Abstract
Interpretation of gene array data presents many potential pitfalls in adult tissues. Gene array techniques applied to fetal tissues present additional confounding pitfalls. The left lobe of the fetal liver is supplied with blood containing more oxygen than the right lobe. Since synthetic activity and cell function are oxygen dependent, we hypothesized major differences in mRNA expression between the fetal right and left liver lobes. Our aim was to demonstrate the need to evaluate RNA samples from both lobes. We performed whole genome expression profiling on left and right liver lobe RNA from six 90-day gestation baboon fetuses (term 180 days). Comparing right with left, we found 875 differentially expressed genes – 312 genes were up-regulated and 563 down-regulated. Pathways for damaged DNA binding, endonuclease activity, interleukin binding and receptor activity were up-regulated in right lobe; ontological pathways related to cell signalling, cell organization, cell biogenesis, development, intracellular transport, phospholipid metabolism, protein biosynthesis, protein localization, protein metabolism, translational regulation and vesicle mediated transport were down-regulated in right lobe. Molecular pathway analysis showed down-regulation of pathways related to heat shock protein binding, ion channel and transporter activities, oxygen binding and transporter activities, translation initiation and translation regulator activities. Genes involved in amino acid biosynthesis, lipid biosynthesis and oxygen transport were also differentially expressed. This is the first demonstration of RNA differences between the two lobes of the fetal liver. The data support the argument that a complete interpretation of gene expression in the developing liver requires data from both lobes.
Gene array analysis is a powerful tool to understand tissue function. However, interpretation of physiological significance from the data obtained is confounded by many potential pitfalls: lack of mRNA translation to protein, changes in mRNA stability, alternative splicing and heterogeneity between cell types in the tissue sampled. When applying gene array techniques to fetal development additional pitfalls can confound data interpretation.
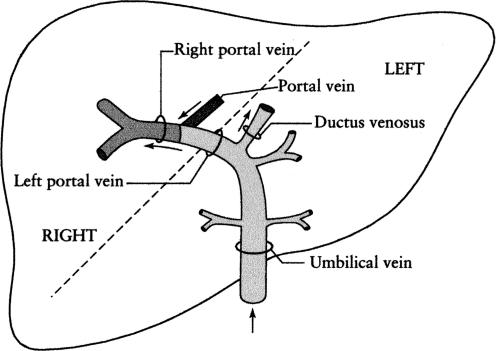
One major difference between the fetus and adult is the presence of the umbilical venous vascular circuit that returns well-oxygenated placental blood to the fetus in the umbilical vein. Umbilical vein blood is partitioned. It either passes through the substance of the fetal liver or is routed through the ductus venosus bypassing the liver. The liver also receives blood from the hepatic arteries and portal venous system. However, the major fetal hepatic blood supply comes from the umbilical vein (Bristow et al. 1983). The variable and changing flow destination of blood in the umbilical vein as it enters the liver influences fetal regional hepatic function. A large and varying proportion of umbilical venous blood traverses the ductus venosus, bypassing the liver. In addition the distribution of well-oxygenated umbilical vein and poorly oxygenated portal vein and hepatic artery blood differs between the right and left lobe (Fig. 1). This difference in vascular supply can lead to marked differences in physiological function (Bristow et al. 1983) as well as pathologies (Gruenwald, 1949) between the two lobes.
Figure 1. Schematic diagram of the fetal umbilical and portal vein circulation.
Arrows indicate direction of blood flow. The vessels with oxygen- and nutrient-rich blood from the umbilical vein circulation are illustrated with light shading. The dark shading represents the portal vein with oxygen- and nutrient-poor blood. The right portal vein with a mixture of blood has intermediate shading. The dashed line represents the division between the right and left lobes of the liver. (From Haugen et al. 2004; © International Society of Ultrasound in Obstetrics & Gynecology (ISUOG); reproduced with permission from John Wiley & Sons Ltd on behalf of the ISUOG.)
Despite the considerable interest in development of fetal liver function, no previous studies have sought to compare mRNA for these two very different parts of the fetal liver in any species. We hypothesized that gene array analysis, correctly applied, would provide valuable initial demonstration of developmental differences related to the differing environments of the hepatic cells in the two lobes. Gene array analysis enables evaluation of multiple genes in the same sample and thus constitutes a powerful approach to demonstrate differences in physiologically important pathways between the two lobes of the liver. This approach provides a wealth of information on differences in single genes as well as important key fetal hepatic pathways such as DNA, lipid, prostaglandin, haemoglobin and oxygen metabolism. To demonstrate the existence and extent of potential differences in cell function between the two lobes we performed whole genome expression profiling on left and right liver lobe RNA from six 90-day gestation (dG) baboon fetuses (term 180 dG). Our aim was to demonstrate the need to evaluate RNA samples from both lobes, and not just one lobe, of the liver. We hypothesized that there would be major differences in mRNA from the right and left lobes.
Methods
Animal care and maintenance
All procedures were approved by the Southwest Foundation for Biomedical Research Institutional Animal Care and Use Committee and conducted in Association for Assessment and Acreditation of Laboratory Animal Care (AAALAC) approved facilities. Housing, group management and feeding in individual cages have been described in detail (Schlabritz-Loutsevitch et al. 2004). Maternal morphometric measurements were made prior to pregnancy to ensure homogeneity of female phenotypes studied. Water was continuously available in individual feeding cages through individual lixits at several locations in group housing. Feed was Purina Monkey Diet 5038 biscuits. Baboons were allowed to feed ad libitum.
Caesarean sections were performed at 90 dG under isoflurane anaesthesia (2%, 2 l min−1) to obtain the fetus. All baboons were premedicated with ketamine hydrochloride (10 mg kg−1). After intubation, isoflurane (2%, 2 l min−1) was administered to maintain a surgical plane of anaesthesia throughout surgery and fetal sampling. The abdomen was shaved and iodine surgical scrub followed by 90% alcohol was applied to the skin of the abdomen. A midline lower abdominal incision was made through the skin and the subcutaneous layer flowed by an incision through the linea alba. The uterus was then gently exteriorized and a hysterotomy incision made in the main body of the uterus. Blunt dissection was used to expose the amnion for fluid sampling. The edges of the incision were carefully manipulated and swabbed to avoid blood contamination of amniotic fluid samples that were taken into a syringe and placed in metal-free vials. The umbilical cord was identified and elevated to the surgical opening for sampling. While retaining the fetus within the body of the uterus, umbilical cord venous blood was taken through a 24-gauge needle directed towards the placenta. Fetuses were exteriorized from the uterus and killed by exsanguination while under anaesthesia. Fetal liver samples were taken as centrally as possible from deep within the right and left lobes and immediately snap frozen in liquid nitrogen and then stored at −80°C until used for RNA extractions. Postoperatively, mothers were maintained in individual cages until returned to the social group in the presence of a vasectomized male to ensure that they did not become pregnant immediately. Analgesia was provided with buprenorphine hydrochloride at 0.015 mg kg−1 day−1 during three postoperative days (Buprenex® Injectable, Reckitt Benckiser Health Care (UK) Ltd, Hull, UK).
RNA isolation from tissue
RNA was isolated from tissue using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, approximately 100 mg section of frozen liver was homogenized in 1 ml Trizol Reagent using a Power General Homogenizer (Omni International, Wilmington, DE, USA). Genomic DNA in the sample was sheared by passing the homogenate three times through a 22-gauge needle attached to a 1 ml syringe. The homogenized samples were incubated for 5 min at 25°C. Two hundred microlitres of chloroform were added to each sample, the samples were shaken vigorously by hand for 15 s and incubated at 25°C for 3 min. Samples were then centrifuged at 4°C and 12 000 g for 15 min. The aqueous phase containing RNA was transferred to a fresh tube and the RNA precipitated by addition of 0.5 ml of isopropyl alcohol. Samples were incubated for 10 min at 25°C and then centrifuged at 4°C and 12 000 g for 10 min. The RNA precipitate was washed with 1 ml of 75% ethanol and centrifuged at 4°C and 7500 g for 5 min. The RNA was resuspended in 100 μl DEPC-treated water and stored at −80°C.
Preparation of cRNA probe for gene chip interrogation
Total RNA samples were shipped on dry ice to Genome Explorations, Inc. (Memphis, TN) for RNA quality check, cRNA synthesis, and determination of gene expression profiles for each RNA sample by interrogation of the Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA).
Gene chip data collection and pathway analyses
Gene expression was detected using GCOS software (Affymetrix). Expression data were analysed using GeneSifter software (GeneSifter.Net, VizX Laboratories, Seattle, WA, USA). To perform pathway analyses, we first created a ‘custom baboon array’ using GeneSifter. z-Score calculations defining significant gene categories and pathways are based on the total number of genes on the array. Thus, to accurately calculate z-scores using GeneSifter software, the array of baboon genes for which expression was detected on the human gene chip had to be defined. To do so, we merged expression array data from five baboon tissues at three fetal time points and three adult baboon tissues. Any gene from any baboon RNA sample with a marginal or present call on the human genechip (Affymetrix U133A 2.0) was considered expressed and included in the ‘custom baboon array’. Using this method, 16 186 of the 22 227 genes on the genechip were detected using baboon RNA. Thus, these 16 186 genes comprise the ‘custom baboon array’ from which z-scores were calculated.
Statistical analysis
Array data were all-median normalized and log2 transformed using GeneSifter software (GeneSifter.Net, VizX Laboratories). Pairwise statistical analyses of array data were performed by Student's t test using GeneSifter software. Array data for significantly differently expressed genes were overlaid onto Ontological Pathways (http://www.geneontology.org/) (Ashburner et al. 2000) using GeneSifter software. z-Scores were calculated in GeneSifter using the following formula:
where R = total number of genes meeting selection criteria, N = total number of genes measured, r = number of genes meeting selection criteria with the specified GO term, and n = total number of genes measured with the specific GO term. z-Scores greater than 2 or less than −2 are considered significant (Doniger et al. 2003).
Results
Differentially expressed genes between fetal left and right liver lobes
Whole genome expression profiling was performed on left and right liver lobe RNA from six 90-day gestation (dG) baboon fetuses. All comparisons between fetal left and right lobe liver RNA expression were based on designating the left lobe as ‘control’ and comparing the right lobe against the left lobe. Pairwise comparisons showed 875 statistically differently expressed genes between left and right liver lobe RNA samples: 312 genes were up-regulated and 563 were down-regulated in right versus left liver lobe RNA.
Biological ontological pathway analysis of differentially expressed genes
Differentially expressed genes were analysed by ontological groups. Biological ontological pathways related to RNA metabolism, RNA processing, RNA export, DNA repair, nuclear transport, protein nuclear import and depolymerization were all up-regulated in right versus left liver lobe RNA (Table 1). Consistent with the biological ontological pathway analysis, molecular ontological pathway analysis showed up-regulation of pathways for damaged DNA binding, endonuclease activity, and interleukin binding and receptor activity (z-scores = 2.64, 2.17, 3.45 and 3.24, respectively).
Table 1.
Ontological Pathways Up-regulated in Right liver lobe RNA compared with left lobe liver RNA
| Ontology: liver left vs. right lobe | Tot diff genes | Genes up | Genes down | Tot genes on array | z-Up |
|---|---|---|---|---|---|
| Acute-phase response | 4 | 2 | 2 | 18 | 2.09 |
| DNA repair | 19 | 10 | 9 | 161 | 2.56 |
| Establishment of RNA localization | 6 | 3 | 3 | 32 | 2.2 |
| Mitochondrial electron transport, NADH to ubiquinone | 4 | 2 | 2 | 19 | 2 |
| mRNA export from nucleus | 5 | 3 | 2 | 26 | 2.64 |
| mRNA metabolism | 23 | 11 | 12 | 174 | 2.75 |
| mRNA processing | 21 | 10 | 11 | 154 | 2.71 |
| mRNA transport | 5 | 3 | 2 | 26 | 2.64 |
| Nuclear export | 7 | 4 | 3 | 34 | 3.11 |
| Nuclear import | 7 | 4 | 3 | 50 | 2.17 |
| Nuclear transport | 13 | 7 | 6 | 81 | 3.12 |
| Nucleic acid transport | 6 | 3 | 3 | 32 | 2.2 |
| Nucleocytoplasmic transport | 15 | 7 | 8 | 87 | 2.9 |
| Organelle localization | 4 | 1 | 3 | 6 | 2.02 |
| Phosphorylation | 40 | 20 | 20 | 432 | 2.24 |
| Protein depolymerization | 6 | 3 | 3 | 24 | 2.82 |
| Protein import into nucleus | 7 | 4 | 3 | 49 | 2.22 |
| Response to DNA damage stimulus | 20 | 10 | 10 | 178 | 2.21 |
| Response to endogenous stimulus | 21 | 10 | 11 | 183 | 2.12 |
| RNA export from nucleus | 6 | 3 | 3 | 32 | 2.2 |
| RNA localization | 6 | 3 | 3 | 33 | 2.14 |
| RNA metabolism | 36 | 17 | 19 | 307 | 2.84 |
| RNA processing | 32 | 14 | 18 | 255 | 2.54 |
| RNA transport | 6 | 3 | 3 | 32 | 2.2 |
| Transcription | 76 | 25 | 51 | 1243 | −2.01 |
In contrast, biological ontological pathways related to cell signalling, cell organization cell biogenesis, development, intracellular transport, phospholipid biosynthesis, phospholipid metabolism, protein biosynthesis, protein localization, protein metabolism, translational regulation and vesicle mediated transport were down-regulated (Table 2). Similarly, molecular ontological pathway analysis showed down-regulation of pathways related to heat shock protein binding, ion channel and transporter activities, oxygen binding and transporter activities, and translation initiation and translation regulator activities (z-scores = 3.20, −3.02, −2.11, 2.42, 5.18, 3.75, and 4.14, respectively.
Table 2.
Ontological Pathways down-regulated in Right liver lobe RNA compared with left lobe liver RNA
| Ontology: Liver left v right lobe | Tot diff genes | Genes up | Genes down | Tot genes on array | z-Down |
|---|---|---|---|---|---|
| Biopolymer catabolism | 18 | 6 | 12 | 136 | 2.12 |
| Cation transport | 13 | 8 | 5 | 263 | −2.3 |
| Cell organization and biogenesis | 101 | 33 | 68 | 1034 | 2.65 |
| Cell-cell signalling | 11 | 4 | 7 | 324 | −2.35 |
| Cellular localization | 62 | 20 | 42 | 464 | 4.25 |
| Cellular macromolecule metabolism | 177 | 60 | 117 | 1903 | 2.87 |
| Cellular physiological process | 482 | 174 | 308 | 5929 | 2.19 |
| Cellular protein metabolism | 174 | 60 | 114 | 1873 | 2.7 |
| Development | 68 | 34 | 34 | 1196 | −3.63 |
| Endocytosis | 14 | 4 | 10 | 103 | 2.26 |
| Establishment of cellular localization | 62 | 20 | 42 | 460 | 4.31 |
| Establishment of protein localization | 59 | 19 | 40 | 436 | 4.23 |
| Gas transport | 4 | 0 | 4 | 10 | 5.13 |
| Glycerophospholipid biosynthesis | 4 | 0 | 4 | 15 | 3.9 |
| Glycerophospholipid metabolism | 5 | 0 | 5 | 19 | 4.32 |
| Intracellular protein transport | 43 | 15 | 28 | 285 | 3.9 |
| Intracellular transport | 59 | 20 | 39 | 456 | 3.7 |
| Ion transport | 17 | 10 | 7 | 396 | −2.98 |
| Macromolecule biosynthesis | 44 | 11 | 33 | 417 | 2.91 |
| Macromolecule metabolism | 246 | 92 | 154 | 2633 | 2.74 |
| Metal ion transport | 8 | 5 | 3 | 191 | −2.17 |
| Negative reg. of cell organization & biogenesis | 6 | 2 | 4 | 24 | 2.66 |
| Negative regulation of protein metabolism | 9 | 2 | 7 | 44 | 3.38 |
| Organelle localization | 4 | 1 | 3 | 6 | 5.11 |
| Organismal physiological process | 65 | 31 | 34 | 1078 | −2.9 |
| Oxygen transport | 4 | 0 | 4 | 10 | 5.13 |
| Phospholipid biosynthesis | 5 | 1 | 4 | 29 | 2.21 |
| Phospholipid metabolism | 7 | 1 | 6 | 44 | 2.68 |
| Positive regulation of biological process | 38 | 9 | 29 | 407 | 2.12 |
| Positive regulation of cell proliferation | 10 | 2 | 8 | 82 | 2.04 |
| Positive regulation of cellular physiological process | 28 | 6 | 22 | 290 | 2.14 |
| Positive regulation of physiological process | 28 | 6 | 22 | 298 | 2.01 |
| Protein biosynthesis | 41 | 10 | 31 | 370 | 3.16 |
| Protein localization | 60 | 19 | 41 | 448 | 4.27 |
| Protein metabolism | 177 | 61 | 116 | 1891 | 2.83 |
| Protein transport | 58 | 18 | 40 | 424 | 4.43 |
| Regulation of body fluids | 11 | 3 | 8 | 81 | 2.07 |
| Regulation of protein metabolism | 19 | 5 | 14 | 141 | 2.78 |
| Regulation of translation | 9 | 2 | 7 | 59 | 2.48 |
| Regulation of translational initiation | 5 | 0 | 5 | 26 | 3.38 |
| Synaptic transmission | 4 | 3 | 1 | 138 | −2.3 |
| Translation | 17 | 5 | 12 | 114 | 2.79 |
| Translational initiation | 9 | 2 | 7 | 44 | 3.38 |
| Transmission of nerve impulse | 4 | 3 | 1 | 143 | −2.36 |
| Vesicle-mediated transport | 28 | 8 | 20 | 251 | 2.27 |
Differential expression of amino acid biosynthesis related genes
Of the 875 differentially expressed genes between left and right liver lobes, two genes related to amino acid catabolism AU RNA binding protein/enoyl-coenzyme A hydratase (AUH) and histidine ammonia-lyase (HAL) were down-regulated. In addition, two genes related to amino acid biosynthesis, aldehyde dehydrogenase 18 family, member A1 (ALDH18A1) and dihydrofolate reductase (DHFR) were up-regulated in right liver lobe RNA samples compared with left liver lobe RNA samples.
Differential expression of lipid biosynthesis related genes
Sixteen genes related to lipid metabolism were differentially expressed with greater than 1.3-fold change between left and right liver lobe RNA samples. Four of the 16 genes were up-regulated and 12 were down-regulated (Table 3). Five genes related to lipid catabolism were differentially expressed between left and right liver lobe RNA samples. Four (> 1.3-fold) of these genes were down-regulated and one (1.45-fold) was up-regulated in right liver lobe RNA samples compared with left liver lobe RNA samples (Table 3).
Table 3.
Gene expression profiles of lipid metabolism and lipid catabolism genes
| Lipid metabolism genes | Gene ID | Left lobe ± s.e.m. | Right lobe ± s.e.m. | Ratio | Direction | P-value |
|---|---|---|---|---|---|---|
| Apolipoprotein L, 2 | APOL2 | −0.415 ± 0.159 | 0.371 ± 0.116 | 1.72 | Up | 0.003 |
| Peroxisome proliferative activated receptor, α | PPARA | −1.631 ± 0.255 | −0.876 ± 0.155 | 1.69 | Up | 0.039 |
| Phospholipase D2 | PLD2 | 0.496 ± 0.139 | 1.031 ± 0.092 | 1.45 | Up | 0.011 |
| Prostaglandin E synthase 2 | PTGES2 | −1.025 ± 0.469 | 0.227 ± 0.100 | 2.38 | Up | 0.043 |
| 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | HMGCS1 | 3.271 ± 0.202 | 2.650 ± 0.143 | 1.54 | Down | 0.038 |
| Acetyl-Coenzyme A acetyltransferase 2 | ACAT2 | 2.862 ± 0.104 | 2.475 ± 0.107 | 1.31 | Down | 0.025 |
| ATP-binding cassette, subfamily A1 | ABCA1 | 1.106 ± 0.083 | 0.730 ± 0.085 | 1.3 | Down | 0.009 |
| Carnitine O-octanoyltransferase | CROT | 1.840 ± 0.127 | 1.276 ± 0.240 | 1.48 | Down | 0.046 |
| Enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | ECHS1 | 1.347 ± 0.127 | 0.298 ± 0.356 | 2.07 | Down | 0.009 |
| Fatty acid desaturase 2 | FADS2 | 0.425 ± 0.203 | −0.305 ± 0.265 | 1.66 | Down | 0.046 |
| Phosphatidylinositol glycan, class B | PIGB | 0.408 ± 0.096 | −0.427 ± 0.270 | 1.78 | Down | 0.007 |
| Phosphatidylinositol glycan, class F | PIGF | 1.609 ± 0.125 | 1.154 ± 0.159 | 1.37 | Down | 0.041 |
| Platelet-activating factor acetylhydrolase, isoform Ib, α 45 kDa | PAFAH1B1 | 0.668 ± 0.106 | −0.218 ± 0.230 | 1.85 | Down | 0.002 |
| Protein phosphatase 2, catalytic subunit, α | PPP2CA | 2.052 ± 0.051 | 1.615 ± 0.083 | 1.35 | Down | 0.000 |
| Pyruvate carboxylase | PC | 1.810 ± 0.119 | 1.289 ± 0.137 | 1.44 | Down | 0.014 |
| SET binding factor 1 | SBF1 | 1.198 ± 0.077 | 0.816 ± 0.087 | 1.3 | Down | 0.007 |
| Lipid Catabolism Genes | ||||||
| Lysophospholipase 3 | LYPLA3 | −0.506 ± 0.100 | −1.340 ± 0.425 | 1.78 | Down | 0.049 |
| Phospholipase A2, group IVB | PLA2G4B | 1.255 ± 0.107 | 0.881 ± 0.091 | 1.3 | Down | 0.026 |
| Phospholipase A2, group VI | PLA2G6 | 0.690 ± 0.132 | 0.172 ± 0.121 | 1.43 | Down | 0.016 |
| Phospholipase D2 | PLD2 | 0.496 ± 0.139 | 1.031 ± 0.092 | 1.45 | Up | 0.011 |
Differential expression of oxygen transport related genes
Six genes related to oxygen transport and iron availability were differentially expressed. All six genes, expansin (HBG1), haemoglobin α1 (HBA1), haemoglobin α2 (HBA2), haemoglobin ɛ1 (HBE1), haemoglobin, γG (HBG2) and ferritin light chain (FTL) were down-regulated in right liver lobe RNA samples compared with left liver lobe RNA samples.
Discussion
The purpose of this study was to demonstrate the need to evaluate both lobes of the fetal liver separately in studies of fetal development. The percentage of umbilical blood that goes through the liver parenchyma under normal resting conditions increases as pregnancy advances from 60% at 20 weeks to 85% at term (Barbera et al. 1999; Konje et al. 2001; Battaglia, 2004). Thus an increasing percentage of well-oxygenated umbilical blood passes through liver parenchyma as pregnancy progresses.
One recent human study (Haugen et al. 2004) reported that in human gestation at 36 weeks 25% of umbilical venous blood is shunted through the ductus venosus. Of the total umbilical flow, 55% flows to the left hepatic lobe and 20% to the right hepatic lobe. The venous flow to the left lobe came exclusively from the oxygen and nutrient rich umbilical vein while the right lobe received 50% of its venous blood supply from the nutrient poor portal vein. The authors suggest that, ‘This watershed between the portal and umbilical venous flows to the fetal liver suggests a corresponding functional dichotomy; this may be modified by haemodynamic influences, with long-term consequences’ (Haugen et al. 2004).
Relative right and left lobe flows have been shown to respond differently to fetal hypoxia. In a classical in vivo physiological study in fetal sheep, Bristow et al. (1983) observed that during normoxia, right and left lobes consumed the same amount of oxygen, 4 ml min−1 (100 g)−1. However, during acute hypoxia while total liver blood flow and its umbilical venous contributions fell by 20%, the flow to the right lobe to the liver fell twice as much as that of the left lobe of the liver. This finding indicates a potential functional difference between the two lobes of the liver. Livers of human infants who die of birth asphysia show greater anoxic injury in the right than left lobe (Gruenwald, 1949). Others have suggested that these differences may result in long-term programming of hepatic function following in utero challenges (Haugen et al. 2004, 2005). All of these observations indicate that it is not enough to consider the fetal liver as a homogeneous organ.
As we hypothesized, the pronounced differences in the conditions under which the right and left lobes of the liver develop were accompanied by major differences in mRNA expression. The ontological pathway analyses provide detailed data on individual genes in the context of that gene's role in described biological/biochemical pathways. Comparison of the right liver lobe whole genome expression profiles compared with the left lobe shows that 64% of the differentially expressed genes are down-regulated in the right lobe. Up-regulation of pathways related to DNA repair, damaged DNA binding, endonuclease activity, interleukin binding and receptor activities combined with down-regulation of pathways related to cell signalling, organization, and biogenesis, development, protein biosynthesis, localization, and metabolism, and translation initiation and regulator activity suggest that the right lobe of the liver has decreased cell proliferation and increased cell damage. Furthermore, the decrease in genes encoding components of the oxygen binding and transporter pathways indicates that the right lobe of the liver is responding to decreased oxygen environment during development.
Evaluation of genes related to amino acid biosynthesis showed that two amino acid catabolism genes, AUH and HAL, were down-regulated. HAL is the amino acid-degrading enzyme of histidine and is regulated by glucagon; glucagon induces HAL expression in primary hepatocytes (Aleman et al. 2005). Two amino acid biosynthesis genes, ALDH18A1 and DHFR, were down-regulated. ALDH18A1 encodes an enzyme that catalyses the first two steps in proline biosynthesis (Aral et al. 1996). DHFR catalyses an essential step for de novo glycine and purine synthesis, DNA precursor synthesis, and conversion of dUMP to dTMP (Rebhan et al. 1997). Taken together, these data suggest decreased amino acid availability in the right versus left liver lobe.
Among genes involved in lipid metabolism, the changes in the genes for phopholipases which are associated with lipid signalling were pronounced but variable. Peroxisome proliferator activator receptor α (PPARα) is a regulatory gene that has attracted much attention as a regulator of developmental processes involved in developmental programming (Lillycrop et al. 2005). PPARα has been shown to alter fatty acid metabolism by inhibiting key enzymes such as Δ-6 desaturase (Tang et al. 2003). The major prostaglandin produced by the liver is PGE2 (Wernze et al. 1986). Prostaglandins are cytoprotective to hepatic cells (Guarner et al. 1985). The biggest difference in all the lipid related genes between the right and left lobes was in mRNA for prostaglandin E synthase. Up-regulation of PGE2 production occurs during liver regeneration (Tsujii et al. 1993) and thus the observed increased expression in the right lobe may relate to compensation for influences tending to decrease liver growth.
Six genes related to oxygen transport and iron availability (HBA1, HBA2, HBG1, HBG2, HBE1 and FTL) were down-regulated in the right lobe. Two of these genes encode haemoglobin α chains (HBA1 and HBA2) and two encode haemoglobin γ chains (HBG1 and HBG2). Early embryonic haemoglobin is formed by heterotetramers of two α chains, encoded by HBA1 or HBA2, and two epsilon chains form. Fetal haemoglobin is formed by heterotetramers of two α chains, encoded by HBA1 or HBA2, and two γ chains, HBG1 or HBG2. Defects in HBA1 or HBA2 cause α-thalassemia (Rebhan et al. 1997). The liver is an important haematopoetic organ at this stage of development and down-regulation of genes responsible for haemoglobin production in the right versus left lobe is another example of potentially decreased function of the right lobe.
Perspective
These data were obtained from individual gene arrays in six animals and therefore are unlikely to be due to an abnormality in one single individual. The observations indicate molecular genetic differences between left and right liver lobes during primate development which strongly suggest functional cellular differences. Many of the changes observed indicate decreased synthetic and signalling activity in the right lobe – the lobe that has the lower pO2. The findings support our argument that care must be taken in studying and interpreting gene expression in the developing fetal liver. While it is well recognized that it is important to maintain uniformity of the sampling site our data indicate that it is also necessary to compare right and left lobes to evaluate the differential impact of both ontogeny and any challenges imposed on the fetus such as suboptimal nutrition. The proportionate distribution of blood entering the liver and the proportion bypassing it alters under certain situations such as hypoxemia. Fetal hypoxia decreases the total amount of umbilical blood going through the liver (Bristow et al. 1983). Interestingly the proportion of umbilical blood going through the liver parenchyma is increased in fetuses of slimmer mothers with lower body fat stores and mothers eating less well balanced diets. Delivering more blood to the liver in these situations may spare the liver from damage and help protein synthesis when necessary (Haugen et al. 2005). Biochemical evaluation of situations such as this will require close attention to function of metabolism in each lobe individually. Differential metabolism in the two lobes may result in differential susceptibility to oxidative and other forms of damage and altered postnatal predisposition to impaired liver function in adult life (Latini et al. 2004).
Acknowledgments
This work was supported by NICHD HD 21350. We would like to thank Jeremy Glenn, Marie Silva, Antonio Perez Scott Chambers, and the staff at the South-west National Primate Research Center.
References
- Aleman G, Ortiz V, Langley E, Tovar AR, Torres N. Regulation by glucagon of the rat histidase gene promoter in cultured rat hepatocytes and human hepatoblastoma cells. Am J Physiol Endocrinol Metab. 2005;289:E172–E179. doi: 10.1152/ajpendo.00584.2004. [DOI] [PubMed] [Google Scholar]
- Aral B, Schlenzig JS, Liu G, Kamoun P. Database cloning human delta 1-pyrroline-5-carboxylate synthetase (P5CS) cDNA: a bifunctional enzyme catalyzing the first 2 steps in proline biosynthesis. C R Acad Sci. 1996;319:171–178. III. [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera A, Galan HL, Ferrazzi E, Rigano S, Jozwik M, Battaglia FC, et al. Relationship of umbilical vein blood flow to growth parameters in the human fetus. Am J Obstet Gynecol. 1999;181:174–179. doi: 10.1016/s0002-9378(99)70456-4. [DOI] [PubMed] [Google Scholar]
- Battaglia FC. Intrauterine and postnatal growth: Circulatory and metabolic changes accompanying fetal growth restriction. In: Polin R, Fox W, Abman S, editors. Fetal and Neonatal Physiology. 3. Vol. 1. Philadelphia: W. B. Saunders; 2004. pp. 259–265. [Google Scholar]
- Bristow J, Rudolph AM, Itskovitz J, Barnes R. Hepatic oxygen and glucose metabolism in the fetal lamb. Response to hypoxia. J Clin Invest. 1983;71:1047–1061. doi: 10.1172/JCI110855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenwald P. Degenerative changes in the right half of the liver resulting from intrauterine hypoxia. Am J Clin Pathol. 1949;19:801–813. doi: 10.1093/ajcp/19.9.801. [DOI] [PubMed] [Google Scholar]
- Guarner F, Fremont-Smith M, Prieto J. Cytoprotective effect of prostaglandins on isolated rat liver cells. Liver. 1985;5:35–39. doi: 10.1111/j.1600-0676.1985.tb00013.x. [DOI] [PubMed] [Google Scholar]
- Haugen G, Hanson M, Kiserud T, Crozier S, Inskip H, Godfrey KM. Fetal liver-sparing cardiovascular adaptations linked to mother's slimness and diet. Circ Res. 2005;96:12–14. doi: 10.1161/01.RES.0000152391.45273.A2. [DOI] [PubMed] [Google Scholar]
- Haugen G, Kiserud T, Godfrey K, Crozier S, Hanson M. Portal and umbilical venous blood supply to the liver in the human fetus near term. Ultrasound Obstet Gynecol. 2004;24:599–605. doi: 10.1002/uog.1744. [DOI] [PubMed] [Google Scholar]
- Konje JC, Kaufmann P, Bell SC, Taylor DJ. A longitudinal study of quantitative uterine blood flow with the use of color power angiography in appropriate for gestational age pregnancies. Am J Obstet Gynecol. 2001;185:608–613. doi: 10.1067/mob.2001.117187. [DOI] [PubMed] [Google Scholar]
- Latini G, De Mitri B, Del Vecchio A, Chitano G, De Felice C, Zetterstrom R. Foetal growth of kidneys, liver and spleen in intrauterine growth restriction: “programming” causing “metabolic syndrome” in adult age. Acta Paediatr. 2004;93:1635–1639. doi: 10.1080/08035250410023106. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- Schlabritz-Loutsevitch NE, Howell K, Rice K, Glover EJ, Nevill CH, Jenkins SL, et al. Development of a system for individual feeding of baboons maintained in an outdoor group social environment. J Med Primatol. 2004;33:117–126. doi: 10.1111/j.1600-0684.2004.00067.x. [DOI] [PubMed] [Google Scholar]
- Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human Δ-6 desaturase gene transcription: identification of a functional direct repeat-1 element. J Lipid Res. 2003;44:686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- Tsujii H, Okamoto Y, Kikuchi E, Matsumoto M, Nakano H. Prostaglandin E2 and rat liver regeneration. Gastroenterology. 1993;105:495–499. doi: 10.1016/0016-5085(93)90725-r. [DOI] [PubMed] [Google Scholar]
- Wernze H, Tittor W, Goerig M. Release of prostanoids into the portal and hepatic vein in patients with chronic liver disease. Hepatology. 1986;6:911–916. doi: 10.1002/hep.1840060517. [DOI] [PubMed] [Google Scholar]