20 | 2.87 |
| Cation channel activity | 4 | 3 | 1 | 129 | −2.36 |
| Coenzyme binding | 3 | 0 | 3 | 16 | 2.35 |
| Cytochrome-c oxidase activity | 4 | 0 | 4 | 17 | 3.29 |
| Cytoskeletal protein binding | 24 | 3 | 21 | 221 | 2.69 |
| Defense response | 17 | 3 | 14 | 510 | −2.8 |
| Endosome to lysosome transport | 3 | 0 | 3 | 5 | 5.37 |
| Endosome transport | 3 | 0 | 3 | 16 | 2.34 |
| Extracellular matrix organization and biogenesis | 4 | 0 | 4 | 21 | 2.74 |
| Extracellular matrix structural constituent | 8 | 0 | 8 | 59 | 2.76 |
| Extracellular structure organization and biogenesis | 4 | 0 | 4 | 21 | 2.74 |
| G2/M transition of mitotic cell cycle | 2 | 0 | 2 | 9 | 2.21 |
| Glutathione transferase activity | 4 | 0 | 4 | 15 | 3.62 |
| Glycerophospholipid biosynthesis | 4 | 0 | 4 | 20 | 2.86 |
| Glycerophospholipid metabolism | 4 | 0 | 4 | 25 | 2.32 |
| G-protein coupled receptor activity | 6 | 2 | 4 | 207 | −2.26 |
| Haeme-copper terminal oxidase activity | 4 | 0 | 4 | 17 | 3.29 |
| Hexose transport | 3 | 0 | 3 | 13 | 2.8 |
| Hydrogen ion transporter activity | 14 | 1 | 13 | 107 | 3.08 |
| Hydrolase activity | 62 | 13 | 49 | 1187 | −2.17 |
| Immune response | 16 | 3 | 13 | 455 | −2.52 |
| Insulin-like growth factor binding | 4 | 0 | 4 | 15 | 3.62 |
| Lipid transport | 6 | 0 | 6 | 48 | 2.15 |
| Lipoprotein biosynthesis | 3 | 0 | 3 | 15 | 2.48 |
| Lysosomal transport | 3 | 0 | 3 | 6 | 4.8 |
| Lysosome organization and biogenesis | 4 | 0 | 4 | 9 | 5.15 |
| Membrane lipid metabolism | 10 | 0 | 10 | 84 | 2.61 |
| Metabolism | 318 | 44 | 274 | 4632 | 2.13 |
| Monosaccharide transport | 3 | 0 | 3 | 13 | 2.8 |
| Monovalent inorganic cation transporter activity | 14 | 1 | 13 | 115 | 2.79 |
| Mrna metabolism | 16 | 1 | 15 | 157 | 2.28 |
| Negative regulation of cell proliferation | 12 | 1 | 11 | 104 | 2.31 |
| Negative regulation of microtubule depolymerization | 2 | 0 | 2 | 6 | 3 |
| Nucleotide metabolism | 13 | 1 | 12 | 125 | 2.05 |
| Oxidoreductase activity | 4 | 0 | 4 | 17 | 3.29 |
| Phosphatase regulator activity | 6 | 1 | 5 | 34 | 2.38 |
| Phosphoinositide biosynthesis | 3 | 0 | 3 | 15 | 2.48 |
| Phosphoinositide metabolism | 3 | 0 | 3 | 16 | 2.34 |
| Polyamine biosynthesis | 3 | 0 | 3 | 6 | 4.8 |
| Polyamine metabolism | 4 | 0 | 4 | 8 | 5.54 |
| Primary active transporter activity | 15 | 1 | 14 | 131 | 2.66 |
| Protein complex assembly | 11 | 0 | 11 | 93 | 2.72 |
| Protein lipidation | 3 | 0 | 3 | 15 | 2.48 |
| Protein phosphatase type 2A regulator activity | 3 | 0 | 3 | 15 | 2.49 |
| Purine nucleotide biosynthesis | 8 | 0 | 8 | 66 | 2.39 |
| Purine nucleotide metabolism | 8 | 0 | 8 | 69 | 2.25 |
| Purine ribonucleotide biosynthesis | 8 | 0 | 8 | 63 | 2.54 |
| Purine ribonucleotide metabolism | 8 | 0 | 8 | 66 | 2.39 |
| Receptor activity | 38 | 12 | 26 | 802 | −2.9 |
| Regulation of actin polymerization | 2 | 0 | 2 | 10 | 2.02 |
| Regulation of cell migration | 2 | 0 | 2 | 9 | 2.21 |
| Response to biotic stimulus | 19 | 3 | 16 | 608 | −3.2 |
| Response to external stimulus | 30 | 5 | 25 | 865 | −3.54 |
| Response to hypoxia | 3 | 0 | 3 | 6 | 4.8 |
| Response to stress | 26 | 4 | 22 | 631 | −2.28 |
| Ribonucleotide biosynthesis | 9 | 0 | 9 | 69 | 2.78 |
| Ribonucleotide metabolism | 9 | 0 | 9 | 73 | 2.59 |
| RNA splicing factor activity | 3 | 0 | 3 | 16 | 2.35 |
| Secondary metabolism | 4 | 0 | 4 | 8 | 5.54 |
| Serine family amino acid biosynthesis | 3 | 0 | 3 | 12 | 2.98 |
| Signal transducer activity | 65 | 16 | 49 | 1367 | −3.33 |
| Sphingoid metabolism | 3 | 0 | 3 | 14 | 2.63 |
| Sphingolipid metabolism | 6 | 0 | 6 | 30 | 3.51 |
| Sulphur amino acid metabolism | 4 | 0 | 4 | 21 | 2.74 |
| Transcriptional repressor activity | 6 | 0 | 6 | 48 | 2.16 |
| Transferase activity | 8 | 1 | 7 | 33 | 4 |
| Transmembrane receptor activity | 21 | 8 | 13 | 487 | −2.79 |
| Vesicle-mediated transport | 24 | 4 | 20 | 224 | 2.31 |
| Wnt receptor signalling pathway | 11 | 2 | 9 | 66 | 2.93 |
‘Diff exp genes’ indicates the total number of genes differentially expressed on the array in that category; ‘Up-reg’ indicates the total number of up-regulated genes; ‘Down-reg’ Indicates the total number of down-regulated genes; ‘Tot on array’ indicates the total number of genes on the array in that ontological category; and ‘z-Score down’ indicates the z-score for that category.
Table 6.
Expression profiles of genes encoding ubiquitin-mediated proteolysis and cell cycle genes
| Gene name | Gene ID | Fold change | Direction of change |
|---|---|---|---|
| Beta-transducin repeat containing | BTRC | 1.41 | Up |
| Cyclin B1 | CCNB1 | 1.35 | Down |
| Cyclin G2 | CCNG2 | 1.55 | Down |
| Cyclin-dependent kinase inhibitor 3 | CDKN3 | 1.31 | Up |
| Chromodomain helicase DNA BP3 | CHD3 | 1.54 | Down |
| Connective tissue growth factor | CTGF | 1.63 | Down |
| Cullin 4B | CUL4B | 1.61 | Up |
| Cullin 4B | CUL4B | 1.61 | Up |
| Dynactin 1 | DCTN1 | 1.28 | Down |
| Damage-specific DNA BP1 | DDB1 | 1.24 | Up |
| E1A binding protein p300 | EP300 | 1.26 | Down |
| Exostoses 2 | EXT2 | 1.38 | Down |
| F-box protein 21 | FBXO21 | 1.19 | Down |
| F-box protein 38 | FBXO38 | 1.47 | Down |
| F-box protein 42 | FBXO42 | 1.26 | Down |
| F-box protein 7 | FBXO7 | 1.22 | Down |
| F-box and WD-40 domain protein 11 | FBXW11 | 1.72 | Up |
| Growth arrest-specific 1 | GAS1 | 1.35 | Down |
| HECT, C2 and WW domain E3 ubiquitin pro lig 1 | HECW1 | 1.47 | Up |
| Katanin subunit A 1 | KATNA1 | 1.26 | Down |
| KH domain containing, RNA bind, sig trans 1 | KHDRBS1 | 1.49 | Down |
| Karyopherin alpha 2 | KPNA2 | 1.46 | Down |
| Ring finger protein 187 | LOC149603 | 1.49 | Down |
| Mitogen-activated protein kinase 7 | MAPK7 | 1.59 | Down |
| Membrane-associated ring finger 6 | MARCH-VI | 1.47 | Down |
| Minichromosome maintenance deficient 6 | MCM6 | 1.18 | Down |
| Makorin, ring finger protein, 1 | MKRN1 | 1.21 | Down |
| Nuclear protein localization 4 | NPL4 | 1.71 | Down |
| Nuclear distribution gene C | NUDC | 1.35 | Down |
| Polymerase epsilon 3 | POLE3 | 1.44 | Down |
| Protein phosphatase 1a | PPP1CA | 1.51 | Down |
| Protein phosphatase 1b | PPP1CB | 1.41 | Down |
| Protein phosphatase 1 g | PPP1CC | 1.26 | Down |
| PRP19/PSO4 premRNA processing factor 19 | PRP19 | 1.29 | Down |
| Protein tyrosine phosphatase IVA1 | PTP4A1 | 1.42 | Up |
| RNA binding motif protein 5 | RBM5 | 1.59 | Down |
| Ring-box 1 | RBX1 | 1.44 | Down |
| Ras homologue gene family, member B | RHOB | 1.50 | Down |
| Ring finger protein 103 | RNF103 | 1.33 | Down |
| Ring finger protein 111 | RNF111 | 1.41 | Down |
| Ring finger protein 34 | RNF34 | 1.19 | Down |
| Ring finger protein 4 | RNF4 | 1.65 | Down |
| SET translocation (myeloid leukaemia-associated) | SET | 1.45 | Down |
| Seven in absentia homologue 1 (Drosophila) | SIAH1 | 1.30 | Down |
| Structural maintenance chromosomes 4L1 | SMC4L1 | 1.77 | Up |
| Stromal antigen 2 | STAG2 | 1.82 | Up |
| STIP1 homology, U-box containing protein 1 | STUB1 | 1.29 | Down |
| Suppressor of mif two 3 homologue 2 | SUMO2 | 1.33 | Down |
| Transcription elongation factor B1 | TCEB1 | 1.56 | Down |
| Tousled-like kinase 1 | TLK1 | 1.35 | Down |
| Tumor suppressor candidate 4 | TUSC4 | 1.69 | Down |
| Ubiquitin-conjugating enzyme E2E3-UBC4/5 homologue | UBE2E3 | 1.49 | Down |
| Ubiquitin specific protease 4 | USP4 | 1.22 | Down |
| Ubiquitin specific protease 46 | USP46 | 1.34 | Down |
| Wilms tumour 1 | WT1 | 2.01 | Down |
| Kinetochore associated homologue | ZW10 | 1.22 | Down |
Genes previously identified that encode components of ubiquitin-mediated proteolysis and cell cycle pathways and shown to be differentially expressed in 90 dG kidneys from fetuses of nutrient restricted mothers versus controls are listed. ‘Gene ID’ indicates the assigned gene identification, ‘Fold change’ indicates the fold change between fetal kidneys from C and NR mothers, and ‘Direction of change’ indicates the direction of change for NR kidney RNA compared with C kidney RNA.
Evaluation of gene expression based on KEGG pathways
We performed a more detailed analysis of the differently expressed genes to provide insight into cellular functions influenced by these changes in gene expression. Differentially expressed genes were overlaid onto KEGG Pathways to identify pathways impacted by changes in expression for these genes. From the KEGG pathway analysis of the 685 significantly differently expressed genes, we found a number of pathways with significant z-scores.
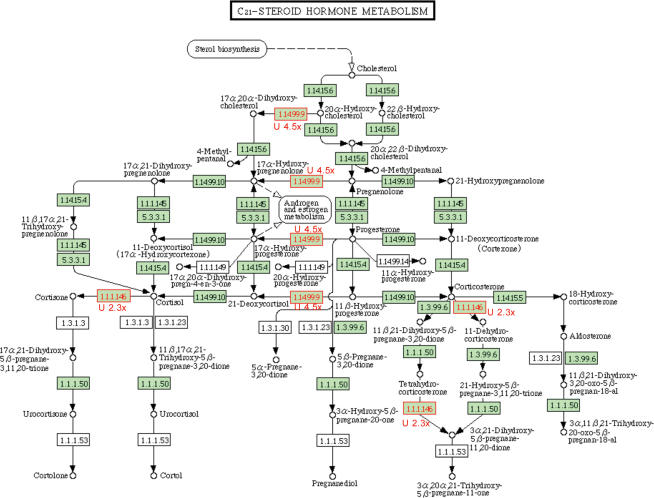
KEGG pathways in which all or most statistically different genes, based on t test analyses, were up-regulated in the NR kidneys include: C21-steroid hormone metabolism (z-score = 3.47), complement and coagulation cascades (z-score = 2.88), cyanoamino acid metabolism (z-score = 2.82), d-glutamine and d-glutamate metabolism (z-score = 2.82), terpenoid biosynthesis (z-score = 2.45), and O-glycan biosynthesis (z-score = 2.19). One example, presented in Fig. 2, shows a graphical representation of the C21-steroid hormone metabolism KEGG pathway, with genes encoding HSD11B1 (hydroxysteroid (11-β) dehydrogenase) and CYP17A1 (cytochrome P450, family 17, subfamily A, polypeptide 1) significantly up-regulated.
Figure 2. Differential expression of genes encoding factors in the C21-steroid hormone metabolism pathway.
Both significantly differentially expressed genes in the C21-steroid hormone metabolism pathway are up-regulated. Homo sapiens specific genes in the pathway are indicated by black font in green boxes. Differentially expressed genes are indicated by red font in a green box. The fold change in expression is indicated numerically with the direction of change indicated by ‘U’ for up-regulated genes. 1.1.1.146 (EC number) denotes HSD11B (gene ID), hydroxysteroid (11-β) dehydrogenase and 1 and 1.14.99.9 denotes CYP17A1, cytochrome P450, family 17, subfamily A, polypeptide 1.
KEGG pathways in which all or most statistically different genes are down-regulated in the nutrient restricted kidneys include: Wnt signalling pathway (z-score = 4.03), peptidoglycan biosynthesis (z-score = 3.59), oxidative phosphorylation (z-score = 3.13), focal adhesion (z-score = 2.91), adherens junction (z-score = 2.66), neuroactive ligand–receptor interaction (z-score = −2.63), tight junction (z-score = 2.38), C5-branched dibasic acid metabolism (z-score = 2.34), ATP synthesis (z-score = 2.29), arginine and proline metabolism (z-score = 2.19), and regulation of actin cytoskeleton (z-score = 1.94).
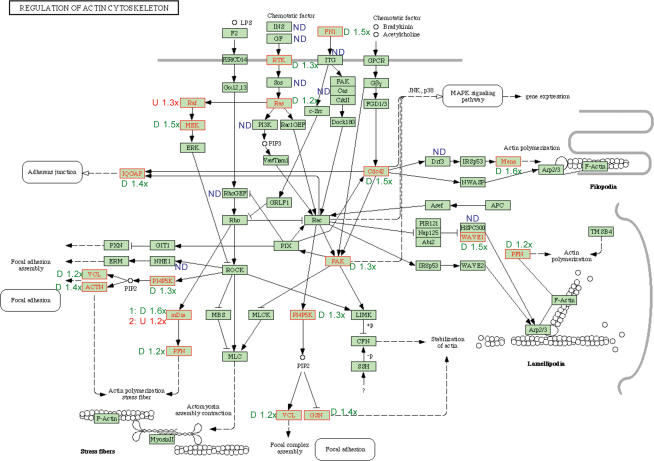
Both ontological categorization and KEGG pathway analysis indicate down regulation of genes encoding actin cytoskeleton assembly and polymerization components in NR kidneys. Figure 3 provides a graphical representation of the KEGG pathway for genes in the actin cytoskeleton assembly and polymerization pathway. Of the 16 significantly differently expressed genes, 14 are down-regulated in the nutrient restricted group. Signal was not detected for 10 genes in this pathway (indicated by ‘ND’ in Fig. 3).
Figure 3. Differential expression of genes encoding actin cytoskeleton assembly.
Fourteen of 16 differentially expressed genes are down-regulated. One gene ‘mDia’ has one family member up-regulated (DIAPH1) and one family member up-regulated (DIAPH2). Homo sapiens specific genes in the pathway are indicated by black font in green boxes. Genes that showed expression on the array are indicated by black font in blue boxes. Differentially expressed genes are indicated by red font in a green box. The fold change in expression is indicated numerically with the direction of change indicated by ‘D’ for down-regulated genes, ‘U’ for up-regulated genes, and ‘ND’ for genes where no signal was detected. Gene names, gene identifications in parentheses, and KEGG gene abbreviations for differentially expressed genes in this pathway are: actinin, α1 (ACTN1), ACTN; cell division cycle 42 (CDC42), CDC42; diaphanous homologue 1 (DIAPH1), mDia; diaphanous homologue 1 (DIAPH1), Mena; early lymphoid activating protein (DIAPH2), mDia; fibronectin 1 (FN1), FN1; gelsolin (GSN), GSN; IQ motif containing GTPase activating protein 1 (IQGAP1), IQGAP; mitogen-activated protein kinase kinase 1 (MAP2K1), MEK; P21(CDKN1A)-activated kinase 4 (PAK4), PAK; phosphatidylinositol-4-phosphate 5-kinase I gamma (PIP5K1C), PI4P5K; platelet-derived growth factor receptor, α polypeptide (PDGFRA), RTK; profilin 2 (PFN2), PFN; vinculin (VCL), VCL; V-Ki-ras2 Kirsten rat sarcoma viral oncogene (KRAS2), RAS; V-raf murine sarcoma viral oncogene homologue B1 (BRAF), RAF; WAS protein family, member 1 (WASF1), WAVE1.
Some KEGG pathways and ontological groups showing significant z-scores include genes that are significantly up-regulated as well as genes that are significantly down-regulated. For example, the dephoshorylation gene cascade is another example of a pathway in which genes were both up- and down-regulated in the kidneys of fetuses from nutrient restricted mothers. Of the five genes, three were significantly up-regulated (cyclin-dependent kinase inhibitor 3, protein tyrosine phosphatase type IVA1, and protein tyrosine phosphatase, receptor type E) and two were down-regulated (protein phosphatase 1D magnesium-dependent, delta isoform and protein phosphatase 2A, regulatory subunit B′ (PR 53)).
Evaluation of expression for kidney development-related genes
Twelve genes that play a role in rodent kidney development (Gazit et al. 1999; Latres et al. 1999; Farrell & Munsterberg, 2000; Stuart et al. 2001; Gasteiger et al. 2003; Sampogna & Nigam, 2004) are significantly differently expressed in the fetal kidneys from NR mothers compared to C. The name, gene ID, role in kidney development, and expression profile of developmental kidney genes from this study are presented in Table 7.
Table 7.
Expression profiles of genes that control branching morphogenesis
| Gene | Gene ID | Role | Fold change | Direction of change |
|---|---|---|---|---|
| Beta-transducin repeat containing | BTRC | Component of the SCF ubiquitin ligase complex in Wnt signalling (Latres et al. 1999) | 1.41 | Up |
| Frizzled homologue 1 (Drosophila) | FZD1 | Wnt protein receptor (Gazit et al. 1999) | 1.41 | Down |
| Frizzled homologue 7 (Drosophila) | FZD7 | Metanephric Kidney Formation (candidate) (Farrell & Munsterberg, 2000) | 1.46 | Down |
| Glypican-1 | GPC3 | Promotes UB branching and patterning (Sampogna & Nigam, 2004) | 2.46 | Down |
| Homeo box A11 | HOXA11 | Cell Proliferation in early metanephric development (Stuart et al. 2001) | 1.51 | Down |
| Integrin-aL | ITGA6 | Kidney tubule formation (Sampogna & Nigam, 2004) | 2.70 | up |
| Matrix metalloproteinase 2 | MMP2 | Promotes branching and remodeling (Sampogna & Nigam, 2004) | 1.35 | Down |
| Sal-like 2 | SALL2 | Promotes UB Growth (SaLL1) (Sampogna & Nigam, 2004) | 1.3 | Down |
| Secreted frizzled-related protein 1 | SFRP1 | Inhibits UB branching and tubule formation (Gasteiger et al. 2003) | 1.5 | Down |
| Tissue inhibitor of metalloproteinases 1 | TIMP1 | Inhibits UB branching (Sampogna & Nigam, 2004) | 2.98 | Down |
| Wilms tumour 1 | WT1 | Promotes UB outgrowth (Sampogna & Nigam, 2004) | 2.00 | Down |
| Wingless-type MMTV integration site 5 A | WNT5A | Wnt's promote UB branching (Sampogna & Nigam, 2004) | 1.53 | Down |
Genes previously identified that control branching morphogenesis and shown to be differentially expressed in 90 dG kidneys from fetuses of nutrient restricted mothers versus controls are listed. ‘Gene ID’ indicates the assigned gene identification, ‘Role’ is the gene function defined from previous work (Gazit et al. 1999; Latres et al. 1999; Farrell & Munsterberg, 2000; Stuart et al. 2001; Gasteiger et al. 2003; Sampogna & Nigam, 2004), ‘Fold change’ indicates the fold change between fetal kidneys from C and NR mothers, and ‘Direction of change’ indicates the direction of change between kidneys from NR and C mothers.
Evaluation of apoptosis-related genes and gene expression profile validation
Apoptosis-related differentially expressed genes from array interrogation were quantified by QRT-PCR. Thirty-nine genes related to apoptosis were significantly differently expressed between C and NR kidney RNA samples: 11 genes were up-regulated and 29 were down-regulated. Five of these genes, BCL2L13, CREBBP, RHOB, SGK, VEGF, were selected for QRT-PCR validation. Gene expression values from arrays and QRT-PCR are presented in Table 8.
Table 8.
Gene expression profiling data for apoptosis-related genes
| Array data (intensity) | QRT-PCR data (ΔCt) | |||||
|---|---|---|---|---|---|---|
| Gene ID | Control | Nutrient restricted | P-value | Control | Nutrient restricted | P-value |
| BCL2L13 | 1.0754 ± 0.0973 | 0.4951 ± 0.1718 | 0.0148 | 19.0452 ± 0.0398 | 18.7380 ± 0.0441 | 0.0003 |
| CREBBP | 2.0744 ± 0.0742 | 1.3647 ± 0.2566 | 0.0240 | 20.0298 ± 0.0242 | 19.6819 ± 0.0546 | 0.0001 |
| RHOB | 3.1924 ± 0.1216 | 2.6060 ± 0.0756 | 0.0022 | 23.1803 ± 0.1422 | 23.6845 ± 0.2301 | 0.0804 |
| SGK | 1.1186 ± 0.0986 | 0.6988 ± 0.0989 | 0.0132 | 19.1889 ± 0.0804 | 18.6509 ± 0.1185 | 0.0027 |
| VEGF | 4.4561 ± 0.0699 | 4.0797 ± 0.1253 | 0.0254 | 13.2016 ± 0.0398 | 12.6847 ± 0.0515 | 0.0294 |
Data are means ± s.e.m. Array Data were all-median normalized, log2 transformed and are presented as intensity values. QRT-PCR data were normalized against 18 s rRNA and are presented as ΔCt values. See QRT-PCR quantification of target gene abundance in Methods for a detailed description of ΔCt value calculations. Gene identification abbreviations are: BCL2L13 – BCL2-like 13 (apoptosis facilitator); CREBBP – CREB binding protein (Rubinstein–Taybi syndrome), RHOB – Ras homologue gene family, member B, SGK – serum/glucocorticoid regulated kinase, VEGF – vascular endothelial growth factor.
Kidney morphology
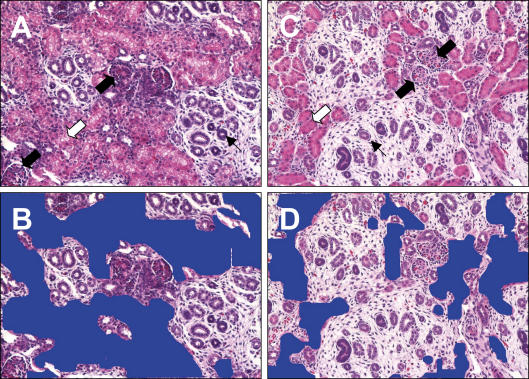
Histological sections of C and NR kidneys are presented in Fig. 4. Mean counts of the number of tubule, duct and glomerular cross-sections, and the percentage area occupied by tubules, in tissue sections from C and NR kidneys, are presented in Table 9. No differences in cross-section counts between groups were found; the area occupied by tubules as indicated in Fig. 4 was significantly decreased in NR kidneys compared to C (31.7 ± 2.0% versus 40.5 ± 2.2%, P = 0.015).
Figure 4. Histological analysis of 90 dG C and NR kidney sections.
Representative photomicrographs (20× magnification) of 5 μm kidney sections from ad libitum fed controls (A) and 30% nutrient restricted (C) baboon fetuses at 90 days of gestation. The area occupied by tubules is represented in blue in the same photomicrographs in control (B) and nutrient restricted (D) animals.
Table 9.
Morphometric analysis of 90 dG kidney sections
| Control fed | Nutrient restricted | P | |
|---|---|---|---|
| Glomeruli (per mm2) | 0.09 ± 0.01 | 0.10 ± 0.00 | 0.166 |
| Convoluted tubules (cross-sections/mm2) | 0.67 ± 0.03 | 0.57 ± 0.04 | 0.115 |
| Ducts (cross-sections/mm2) | 0.38 ± 0.08 | 0.30 ± 0.03 | 0.331 |
| Convoluted tubule density (% field of view) | 40.5 ± 2.2% | 31.7 ± 2.0% | 0.015 |
Discussion
The present study reports several important findings. Foremost, we present evidence of gene expression changes in the developing fetal primate kidney that result from moderate maternal NR. In addition, we have used the power of ontological and pathway analysis to demonstrate changes in biological and biochemical pathways impacted by maternal NR. Screening six individual RNA samples in each group allowed us to evaluate differential expression based on statistical significance in addition to the more commonly used 2-fold expression differences for pooled samples (Quackenbush, 2002). Selection of genes using differential expression and statistical criteria demonstrated that inclusion of genes with small but statistically significant expression differences highlights ontological pathways significantly altered by nutrient restriction that might be overlooked using a minimum twofold expression difference approach.
Further evidence that maternal NR impacts kidney development is provided by placing our expression data in the context of ontological categories and KEGG pathways. The pathway analysis data strongly suggest that NR impacts specific cellular pathways even for genes with small changes in expression. For example, in the oxidative phosphorylation KEGG pathway analysis, 15 of 16 genes that are differently expressed are down-regulated in the NR fetal kidney at 90 dG. The difference in expression for 13 of these genes ranges from 1.1× to 1.4×. Evaluating these expression patterns in the context of oxidative phosphorylation proteins suggests that protein modification is down-regulated in the NR kidneys resulting in decreased phosphorylation events in phosphorylation-dependent cascades and pathways. It is important to note that although 589 of the 685 differently expressed genes are down-regulated, pathways such as C21 steroid metabolism are up-regulated suggesting that NR effects are more complex than global down-regulation of all systems in the developing kidney.
Nucleic Acids, ubiquitin mediated proteolysis and cell cycle regulation
The ontological analysis of gene expression indicates that pathways relevant to nucleic acid component biosynthesis and metabolism, modification and packaging, RNA translation, protein biosynthesis and metabolism, and ubiquitin-dependent protein catabolism contain significant numbers of pathway genes that are down-regulated in the fetal kidneys of NR mothers.
Ubiquitin-mediated proteolysis, through various renal specific complexes, is thought to target several cell-cycle regulators that inhibit G1 to S progression for proteasomal degradation during renal growth (Franch, 2002). In the NR kidney RNA samples, ubiquitin-mediated proteolysis components such as ring-box 1, ubiquitin-conjugating enzyme E2E3, and ubiquitin-conjugating enzyme E2D are all down-regulated. Furthermore, cell cycle inhibitors such as cyclin-dependent kinase inhibitor 3 is up-regulated, whereas cell cycle progression factors such as cyclin B1 and cyclin G2 are down-regulated. Evaluating gene expression profiles in the context of KEGG pathways reveals a consistent pattern where cell cycle progression is inhibited and the availability of cellular components necessary for cell division, such as DNA, is reduced.
As shown in Table 4, a number of Gene Ontology pathways contain significantly up-regulated genes. Up-regulated pathways include: electron, Golgi vesicle and nucleocytoplasmic transport, carbohydrate, sterol and steroid metabolism, metal ion binding, protein transport and targeting, protein tyrosine phosphatase and heterodimerization activity, regulation of cyclin-dependent protein kinase activity, and voltage-gated potassium channel activity. These data suggest, consistent with the cell cycle inhibition evidence, that the NR fetal kidneys are more differentiated than those from C fetuses. It is important to note that the array data do not present a clear picture regarding cellular activity. For example, analysis of the KEGG mitogen activated protein kinase (MAPK) pathway shows large changes in gene expression; however, the direction of change for each gene in the pathway is not consistent with an increased rate of MAPK activity in the NR fetal kidneys. Our data show that adrenomedullin is up-regulated in the kidneys of fetuses from NR mothers, suggesting that nutrient restriction inhibits cell proliferation. Adrenomedullin inhibits proliferation and enhances apoptosis of kidney mesangial cells, through the modulation of MAPK cascades (Belloni et al. 2001). However at this time, the impact of these gene expression changes in the apoptosis and MAPK signalling pathways on cell proliferation and differentiation in the developing primate kidney is not clear. Further experiments will be necessary to ascertain the cellular activities of NR fetal kidneys compared with C kidneys. It will be of interest to see if the inconsistent gene expression profiles are due to differences between gene expression and gene product activity or if these factors play different roles in developing primate kidneys than the functions elucidated in other systems.
Expression of kidney development-related genes
Several genes have been identified that control different aspects of branching morphogenesis in the developing kidney. At least 30 genes have been described that play specific roles in rodent kidney development (Gazit et al. 1999; Latres et al. 1999; Farrell & Munsterberg, 2000; Stuart et al. 2001; Gasteiger et al. 2003; Sampogna & Nigam, 2004). Of these 30 genes, 12 are significantly differently expressed in the baboon fetal kidneys from nutrient restricted mothers versus controls. Beta-transducin repeat containing gene (BTRC), a component of the ubiquitin ligase complex and a factor in promotion of branching through Wnt signalling (Latres et al. 1999), is up-regulated (1.4×) in NR fetal kidney. Secreted frizzled-related protein 1 (SFRP1) and tissue inhibitor of metalloproteinases-1 (TIMP1), inhibitors of ureteric bud branching (Gasteiger et al. 2003; Sampogna & Nigam, 2004), are both down-regulated (1.5× and 3.0×, respectively) in the NR fetuses. The changes in expression of these genes suggest that branching activity is increased following maternal NR. Conversely, wingless-type MMTV integration site 5A (WNT5A), a member of the Wnt gene family known to promote ureteric bud branching (Sampogna & Nigam, 2004), is down-regulated (1.5×). Frizzled homologue 1 (FZD1), a Wnt protein receptor that promotes branching (Gazit et al. 1999), Sal-like 2 (SALL2), which promotes ureteric bud growth (Sampogna & Nigam, 2004), and Wilms tumour 1 (WT1), which promotes ureteric bud outgrowth (Sampogna & Nigam, 2004), are all down-regulated in the NR group (1.4×, 1.3× and 2.0×, respectively). In addition, frizzled homologue 7 (FZD7), which is thought to play a role in metanephric kidney formation (Farrell & Munsterberg, 2000), homeo box A11 (HOXA11), known to play a role in cell proliferation during early metanephric kidney development (Stuart et al. 2001), matrix metalloproteinase 2 (MMP2), which promotes branching and remodelling during kidney development (Sampogna & Nigam, 2004) and glypican-3, which plays a role in late stage branching and tubule maturation, are also all down-regulated in the NR compared to the C kidney (1.5×, 1.5×, 1.4× and 2.5×, respectively). The down-regulation of genes such as these is suggestive of the development of renal hypoplasia in the NR group and is consistent with other models of maternal NR that result in an impaired nephron endowment (Langley-Evans, 1996; Vehaskari et al. 2001; Woods et al. 2001).
Based on previous studies in characterizing molecular mechanisms of kidney development in rodents, these data present a conflicting picture for the impact of maternal NR on the developing primate kidney. Previous work has shown that complex systems, including gene systems, are often error tolerant for some components of a network (Jeong et al. 2001). Studies have also shown that some molecules play contrasting roles at different stages in development. For example, BMP7 can play an activator role at one stage of kidney development and an inhibitory role at another stage (Gupta et al. 1999). From this perspective, the apparent inconsistencies in gene expression for the NR baboon kidney at 90 dG compared to C kidney, at least when compared to previous studies on developing kidneys and differentiating kidney cells in vitro, may be the result of compensatory responses to NR. It is not yet clear which molecules in the network of molecules for kidney development may fluctuate in expression without phenotypic impact, which have detrimental effects on development, and which demonstrate compensation for environmental deficiencies such as NR.
Expression for apoptosis-related genes
Of the 39 differentially expressed apoptosis-related genes, 11 were up-regulated and 28 were down-regulated. Five genes known to play a role in apoptosis were selected for QRT-PCR validation of the array data: BCL2L13 (B-cell CLL/lymphoma 2 like protein 13) promotes apoptosis via the activation of caspase-3; CREBBP (cAMP responsive element binding protein bind protein) mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein, thus functioning as a coactivator of cAMP-responsive genes transcription and consequently playing a role in TGF β (transforming growth factor β) signalling, Wnt signalling and apoptosis pathways; RHOB (ras homologue gene family, member B) promotes endothelial cell survival during vascular development; SGK (serum/glucocorticoid regulated kinase) mediates cell survival signals and negatively regulates pro-apoptotic factors; SGK also initiates a cascade that inactivates various channels and transporters such as ENaC (epithelial sodium channel); and VEGF (vascular endothelial growth factor), a growth factor active in angiogenesis, vasculogenesis and endothelial cell growth, induces endothelial cell proliferation, promotes cell migration, and induces permeabilization of blood vessels. All five genes were found to be down-regulated by QRT-PCR, thereby confirming the validity of the array analysis for these particular genes. Given the mix of expression changes found for both pro- and antiapoptotic genes, the impact of maternal NR on programmed cell death in the 90 dG primate kidney remains to be fully elucidated.
Actin cytoskeleton assembly and histological analysis of 90 dG kidney sections
Pathway analysis of C versus NR kidney gene expression profiles indicated down-regulation of actin cytoskeleton polymerization and assembly. Consistent with this array data as well as that for genes involved in tubular branching discussed above, histological analysis of 90 dG kidney sections shows a reduction in the percentage of cortex occupied by tubule cross-sections in NR versus controls kidneys. The reduction of genes encoding components for actin cytoskeleton assembly and polymerization suggest that nutrient restriction may decrease the number of kidney structural components. These changes in gene expression may also lead to decreased tortuosity and/or decreased length of tubules which manifest as a decrease in cross-sections per unit area.
Perspective
The ability to apply gene array analysis to questions focusing on fetal development exemplifies the utility and importance of our baboon model. Gene array analysis provides a powerful synthesis of system-wide levels of mRNA expression at a given point in development and, as reported here, following perturbation of the intrauterine nutritional milieu. Of the data presented the least clear are those involving cellular activity pathways. KEGG pathways show what is known about Homo sapiens; however, some genes are expressed in a tissue specific and/or temporal specific manner. In addition, gene regulation may vary as well. Therefore, pathways such as the MAPK cascade may vary by cell type within a tissue and also between tissues. Thus the apparent contradictions in gene expression from our study may not be contradictory when more is known about gene expression relevant to developing primate kidneys. Our data strongly support the observation that maternal nutrient restriction is associated with decreases in gene expression for components of RNA, DNA, and protein biosynthesis and metabolism, as well as decreases in gene expression of cell cycle promoters and increases in gene expression of cell cycle inhibitors. These observations, in conjunction with the decreases in tubular density evident in our histological analysis of the fetal renal cortex, therefore suggest that maternal nutrient restriction inhibits cell division, promotes differentiation and results in a global decrease in structural components within the developing NR kidney. Although the fetal morphometry did not indicate differences between C and NR 90 dG kidneys, gene array, QRT-PCR and histological data all indicate differences in kidney development due to NR, confirming that fetal weight is a poor measure of fetal compromise at mid gestation.
Acknowledgments
This work was supported by NICHD grant HD21350 and NHLBI grant HL5263.
References
- Abramoff M, Magelhaes P, Ram S. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Antonow-Schlorke I, Schwab M, Li C, Nathanielsz PW. Glucocorticoid exposure at the dose used clinically alters cytoskeletal proteins and presynaptic terminals in the fetal baboon brain. J Physiol. 2003;547:117–123. doi: 10.1113/jphysiol.2002.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni AS, Albertin G, Forneris ML, Nussdorfer GG. Proadrenomedullin-derived peptides as autocrine-paracrine regulators of cell growth. Histol Histopathol. 2001;16:1263–1274. doi: 10.14670/HH-16.1263. [DOI] [PubMed] [Google Scholar]
- De Vos K. Department of Biological Sciences, Columbia University. 2006. http://rsb.info.nih.gov/ij/plugins/cell-counter.html.
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducsay CA, Hess DL, McClellan MC, Novy MJ. Endocrine and morphological maturation of the fetal and neonatal adrenal cortex in baboons. J Clin Endocrinol Metab. 1991;73:385–395. doi: 10.1210/jcem-73-2-385. [DOI] [PubMed] [Google Scholar]
- Farrell ER, Munsterberg AE. csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Dev Biol. 2000;225:447–458. doi: 10.1006/dbio.2000.9852. [DOI] [PubMed] [Google Scholar]
- Franch HA. Pathways of proteolysis affecting renal cell growth. Curr Opin Nephrol Hypertens. 2002;11:445–450. doi: 10.1097/00041552-200207000-00012. [DOI] [PubMed] [Google Scholar]
- Galaverna O, Nicolaidis S, Yao S, Sakai R, Epstein A. Endocrine consequences of prenatal sodium depletion prepare rats for high need-free NaCl intake in adulthood. Am J Physiol. 1995;269:R578–R583. doi: 10.1152/ajpregu.1995.269.3.R578. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucl Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit A, Yaniv A, Bafico A, Pramila T, Igarashi M, Kitajewski J, et al. Human frizzled 1 interacts with transforming Wnts to transduce a TCF dependent transcriptional response. Oncogene. 1999;18:5959–5966. doi: 10.1038/sj.onc.1202985. [DOI] [PubMed] [Google Scholar]
- Gilbert J, Lang A, Grant A, Nijland M. Maternal nutrient restriction in sheep: hypertension and decreased nephron number in offspring at 9 months of age. J Physiol. 2005;565:137–147. doi: 10.1113/jphysiol.2005.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta IR, Piscione TD, Grisaru S, Phan T, Macias-Silva M, Zhou X, et al. Protein kinase A is a negative regulator of renal branching morphogenesis and modulates inhibitory and stimulatory bone morphogenetic proteins. J Biol Chem. 1999;274:26305–26314. doi: 10.1074/jbc.274.37.26305. [DOI] [PubMed] [Google Scholar]
- Hay WW, Jr, Wilkening RB. Metabolic activity of the placenta. In: Thorburn GD, Harding R, editors. Textbook of Fetal Physiology. Oxford: Oxford University Press; 1994. pp. 30–47. [Google Scholar]
- Hendrickx AG. The menstrual cycle of the baboon as determined by vaginal smear, vaginal biopsy, and perineal swelling. Baboon Med Res. 2001;2:437–459. [Google Scholar]
- Hennessy A, Whitworth JA, Raymond CJ, Phippard AF, Thompson JF, Horvath JS. Haemodynamic actions of a nitric oxide (EDRF) synthesis inhibitor in conscious baboons (Papio hamadryas) Clin Exp Pharmacol Physiol. 1994;21:695–700. doi: 10.1111/j.1440-1681.1994.tb02572.x. [DOI] [PubMed] [Google Scholar]
- Ingelfinger JR, Woods LL. Perinatal programming, renal development, and adult renal function. Am J Hypertens. 2002;15:46S–49S. doi: 10.1016/s0895-7061(01)02302-0. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen SV, Mecenas CA, Smith GS, Jenkins S, Nathanielsz PW. Effects of maternal betamethasone administration on fetal and maternal blood pressure and heart rate in the baboon at 0.7 of gestation. Am J Obstet Gynecol. 2002;186:812–817. doi: 10.1067/mob.2002.121654. [DOI] [PubMed] [Google Scholar]
- Langley-Evans S. Intrauterine programming of hypertension in the rat: nutrient interactions. Comp Biochem Physiol A Physiol. 1996;114:327–333. doi: 10.1016/0300-9629(96)00018-7. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC, Phillips GJ, Gardner DS, Jackson AA. Role of glucocorticoids in programming of maternal diet-induced hypertension in the rat. J Nutr Biochem. 1996;7:173–178. [Google Scholar]
- Latres E, Chiaur DS, Pagano M. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- Pepe GJ, Ballard PL, Albrecht ED. Fetal lung maturation in estrogen-deprived baboons. J Clin Endocrinol Metab. 2003;88:471–477. doi: 10.1210/jc.2001-010228. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl.):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Rasch R, Skriver E, Woods LL. The role of the RAS in programming of adult hypertension. Acta Physiol Scand. 2004;181:537–542. doi: 10.1111/j.1365-201X.2004.01328.x. [DOI] [PubMed] [Google Scholar]
- Sampogna RV, Nigam SK. Implications of gene networks for understanding resilience and vulnerability in the kidney branching program. Physiology (Bethesda) 2004;19:339–347. doi: 10.1152/physiol.00025.2004. [DOI] [PubMed] [Google Scholar]
- Schlabritz-Loutsevitch NE, Howell K, Rice K, Glover EJ, Nevill CH, Jenkins SL, et al. Development of a system for individual feeding of baboons maintained in an outdoor group social environment. J Med Primatol. 2004;33:117–126. doi: 10.1111/j.1600-0684.2004.00067.x. [DOI] [PubMed] [Google Scholar]
- Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc Natl Acad Sci U S A. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehaskari VM, Aviles DH, Manning J. Prenatal programming of adult hypertension in the rat. Kidney Int. 2001;59:238–245. doi: 10.1046/j.1523-1755.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- Welham S, Riley P, Wade A, Hubank M, Woolf A. Maternal diet programs embryonic kidney gene expression. Physiol Genomics. 2005;22:48–56. doi: 10.1152/physiolgenomics.00167.2004. [DOI] [PubMed] [Google Scholar]
- Woods LL, Ingelfinger JR, Nyengaard JR, Rasch R. Maternal protein restriction suppresses the newborn renin-angiotensin system and programs adult hypertension in rats. Pediatr Res. 2001;49:460–467. doi: 10.1203/00006450-200104000-00005. [DOI] [PubMed] [Google Scholar]