Abstract
In this study we utilize fluorescent activated cell sorting (FACS) of cells from transgenic zebrafish coupled with microarray analysis to globally analyze expression of cell type specific genes. We find that it is possible to isolate cell populations from Tg(fli1:egfp)y1 zebrafish embryos that are enriched in vascular, hematopoietic and pharyngeal arch cell types. Microarray analysis of GFP+ versus GFP− cells isolated from Tg(fli1:egfp)y1 embryos identifies genes expressed in hematopoietic, vascular and pharyngeal arch tissue, consistent with the expression of the fli1:egfp transgene in these cell types. Comparison of expression profiles from GFP+ cells isolated from embryos at two different time points reveals that genes expressed in different fli1+ cell types display distinct temporal expression profiles. We also demonstrate the utility of this approach for gene discovery by identifying numerous previously uncharacterized genes that we find are expressed in fli1:egfp positive cells, including new markers of blood, endothelial and pharyngeal arch cell types. In parallel, we have developed a database to allow easy access to both our microarray and in situ results. Our results demonstrate that this is a robust approach for identification of cell type specific genes as well as for global analysis of cell type specific gene expression in zebrafish embryos.
Keywords: zebrafish, hematopoietic, vascular, endothelial, microarray
Introduction
The zebrafish has become an ideal model organism for the study of vertebrate embryogenesis. The transparency and accessibility of zebrafish embryos allow for a variety of detailed manipulations and phenotypic analyses, while its small size, fecundity, and ease of maintenance have helped to establish it as a powerful genetic system. Numerous mutants have been described in the course of several large- and small-scale forward genetic screens (Driever et al., 1996; Haffter et al., 1996; Patton and Zon, 2001). Increasingly dense genetic (Gates et al., 1999; Kelly et al., 2000; Knapik et al., 1998; Postlethwait et al., 1994) and physical maps (Geisler et al., 1999; Hukriede et al., 1999) have greatly aided the subsequent positional or candidate cloning of the genes responsible for these mutant phenotypes and large scale screens utilizing retroviral mutagenesis now allow rapid cloning of affected genes (Amsterdam, 2003). In addition to classical forward genetic approaches, the advent of modified antisense oligonucleotides that block translation or splicing in a gene specific manner (referred to as Morpholinos) has allowed reverse genetic approaches as well (Nasevicius and Ekker, 2000). The current availability of genomic sequence has greatly benefited both forward and reverse genetic approaches by facilitating the identification of candidate genes of interest.
The transparency of the zebrafish embryo and the ability to collect hundreds of embryos at specific time points has also allowed detailed temporal and spatial analysis of gene expression through whole mount in situ hybridization. Several groups have initiated large scale in situ screens in order to provide tissue specific expression data in parallel to current expressed sequence tag (EST) projects. In some cases, these screens are unbiased and involve random selection of clones from normalized libraries derived from different embryonic stages (Kudoh et al., 2001; Thisse et al., 2004). Thus far, several thousand expression patterns have been generated from these efforts and curated by the Zebrafish Information Network (ZFIN). In other cases, researchers have focused their expression screens on genes containing particular motifs (Crosier et al., 2001), or involved in specific developmental processes (Yoda et al., 2003). In some cases, these approaches have proven to be useful at identifying new components of known signaling pathways since genes with common expression patterns often have functional relationships that are important for development. For example, both the sef gene and dusp6 were identified in an in situ screen based on the similarity of their expression patterns to genes known to be involved in fibroblast growth factor (FGF) signaling pathway, such as fgf8 (Furthauer et al., 2002; Tsang et al., 2002; Tsang et al., 2004). Subsequent functional analysis of both sef and dusp6 revealed their role as negative regulators of FGF signaling.
In addition to whole mount in situ hybridization, researchers have begun to utilize microarrays for global analysis of gene expression in zebrafish embryos. Several studies have used microarray analysis of zebrafish mutants in order to characterize changes in gene expression associated with defects in hematopoietic and vascular development (Qian et al., 2005; Sumanas et al., 2005; Weber et al., 2005). Thus far, this work has proven useful at identifying a number of new hematopoietic and vascular markers and new genes required for development of these cell types (Sumanas and Lin, 2006). Additionally, microarray analysis has allowed the identification of pathway-specific molecular signatures through the combinatorial comparison of multiple related mutant lines (Weber et al., 2005). In most of the microarray studies to date, researchers relied on whole embryos as a source for RNA for microarray analysis. However, there are drawbacks to the use of whole embryo array profiling. First, the primary cell type affected in a particular mutant may only represent a small proportion of the embryo. This is certainly the case for many of the mutants that affect development of endothelial or hematopoietic cell types. Second, given the complexity of transcripts obtained from a whole embryo, it is likely that moderate and low abundant transcripts in a cell type of interest will be poorly detected in this context. Finally, transcript levels for many genes expressed in multiple tissues may change in only the affected cell types in a particular mutant. These same genes would still be expressed normally in other tissue type and would likely appear to be unchanged by microarray analysis using whole embryos.
In this study, we describe an approach to globally analyze gene expression in specific cell types in the zebrafish embryo. We have taken advantage of the Tg(fli1:egfp) transgenic zebrafish line in which blood, endothelial, and pharyngeal arch cells express enhanced green fluorescent protein. We demonstrate that it is possible to isolate these cells types from Tg(fli1:egfp)y1 embryos using fluorescence activated cell sorting (FACS). We show that comparison of expression profiles from GFP+ and GFP− cells by microarray analysis allows identification of genes expressed in fli1+ cell types. Furthermore, by isolating and comparing expression profiles in GFP+ cells from embryos at two different developmental time points, we are able to globally observe dynamic changes in genes within these specific cell types. We also demonstrate the utility of this approach to identify new genes expressed in fli1+ cell types. We have established a database of these microarray data, including in situ results, that is easily navigated with a web-browser interface and allows access to the general scientific community.
Methods
Zebrafish
Zebrafish and their embryos were handled according to standard protocols. Tg(fli1:egfp)y1, Tg(hsp70:gal4)kca4, Tg(uas:notch1intra)kca3 and mibta52b lines have been described elsewhere (Lawson et al., 2001) and are available through the Zebrafish International Resource Center (ZIRC). Heat shock treatment of Tg(hsp70:gal4)kca4; Tg(uas:notch1intra)kca3 embryos was performed as described (Lawson et al., 2001). The y18 allele of phospholipase c gamma 1 is a splice mutation that truncates Plcg1 protein in the X catalytic domain (unpublished observations).
Embryo dissociation and fluorescence activated cell sorting (FACS)
Wild type Tg(fli1:egfp)y1 embryos were grown in egg water to indicated stage and were dechorionated by pronase treatment. Embryos were rinsed for 15 minutes in calcium free Ringer and passed several times through a 200μL pipet tip to remove their yolk. Embryos were transferred into a 35 mm culture dish with 2 mL phosphate buffered saline (PBS, pH 8) containing 0.25% trypsin and 1mM EDTA and incubated for 30 to 60 min at 28.5°C during which they were triturated with a 200μL pipet tip every 10 minutes. The digest was stopped by adding CaCl2 to a final concentration of 1 mM and fetal calf serum to 10%. Cells were centrifuged for 3 min. at 3000 rpm, rinsed once with PBS and resuspended at 107 cells/mL with Leibovitz medium L-15 without phenol red, 1% fetal calf serum, 0.8mM CaCl2, penicillin 50U/mL, streptomycin 0.05 mg/mL. FACS of single cell suspensions was performed at room temperature under sterile conditions using a FACSVantage SE /DIVA (Becton Dickinson) with a Coherent Innova 70 laser at 488nm and 200mW power. GFP+ and GFP− cells were separately collected in L15, 0.8mM CaCl2, 10% fetal calf serum, 10% zebrafish embryo extract, penicillin 50U/mL, streptomycin 0.05 mg/mL. Following sorting, cell viability was greater than 90%. We were routinely able to obtain 7x105 GFP+ from approximately 300 Tg(fli1:egfp)y1 embryos which was usually sufficient to obtain 1 μg of total RNA.
RNA isolation and quantitative RT-PCR
Equal numbers of GFP+ and GFP− cells were centrifuged, resuspended in 250μL of Trizol Reagent (Invitrogen) and stored at –80°C. Total RNA was isolated according to manufacturer’s instructions. RNA pellets were resuspended in 20μL nuclease-free water (Ambion). RNA integrity was confirmed by separation and visualization in ethidium stained formaldehyde/agarose gels according to standard protocols. For quantitative PCR, first strand cDNA was generated using 1μg total RNA, 200 U Superscript III reverse transcriptase (Invitrogen), and 2.5μM oligo dT primer in a 20μL reaction. Both cDNA concentrations were adjusted to 200ng/μL and QPCR was performed using iTaq SYBR Green Supermix with ROX (Biorad) using 500ng of cDNA and 0.6μM gene specific-primers in a 25μL reaction and detected in ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. PCR primers were as follows: gapdh, 5’-TGCTGTAACCGAACTCATTGTC and 5’-CAAGCTTACTGGTATGGCCTTC; ef1a, 5’-ATACATCAAGAAGATCGGCTACAAC and 5’-AATACCTCCAATTTTGTACACAT; bactin, 5’-TGGCCCCTAGCACAATGAAG and 5’-GCCTCCGATCCAGACAGAGTAT; fli1a, 5’-CCGAGGTCCTGCTCTCACAT and 5’-GGGACTGGTCAGCGTGAGAT; kdra 5’-ACTTTGAGTGGGAGTTTCATAAGGA and 5’-TTGGACCGGTGTGGTGCTA; flt4 5’-CTGTCGGATTTGGATTGGGA and 5’-GGTGGACTCATAGAAAACCCATTC; dlx2a, 5’-TCTGGGCCTCACGCAAAC and 5’-ACTTTGAACGACGATTCTGGAA; tbx6, 5’-GCCAAAGGCTTTCGTGACAAT and 5’-ACATTCCAGCTGCACACTCGTT; ptprn2, 5’- GGGCAGATCTTGTCCAATAATTGT and 5’-TGTCAATCAGGATGTAGGTTCCA; dlb, 5’-CGCTGTAGCCACAAACCATGT and 5’-CGAGAGCCGCTGAAACCA; acp5, 5’-ATGATGGCCAAAACTGCTTCTC and 5’-CGTCTTCAAAGGTTTCCTGGAA; bnip3l, 5’-CGCTGCTTCTGTCCCACAT and 5’-GGAGTTGTCAGTCTTTTCCCGATA; cbr1l, 5’-GACCAAGGAAGCTGCATGAA and 5’-CAAAAGGCTCTGTTGCTGCATT; gpx, 5’-GCGCTCCCTGCAACCA and 5’-TATTTCAGAGACTGCAGGATTTCTTC; krml2, 5’-CATGTCGGCGGAGCAGCACG and 5’-CTGCAGCCTCCTTCTTCACATC, pik3cd, 5’-TCCTGGAGATTCATGGTGCATA and 5’-GTGAACAACTTGAGGGTCAG. Expression levels were determined by comparison to a standard curve from total RNA isolated from whole embryo. Values from GFP- and GFP+ were then normalized to ef1a to obtain relative expression levels. Standard deviations were deduced from triplicate measurements.
Microarray analysis and Databases
Microarray analysis was performed by the Kimmel Cancer Center microarray facility at Thomas Jefferson University. The microarray slides were spotted with 16,399 65-mer oligonucleotides (Compugen/Sigma-Genosys) and hybridized with a biotinylated cRNA probes synthesized by linear amplification from 1 μg of total RNA. Normalized data received from the TJU core facility were log-transformed and GFP+ and GFP- values were separately averaged over the three experiments. Fold differences were calculated from log averages and a t-test was used to determine statistical significance. In order to compare datasets and generate tables, we constructed a relational database using FileMaker Pro 8. Datasets containing information on expression patterns, Gene name, and Unigene numbers were downloaded from ZFIN. Gene “Descriptions” were obtained for ZGC and other clones from ENSEMBL and cross-referenced by Unigene number. A Unigene dataset with corresponding clustered Genbank accession numbers was downloaded from NCBI. The data shown here are from Unigene build #90, 3/10/06. Records were related by Accession number since each sequence on the microarray corresponds to an Accession number. A similarly based web-accessible relational database containing these datasets was written in MySQL. Further details on the establishment of these databases are available upon request.
Whole mount in situ hybridization. Plasmids containing corresponding EST sequences were obtained from OpenBiosystems (Huntsville, AL). Inserts from plasmids were PCR amplified using M13F and M13R, purified using Qiaquick columns (Qiagen) and sequenced. Antisense digoxigenin-labeled riboprobes were synthesized from PCR products using the appropriate bacteriophage polymerase. Whole mount in situ hybridization was performed according to standard protocols (Hauptmann, 1999).
Results
Isolation of enriched cell populations from zebrafish embryos
Our primary interest is the development and differentiation of the endothelial cells that line arteries and veins. However, endothelial cells represent a small fraction of the embryo making global expression analysis of these cells difficult. In addition, enrichment of these cells would aid in the identification of low abundant endothelial genes that may be poorly represented in currently available cDNA libraries. Therefore, we investigated the possibility of utilizing transgenic zebrafish expressing Egfp in endothelial cells to obtain enriched cell populations for expression analysis. For this purpose, we utilized transgenic zebrafish in which the fli1 promoter drives expression of an enhanced green fluorescent protein transgene (referred to hereafter as Tg(fli1:egfp)y1). Tg(fli1:egfp)y1 embryos recapitulate endogenous fli1 expression and express Egfp in endothelial cells, as well as hematopoietic cells and pharyngenal arch tissue (Lawson and Weinstein, 2002). Following proteolytic dissociation of embryos, diagnostic fluorescence activated cell sorting (FACS) revealed that approximately 10 percent of cells in Tg(fli1:egfp)y1 embryos display strong fluorescence (Fig 1A), while 0.1 percent of cells from non-transgenic wild type embryos are fluorescent (Fig 1B). To confirm that this population contained cell types known to express the fli1:egfp transgene, we performed FACS to separate GFP+ and GFP− cell populations followed by quantitative RT-PCR to assay the expression levels of a number of cell type specific genes. Diagnostic FACS of the sorted cell populations shows that the GFP− and GFP+ cell display 99.8% and 95.7% purity, respectively (Fig 1C, 1D). Quantitative PCR analysis revealed that gapdh was expressed slightly higher in GFP− than in GFP+ cells, while the converse was true for bactin (Fig. 1E). We find that several genes known to be expressed in endothelial cells, including fli1 itself, the Vegf receptor-2 ortholog, kdra, and the Vegf receptor-3 ortholog flt4, all display much higher expression in GFP+ cells than in GFP− cells (Fig. 1E). Similarly, dlx2a, a known pharyngeal arch marker, and tbx6, which is expressed in lateral mesoderm and blood cells, show higher expression in GFP+ cells. By contrast, two genes that are not expressed in fli1+ cell types – ptprn2 and dlb – are expressed at higher levels in GFP− cells. These results indicate that it is possible to isolate known fli1:egfp cell types from Tg(fli1:egfp)y1 transgenic embryos.
Figure 1.
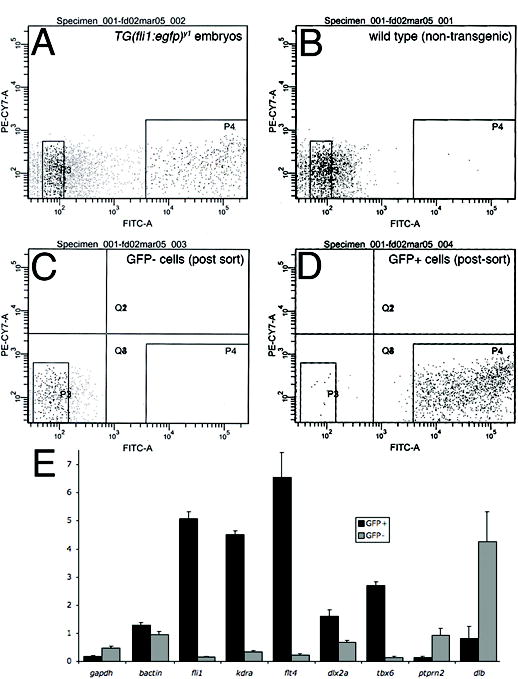
Enrichment of GFP+ cells from Tg(fli1:egfp)y1 zebrafish embryos. A-D. Diagnostic fluorescence activated cell sorting of dissociated Tg(fli1:egfp)y1 embryos. A. Cells from Tg(fli1:egfp)y1 embryos. B. Cells from non-transgenic wild type embryos. A, B. P3 and P4 demarcate cells sorted as GFP− and GFP+, respectively. C. GFP− cells following FACS isolation. D. GFP+ cells following FACS isolation. E. Relative expression of indicated genes in GFP+ (black bars) and GFP− (grey bars) determined by quantitative RT-PCR. Relative expression levels were determined by normalization to ef1a.
Microarray analysis of fli1:egfp cells
To globally analyze gene expression in fli1+ cells, we isolated RNA from GFP+ and GFP− cells sorted from Tg(fli1:egfp)y1 embryos and performed microarray analysis. We collected and compared the expression profiles of GFP+ and GFP− cells at two separate developmental time points: embryos at 22 to 24 hpf and at 28 to 30 hpf. We identified 315 sequences that were expressed at least 2.5 fold higher in GFP+ versus GFP− cells at 28–30 hpf (p<0.05; see Table 1 for selected genes and Supplementary Table 1 for complete list; normalized data for all arrays are in Supplementary Table 6), while 133 transcripts were similarly enriched at the earlier time point (22–24 hpf; Supplementary Table 2). To determine the efficacy of the FACS/microarray approach, we first cross-referenced the expression patterns available through the Zebrafish Information Network (ZFIN; zfin.org). At the 28–30 hpf time point, gene expression patterns were available for 85 genes out of the 315 GFP+-enriched sequences (Table 1 and Supplementary Table 1). Sixty-three of these genes are known to be expressed in cells that also express the fli1:egfp transgene including markers of pharyngeal arch tissue (e.g. dlx4b, end1, dlx2a), endothelial cells (e.g. plxnd1, tie1, dab2), red blood cells (e.g. gata1, tbx6, alas2), and white blood cells (e.g. lyz, mmp13, mpx, spi). The remaining genes have been classified with a non-specific or non-fli1+ pattern of expression by the ZFIN database. Similarly, thirty-five genes differentially expressed at 24 hpf have known expression patterns and twenty-six of these are expressed in fli1+ cell types (Supplementary Table 2).
Table 1.
Selected genes enriched in fli1:egfp-positive cells (28–30hpf) with known expression patterns
| Unigene | Accession No. | Cell type | Gene name | Fold diff | p-value |
|---|---|---|---|---|---|
| Dr.153 | U67845 | arc | dlx4b | 5.03 | 1.21E-03 |
| Dr.4076 | AF281858 | arc | edn1 | 4.94 | 6.69E-03 |
| Dr.2 | U03875 | arc | dlx2a | 4.22 | 1.66E-03 |
| Dr.19254 | AF219950 | arc | foxc1b | 3.39 | 1.15E-02 |
| Dr.12726 | AF164742 | endo | yrk | 5.07 | 2.17E-03 |
| Dr.12704 | BI878456 | endo | plxnd1 | 4.38 | 3.83E-03 |
| Dr.594 | AF053633 | endo | tie1 | 3.81 | 1.08E-02 |
| Dr.7977 | AW232474 | endo | gpx1 | 3.10 | 1.74E-02 |
| Dr.15055 | AY057095 | endo, arc | cxcr4a | 3.01 | 3.86E-02 |
| Dr.8126 | AJ249590 | endo, rbc, arc | fli1 | 7.10 | 8.49E-03 |
| Dr.9443 | AW077854 | endo | dab2 | 4.37 | 3.47E-05 |
| Dr.20582 | AW826487 | heart | rbpms2 | 2.55 | 9.28E-03 |
| Dr.8328 | AF228334 | heart, endo | hand2 | 3.53 | 5.91E-03 |
| Dr.24250 | BI984435 | rbc | uros | 5.64 | 4.05E-02 |
| Dr.8106 | BG884411 | rbc | cmyb | 3.69 | 2.12E-02 |
| Dr.355 | U18311 | rbc | gata1 | 3.66 | 2.06E-02 |
| Dr.7417 | AW281658 | rbc | sh3bp5 | 2.54 | 3.95E-02 |
| Dr.8064 | AF157109 | rbc,wbc | drl | 6.94 | 7.97E-03 |
| Dr.10314 | AW420822 | wbc | mmp13 | 17.40 | 3.51E-04 |
| Dr.6437 | AW232650 | wbc | ptpn6 | 14.47 | 6.50E-05 |
| Dr.7612 | AW077654 | wbc | spi1 | 14.06 | 1.78E-03 |
| Dr.9665 | BG307536 | wbc | rhog | 5.31 | 3.70E-02 |
| Dr.28437 | AW420718 | wbc | lgals9l1 | 3.80 | 8.25E-03 |
| Dr.10542 | AW133873 | wbc | hsd3b7 | 3.24 | 2.05E-02 |
Listed genes display greater than 2.5 fold enrichment in fli1:egfp versus egfp− cells (p<0.05). Gene expression patterns were obtained from ZFIN. arc – pharyngeal arches; endo – endothelial; rbc – red blood cell; wbc – white blood cell. For complete list of enriched genes, see Supplementary Table 1.
To further validate our microarray data, we determined if genes with expression patterns not assigned to fli1+ tissue by ZFIN were indeed enriched in fli1:egfp+ cells. We performed quantitative RT-PCR analysis on selected genes (Table 2) including BCL2/adenovirus E1B interacting protein 3-like (bnipl) and glutathione peroxidase 1 (gpx1) which are described as ubiquitously expressed, as well as several other genes expressed in non-fli1+ tissues: acid phosphatase 5, tartrate resistant (acp5, expressed in the yolk syncytial layer), carbonyl reductase 1-like (cbr1l, expressed in eye and optic tectum), kreisler maf-related leucine zipper homolog 2 (krml2, somites and motor neurons), and phosphoinositide-3-kinase, catalytic, delta polypeptide (pik3cd, cranial ganglia). For all of these genes, quantitative RT-PCR analysis revealed significant enrichment in fli1:egfp+ cells at the 22–24 hpf time point (Figure 2).
Table 2.
Selected genes enriched in fli1:egfp+ cells without known fli1+ expression patterns
| 28–30 hpf |
22–24 hpf |
||||||
|---|---|---|---|---|---|---|---|
| Unigene | Accession No. | Gene name | expression pattern* | GFP+/GFP− | p-value | GFP+/GFP− | p-value |
| Dr.1508 | AI882824 | acp5 | YSL | 5.44 | 0.0056 | 3.77 | 0.1189 |
| Dr.15862 | AW420488 | bnip31 | ubiquitous | 3.98 | 0.008 | 4.24 | 0.1370 |
| Dr.10648 | AF298898 | cbr1L | eye, optic tectum | 3.65 | 0.0095 | 4.36 | 0.4050 |
| Dr.7977 | AW23247 | gpx1 | ubiquitous | 3.1 | 0.0175 | 2.39 | 0.0844 |
| Dr.8198 | AF109780 | krml2 | somites, motor neurons | 3.49 | 0.0165 | 3.66 | 0.0398 |
| Dr.26647 | AW116052 | pik3cd | cranial ganglia | 9.88 | 0.0001 | 2.97 | 0.048 |
Expression pattern information derived directly from ZFIN except krml2 which was reported by Schvarzstein et al. (1999) Mech Dev 80:223. Fold increase and p-value are from microarray analysis. See Figure 2 for quantitative RT-PCR analysis of these genes in fli1:egfp-positive versus negative cells.
Figure 2.
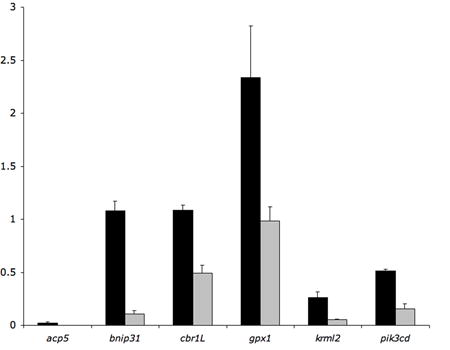
Quantitative RT-PCR analysis of previously described non-fli1+ and ubiquitously-expressed genes (see Table 2). qRT-PCR was performed on RNA derived from fli1:egfp+ (black bars) and egfp− (grey bars) cells from 22–24 hpf embryos.
We next evaluated the expression profiles in GFP+ cells at the two different time points. By directly comparing the levels of expression in GFP+ cells at 22–24 hpf and 28–30 hpf, we identified 218 genes that were up or downregulated at least 2.5 fold (p<0.05; Supplementary Table 3). Sixty-seven of these genes have known expression patterns and 35 are expressed in fli1+ cell types. Interestingly, this analysis revealed an additional 21 genes that were expressed in fli1+ cell types, but were not significantly enriched in GFP+ cells. For the most part, these genes (e.g. meis4.1a, id3) are not restricted only to fli1+ cells and are expressed in multiple tissue types. To determine if markers of different fli1+ cell types may display distinctive temporal expression patterns, we searched our dataset for known genes expressed in pharyngeal arch, white and red blood cells, or blood vessels (based on ZFIN expression patterns) and then determined if they were up or down regulated (>2.5 fold, p<0.08; Supplementary Table 4). This analysis revealed that most red blood cell markers were down regulated in GFP+ cells at 26–30 hpf. We identified 17 erythroid markers with statistically significant regulation and 12 of these were downregulated 2.5-fold or more (Table 3). Only two were upregulated and 3 were unchanged. By contrast, 3 of the 5 white blood cell markers were upregulated, while 1 one was reduced (Table 3). We identified 61 arch markers that displayed statistically significant regulation. Eleven of these were down-regulated from early to late timepoints, while fourteen were upregulated and the remaining genes were relatively unchanged (Supplementary Table 4). For the most part, the levels of blood vessel markers remained the same; 3 out of 16 genes were upregulated, while the remaining genes were unchanged (Supplementary Table 4).
Table 3.
Temporal changes in blood cell gene expression
| Unigene | Accession No. | cell type | Gene name | Fold diff | ttest |
|---|---|---|---|---|---|
| Dr.1458 | AF045432 | rbc, endo | tal1 | −8.83 | 2.19E-02 |
| Dr.208 | AI588172 | rbc | slc11a2 | −8.80 | 6.52E-02 |
| Dr.5053 | AF262978 | rbc | klf4 | −5.75 | 4.33E-02 |
| Dr.8065 | AF191560 | rbc, endo | lmo2 | −5.20 | 1.35E-02 |
| Dr.47214 | AJ011790 | rbc | cldng | −4.39 | 5.72E-03 |
| Dr.4736 | BF158097 | wbc | mpp1 | −4.23 | 1.45E-02 |
| Dr.355 | U18311 | rbc | gata1 | −4.13 | 2.53E-02 |
| Dr.10516 | AI959558 | rbc, other | fam46c | −3.81 | 7.33E-02 |
| Dr.33616 | AI974203 | rbc | sb:cb535 | −3.80 | 6.97E-02 |
| Dr.311 | BG985785 | rbc, vessel | glrx5 | −3.03 | 5.09E-02 |
| Dr.4468 | BM035000 | rbc, arch | zgc:56602 | −2.97 | 4.96E-02 |
| Dr.28283 | BI886864 | rbc | hbbe1 | −2.89 | 2.25E-02 |
| Dr.4989 | BM181762 | rbc, arch | c10orf119 | −2.54 | 7.51E-02 |
| Dr.10516 | BM183817 | rbc, other | fam46c | −2.08 | 5.63E-02 |
| Dr.16822 | BI983488 | rbc | si:ch211–234f20.6 | −1.80 | 5.87E-02 |
| Dr.3804 | AW174333 | wbc | zgc:55891 | 1.62 | 1.26E-02 |
| Dr.43005 | BG985478 | rbc, vessel | ets1 | 1.86 | 4.03E-02 |
| Dr.12986 | BE605310 | rbc | fos | 3.09 | 1.78E-02 |
| Dr.10542 | AW133873 | wbc, arch | hsd3b7 | 3.71 | 1.52E-02 |
| Dr.10314 | AW305943 | wbc | mmp13 | 7.62 | 4.29E-02 |
| Dr.9478 | AF349034 | wbc | mpx | 11.68 | 6.92E-03 |
| Dr.4882 | AF376130 | rbc | cpa | 11.73 | 1.58E-03 |
rbc – expressed in red blood cells; endo – expressed in blood vessels, wbc – expressed in white blood cells. Fold difference is comparison of expression level at 28–30 hpf to levels at 22–24 hpf.
In situ hybridization analysis of putative fli1:egfp+ genes
Our comparison of fli1:egfp microarray data with available ZFIN expression patterns demonstrates that the majority of genes enriched in GFP+ cells are expressed in fli1+ tissue and most are specific to these cell types. However, expression patterns are available for only about one-quarter of fli1:egfp+-enriched sequences. Therefore, we surveyed the expression of 40 additional genes by whole mount in situ hybridization to wild type embryos between 24 hpf and 30 hpf (Supplemental Table 5). Based on our in situ hybridization analysis, we find that 34 out of 40 of these genes display expression in fli1:egfp-positive cell types (Supplemental Table 5). Four of the genes (Dr.10376, Dr. 16151, Dr.10137, Dr.12289) that we analyzed have also been shown to be expressed in blood or endothelial cells in a recent microarray analysis in zebrafish (Weber et al., 2005). Nine of the genes displayed expression in blood cells within the intermediate cell mass. For example, at 26 hpf, Dr.1382 displays specific expression in cells located between the dorsal aorta and posterior cardinal vein within the trunk (Fig. 3A, B), but is not expressed in other tissues at this time point. This expression pattern is the same as known erythroid markers such as gata1 and the morphology and location of these cells are consistent with their identity as red blood cells or their progenitors (Detrich et al., 1995). Dr.1382 displays homology to a hypothetical human transmembrane protein that was identified in an EST analysis of CD34+ stem cells (Zhang et al., 2000). Some presumptive red blood cell markers display expression in other cell types as well. For example, Dr.30834, an ortholog of human chloride intracellular channel 2 (CLIC2), shows expression both in blood cells (Fig. 3C, D) and the heart (Fig. 3E). Dr.919 is expressed in red blood cells (Fig. 3F,G) as well as in numerous cells scattered throughout the surface of the yolk sac (Fig. 3H). This pattern is consistent with the expression of numerous macrophage cell markers (Herbomel et al., 1999). Dr.919 also displays low level expression in the lens (Fig. 3F). Dr.919 is homologous to human SAMSN-1, a SAM domain, SH3 domain and nuclear localization domain containing protein that is expressed in human hematopoietic cells (Claudio et al., 2001). Dr.6172 expression is restricted to a small sub-population of blood cells (Fig. 3I-K). We also observe Dr.6172 expression in the floor plate and hypochord (Fig. 3I, J), two tissues which do not express the fli1:egfp transgene.
Figure 3.
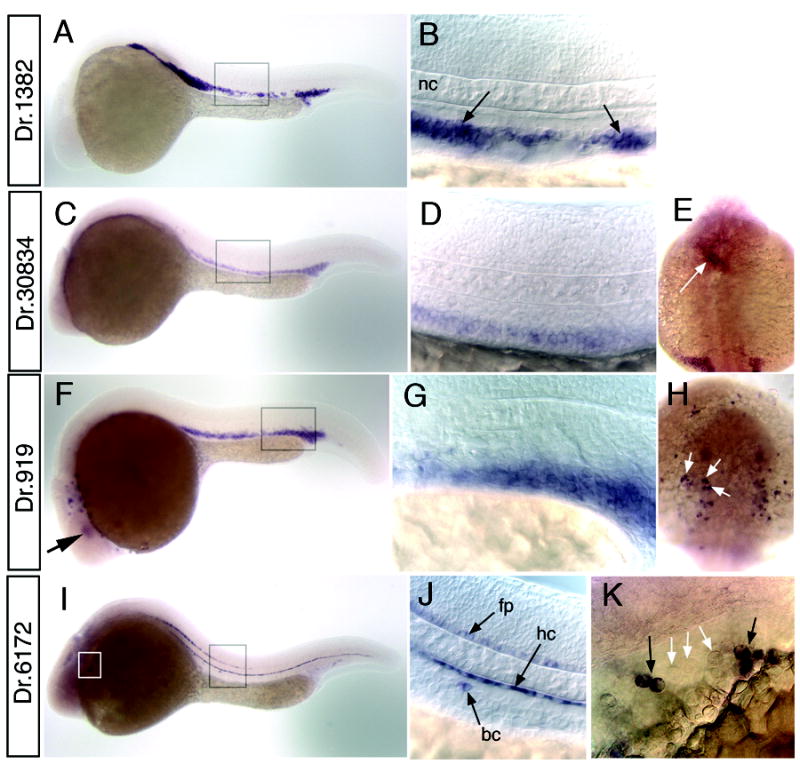
Whole mount in situ hybridization analysis of blood cell markers. A, B. Dr.1382 at 26 hpf; box indicate magnified view in B. B. Arrows indicate expression in erythroid cells; nc – notochord. C-E. Dr.30834 at 26 hpf; box indicates magnified view in D. E. Dorsal view of head, anterior is up; white arrow indicates expression in the heart. F-H. Dr.919 at 24 hpf. F. Box indicates magnified view in G. H. Dorsal view of the head, anterior is up. White arrows denote expression in white blood cells on yolk sac. I-K. Dr.6172 at 24 hpf. I. Black box indicates magnified view in J, white box indicates view in K. J. expression in floor plate (fp), hypochord (hc), and a blood cell (bc). K. Blood cells expressing Dr.6172 indicated with black arrows. White arrows denote adjacent cells with similar morphology that do not express Dr.6172. A-D, F, G, I, J. Lateral view, dorsal is up anterior to the left.
Our in situ analysis also revealed a number of cardiovascular specific genes. Dr.24287, a complement component C1q ortholog, is expressed in all blood vessels at 26 hpf (Fig. 4A, B). Recent microarray analysis of the cloche mutant has identified a similar C1qR ortholog (Sumanas et al., 2005) referred to as crl (Unigene: Dr.23967); however, Dr.24287 appears to be a separate related gene (data not shown). Dr.9655 expression is apparent in blood vessels as well (Fig. 4C), although we observe lower levels of expression in the dorsal aorta when compared to the posterior cardinal vein at 24 hpf (Fig. 4D) and this difference is more pronounced by 30 hpf (Fig. E). Dr.9655 was recently described as vessel specific 1 (vsg1) in a microarray analysis of vascular genes regulated in the zebrafish cloche mutant (Qian et al., 2005) and displays homology to plasmalemma vesical associated protein (PLVAP) which is found in fenestrated endothelial cells (Stan et al., 2004). We find that Dr.33656 is expressed specifically in arteries, but not veins (Fig. 4F, G). In addition to genes expressed in blood vessels, we have also identified several genes expressed in the heart such as Dr.398 (Fig. 4H) a titin ortholog that also displays expression in somite tissue (Fig. 4I). Several genes displayed expression in pharyngeal arch tissue, including Dr.51180 which is also expressed in blood vessels (Fig. 5A, B) and BI671621 (no Unigene cluster) which appears to be restricted to the arches at 26 hpf (Fig. 5C, D).
Figure 4.
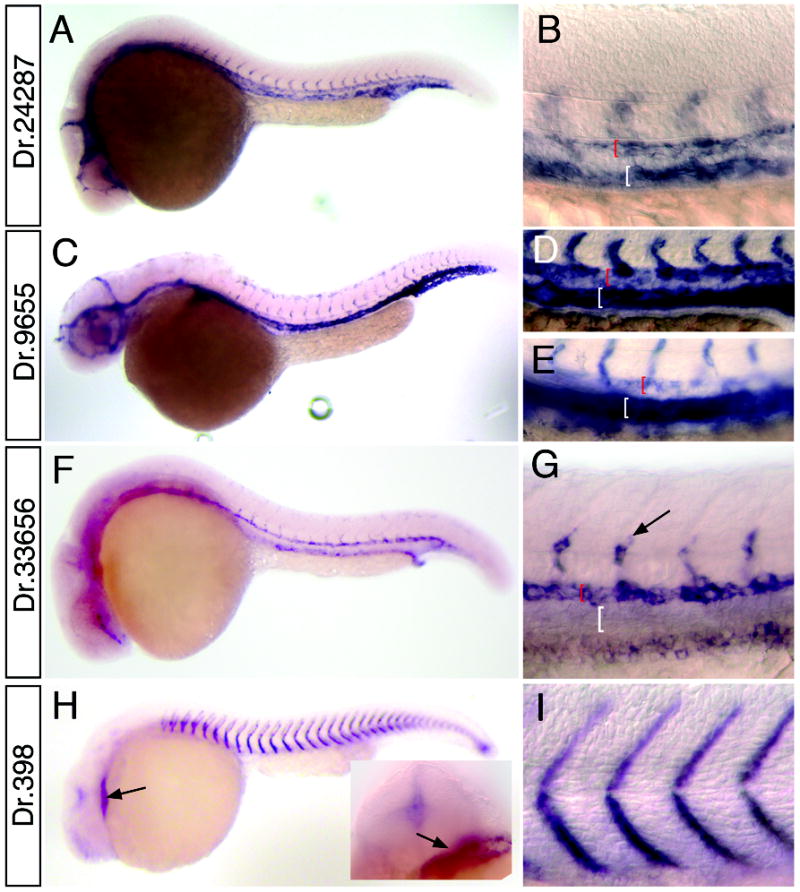
Whole mount in situ hybridization analysis of cardiovascular markers. A, B. Dr.24287 at 24 hpf; box indicates magnified view in B. B. Black bracket indicates dorsal aorta, white bracket shows posterior cardinal vein. C-E. Dr. 9655. C. Embryo at 30 hpf. D. Magnified view of dorsal aorta (red bracket) and posterior cardinal vein (white bracket) at 24 hpf; E. Dorsal aorta (red bracket) and posterior cardinal vein (white bracket) at 30 hpf. F, G. Dr.33656. G. expression in segmental arteries (black arrow) and dorsal aorta (red bracket); expression is absent from posterior cardinal vein (white bracket). H, I. Dr.398 at 24 hpf. H. Arrow indicated expression in heart; inset head-on view showing heart expression (arrow). I. Magnified view of embryo in H.
Figure 5.
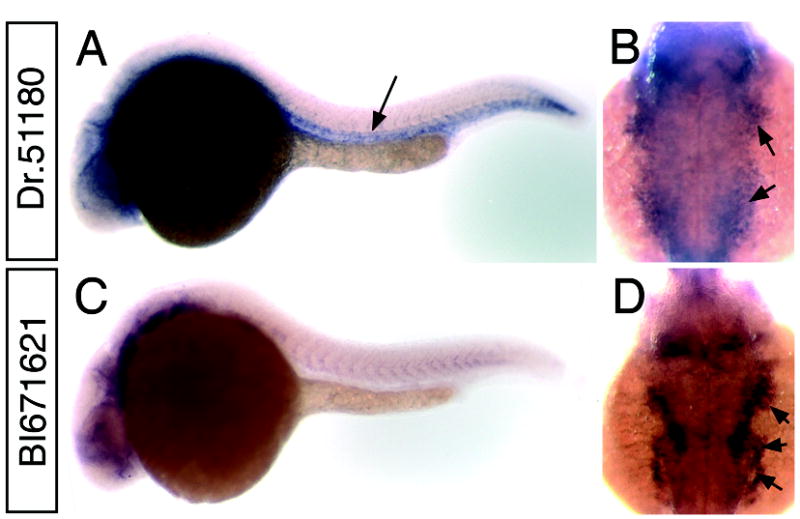
Whole mount in situ hybridization analysis of pharyngeal arch markers. A, B. Dr.51180 at 26 hpf; arrow indicates expression in trunk blood vessels B. Dorsal view of head of embryo in A; arrows indicate expression in pharyngeal arches. C, D. BI671621. D. Dorsal view of head of embryo in C; arrows indicate expression in pharyngeal arches.
Regulation of artery and vein specific genes
We have previously shown that the restricted expression of artery and vein specific genes is controlled through the activity of both the vascular endothelial growth factor and Notch signaling pathways (Lawson et al., 2001; Lawson et al., 2002). For example, artery specific genes such as ephrinb2a and notch5 fail to be expressed in zebrafish mutants lacking Vegf or Notch signaling components. In order to determine if artery and vein restricted genes identified by our microarray analysis responded to these pathways, we analyzed their expression in mutant or transgenic zebrafish embryos in which Vegf or Notch signaling was affected. As shown above, Dr.33656 is expressed specifically in the dorsal aorta and pronephric ducts in wild type sibling embryos (Fig. 6A). Similar to other artery specific markers, we find that Dr.33656 is absent in the dorsal aorta of embryos mutant for phospholipase c gamma 1 (plcg1), an important component of the Vegf signaling pathway (Fig 5B; Lawson et al., 2003). Dr.33656 expression in the pronephric duct is unchanged in plcg1y18 mutant embryos indicating a specific defect in the expression of this gene only in the developing dorsal aorta (Fig. 6B). Dr.33656 expression was similarly lost in the dorsal aorta of embryos lacking Notch activity. In embryos mutant for mindbomb (mibta52b (Itoh et al., 2003) or injected with a Morpholino designed to block the translation of Suppressor of hairless (Rbpsuh; Sieger et al., 2003), we observed loss of Dr.33656 expression in the dorsal aorta (Fig. 6C, D). As noted above for plcg1y18 mutant embryos, in both cases of loss of Notch signaling, Dr.33656 expression is maintained in the pronephric duct (Fig. 6C, D).
Figure 6.
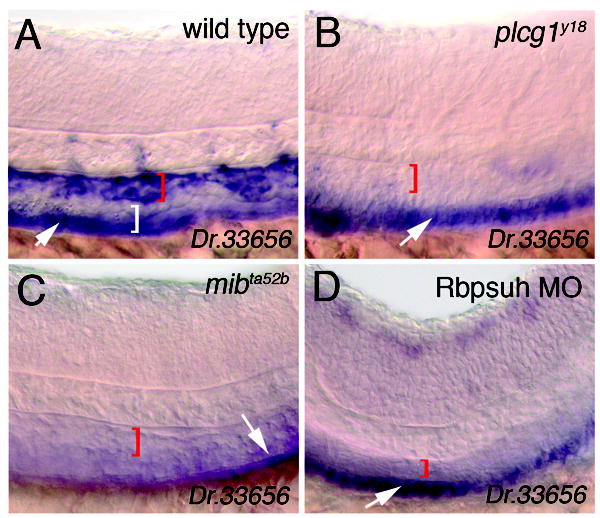
plcg1 and mib are required for artery specific expression of Dr.33656. A. Wild type embryo. B. Embryo mutant for plcg1y18. C. Embryo mutant for mibta52b. D. Embryo injected with 5 ng Rbpsuh morpholino. Lateral views, anterior to the left, dorsal is up. A-D. White arrow indicates expression in pronephric ducts (out of focal plane). Red bracket demarcates dorsal aorta in A.-D. and white bracket indicates posterior cardinal vein in A.
We have previously shown that an important function of the Notch signaling pathway during arterial endothelial cell differentiation is to repress the expression of vein-specific markers in the developing artery (Lawson et al., 2001). Therefore, we determined if vein-specific markers identified in our microarray analysis are similarly regulated by the Notch signaling pathway. In order to activate Notch signaling, we relied on two transgenic lines: one in which the yeast transactivator GAL4 is driven by a heat inducible promoter (Tg(hsp70:gal4)kca4) and one in which the GAL4 responsive upstream activating sequence (uas) drives expression of a myc-tagged Notch1a intracellular domain (Tg(uas:mycnotch1intra)kca3). We find that the disabled2 (dab2) gene is enriched in fli1:egfp+ cells (Supplementary Table 1) and its expression is reduced in the dorsal aorta compared to the posterior cardinal vein in wild type sibling embryos (Fig. 7A) similar to vsg1 (Fig. 7F). In this case, the embryos pictured are siblings that bear either the hsp70:gal4 or uas:mycnotchintra transgenes alone and therefore do not express Myc-Notch1intra following heat shock. By contrast, embryos bearing both transgenes show reduced expression of dab2 (Fig. 7B) and vsg1 (Fig. 7G) after heat shock. Despite the potent downregulation of venous dab2 expression, its expression in pronephric ducts is not affected by activation of the Notch pathway (Fig. 7C, D). Consistent with the downregulation of these vein markers by activation of Notch signaling, we find that both dab2 (Fig. 7E) and vsg1 (Fig. 7H) become ectopically expressed in the dorsal aorta of embryos mutant for mibta52b. Taken together, these observations indicate that the artery and vein markers identified by our microarray analysis respond to Vegf and Notch signaling. Further experiments will be required to determine whether these represent direct and/or functional targets of these pathways during artery and vein development.
Figure 7.
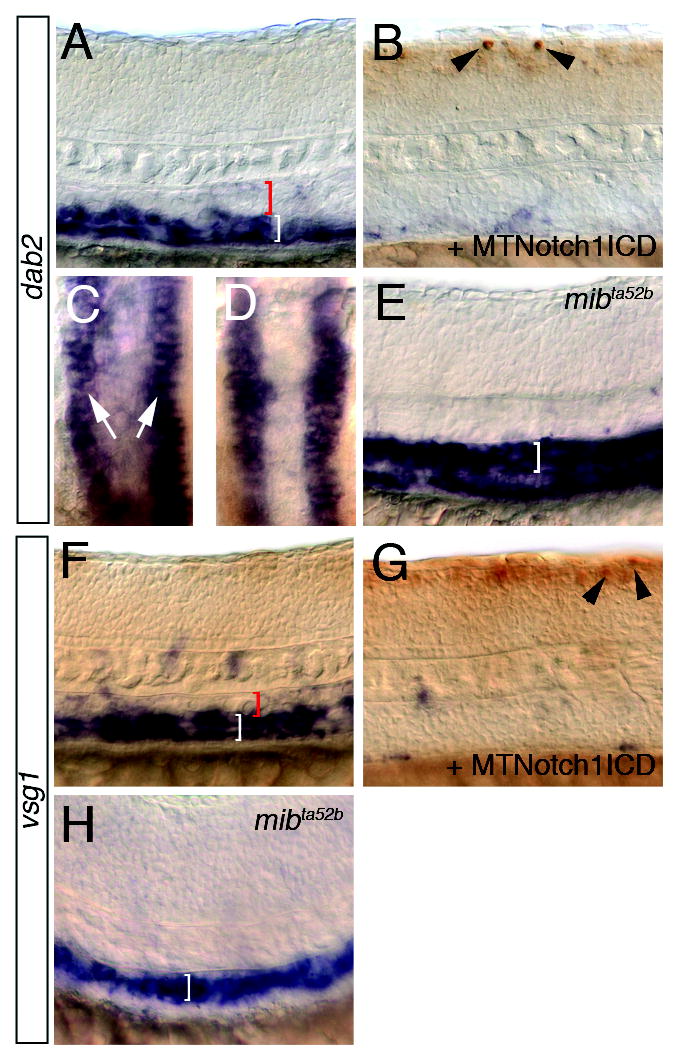
Notch represses dab2 and vsg1 expression. A-E. dab2 expression. A. Wild type sibling embryo following heat shock. Red bracket indicates dorsal aorta, white bracket posterior cardinal vein. B. Tg(hsp70:gal4)kca4;Tg(uas:myc-notch-intra)kca3 embryo following heat shock. Myc-positive cells in the neural tube are indicated by black arrowheads. C, D. Dorsal view of dab2 expression in pronephric ducts (white arrows). C. Same embryo as shown in A; D. same embryos as seen in B. E. Ectopic dab2 expression in the dorsal aorta (white bracket) of a mibta52b mutant embryo. F-H. vsg1 expression. F. Wild type sibling embryo following heat shock. Red bracket indicates dorsal aorta, white bracket posterior cardinal vein. G. Tg(hsp70:gal4)kca4;Tg(uas:myc-notch-intra)kca3 embryo following heat shock. Myc-positive cells in the neural tube are indicated by black arrowheads. H. Ectopic vsg1 expression in the dorsal aorta (white bracket) of a mibta52b mutant embryo.
A Zebrafish Microarray Database
In order to provide access to the zebrafish and other research communities, we have designed and developed a relational database based on MySQL as a repository and query tool for our microarray data. The database stores raw and processed data and is web-based. It can be accessed at https://titlis.umassmed.edu/danio using an internet browser (username: “guest”, password: “password”; once you have logged on, click on “Analysis” in the upper right hand corner). The database can be searched through binary comparisons between available datasets with user-definable values for fold change and P-value; the default is set at fold change of 2.5 and a p-value < 0.05. The resulting output provides a list of genes that displays Accession number, Unigene number, and ZFIN gene symbol. For Accession number and Unigene, clicking on the link will open the corresponding NCBI page in a new window; similarly, the Symbol link will open the corresponding gene page at ZFIN. When applicable we have also included links to images of our whole mount in situ hybridization data. The display also includes normalized average values across replicate experiments (as calculated in Materials and Methods) for the two samples along with fold difference and P-value. Finally, there is a link to view all experimental values for a particular gene. These values are displayed numerically as well as in a histogram (Fig. 8C). It is also possible to search by individual gene name, accession number or Unigene number.
Figure 8.
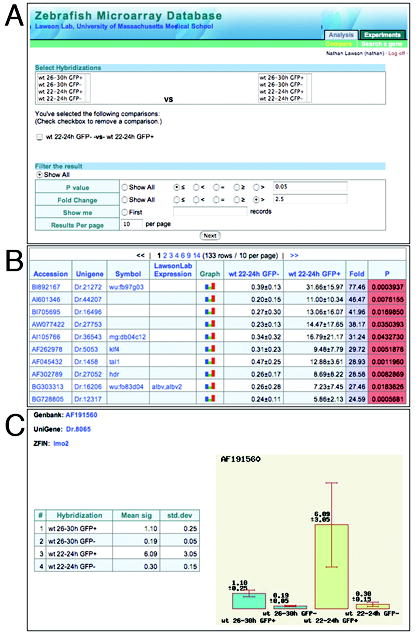
A Zebrafish microarray database. A. Opening page of database with possible pairwise combinations and user defined analysis. B. Results output page showing Accession number, gene name, Unigene, and values from selected microarray experiment. Also includes links to in situ data. C. Example of and individual gene page with values across all available microarray data.
Discussion
In this study we have utilized microarrays to globally analyze gene expression in specific cell types isolated from transgenic zebrafish embryos. We find that it is possible to directly compare the global expression profiles from two experimentally separate populations of FACS isolated cells. In addition, we demonstrate that this approach is ideal for the identification of new cell type specific genes.
The comparison of GFP+ and GFP- cells in our analysis proved to be reliable in identifying cell type specific genes. Nearly 70% of the genes with significantly higher expression in fli1:egfp+ cells are expressed specifically in blood cells, blood vessels or pharyngeal arch tissue. Accordingly, the vast majority of previously uncharacterized genes that we analyzed by whole mount in situ hybridization also display expression in these cell types. These results indicate that this approach can be utilized as an important initial step in performing whole mount in situ screens directed at particular cell types. The growing availability of transgenic zebrafish lines expressing fluorescent proteins driven by different tissue specific promoters will make it possible to perform similar analyses for nearly any cell type in the developing zebrafish embryo. A comprehensive microarray analysis of multiple transgenic lines, especially those labeling tissue types that may be poorly represented in the current ZFIN expression database, would provide an ideal parallel to ongoing in situ sceens.
Despite the robust detection of cell type specific genes in our microarray analysis, a number of genes that are expressed in fli1+ cell types failed to be enriched. In many of these cases, these genes may be expressed abundantly in other tissues. This is certainly the case for many artery-specific genes such as notch3 which is expressed at high levels throughout the developing nervous system (Westin and Lardelli, 1997) and ephrinb2a, which is expressed in developing somites, the forebrain and optic tectum (Durbin et al., 1998). However, several known endothelial cell specific genes failed to be enriched as well, including kdra and flt4, neither of which is expressed in other cell types (Liao et al., 1997; Thompson et al., 1998). One possibility is that fli1:egfp+ endothelial cells begin to phenotypically change soon after dissociation of the embryos. This could lead to corresponding changes in gene expression and the possible loss of cell specific genes. In addition, these cell types may begin to undergo apoptosis due to loss of growth factors and/or cell-cell contact. Although we cannot rule out these possibilities, endothelial cell types appear to be well represented. Analysis of known and newly characterized genes shows relatively equal proportions of enriched genes from each cell type expressing the fli1:egfp transgene. In addition, our qPCR analysis indicates that both kdra and flt4 are expressed at high levels in the GFP+ sorted cell populations. These results suggest that the failure to detect some genes may be due to poor oligo hybridization on the microarray itself.
We identified a number of genes that are widely expressed in multiple cell types or ubiquitously expressed throughout the embryo, yet still are significantly enriched in fli1:egfp+ cell types according to our microarray analysis. In most cases we believe that this may represent actual differences in expression levels. Indeed, our qRT-PCR analysis demonstrates that this is the case for some ubiquitous genes as well as those not previously known to be expressed in fli1+ tissue. The confirmation that these genes are enriched in fli1:egfp+ cells suggests a much higher true positive rate from our microarray analysis than indicated by fli1+ genes alone. The observed differences in the expression of ubiquitous genes could be due, in part, to the fact that we are comparing the level of a particular gene in a select number of cell types to all of the remaining cell types in the embryo. Accordingly, ubiquitously expressed genes, such as bactin, can be expressed at quite variable levels across different cell types (Barrallo et al., 1999). Our results suggest that the combined use of cell sorting and microarray analysis may, in some cases, be more sensitive than in situ hybridization to identify genes that are expressed in particular types.
In addition to the identification of new tissue specific genes, we demonstrate that it is possible to directly and reliably compare the global expression profiles from two separate GFP+ cell populations. In this case, we have compared GFP+ cells isolated from Tg(fli1:egfp)y1 embryos at different time points. The direct comparison of these two datasets allowed us to make several important observations. First, we find distinct expression patterns for each of the cell types that express the fli1:egfp transgene. For example, white blood cell markers are upregulated at later stages, while red blood cell markers are largely down-regulated. A benefit of this observation is that it provides preliminary classification of candidate ESTs for which there is no known expression pattern. This is especially important when using transgenic lines, such as Tg(fli1:egfp)y1, in which the transgene is expressed in multiple cell types (Lawson and Weinstein, 2002). An important caveat here is that the dynamic changes can be due to changes either in the transcript itself, or changes in the level of transgene expression in a particular cell type. This latter scenario is likely the case for red blood cell markers as we have previously shown that fli1:egfp is initially expressed in erythroid cells at 24 hpf, but is subsequently down-regulated in these cells (Lawson and Weinstein, 2002). In contrast to red blood cells, fli1:egfp expression is relatively constant in pharyngeal arch cell types between these two time points and we find that many markers expressed in these cells display either up- or down-regulation. A second observation from our temporal analysis was the identification of genes that are expressed in fli1+ cells but were not enriched when compared to GFP− cells. For the most part, these genes are expressed in numerous cell types throughout the embryo and therefore did not exhibit any enrichment between GFP+ and GFP− cells. Taken together, our results suggest that microarray analysis of FACS isolated cells will be valuable for the analysis of mutant, transgenic or Morpholino-injected zebrafish embryos. This approach will be particularly useful to focus on changes in gene expression in mutants that only affect a specific cell type during development.
With the ease of obtaining numerous zebrafish embryos at multiple time points in development, the use of microarrays can be a powerful tool to globally analyze changes in gene expression in wild type and mutant zebrafish. Furthermore, the availability of multiple mutants in related genetic pathways will allow the use of microarray analysis to identify networks of commonly regulated genes. Given the expense of performing multiple replicates to analyze numerous mutations, it will be important to make all microarray data easily accessible. The database we have established provides straightforward pairwise analysis of our microarray data and also allows searches based on gene name. We hope that this will be a valuable resource for zebrafish researchers, as well as to researchers who are interested in blood and vascular development and may wish to obtain some preliminary data on the expression of their gene of interest in zebrafish tissue.
Supplementary Material
Acknowledgments
This work was supported by 5R03HD047490 from National Institute of Child Health and Human Development awarded to N. D. L. and 5 P30 DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases. We would like to thank Charles Sagerstrom and members of the Lawson Lab for critical reading of this manuscript. We also thank Jennifer Ma at the Thomas Jefferson University microarray facility and Barbara Gosselin at the UMass Medical School FACS facility for their contributions to this work.
References
- Amsterdam A. Insertional mutagenesis in zebrafish. Dev Dyn. 2003;228:523–34. doi: 10.1002/dvdy.10381. [DOI] [PubMed] [Google Scholar]
- Barrallo A, Gonzalez-Sarmiento R, Garcia-Isidoro M, et al. Differential brain expression of a new beta-actin gene from zebrafish (Danio rerio) Eur J Neurosci. 1999;11:369–72. doi: 10.1046/j.1460-9568.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- Claudio JO, Zhu YX, Benn SJ, et al. HACS1 encodes a novel SH3-SAM adaptor protein differentially expressed in normal and malignant hematopoietic cells. Oncogene. 2001;20:5373–7. doi: 10.1038/sj.onc.1204698. [DOI] [PubMed] [Google Scholar]
- Crosier PS, Bardsley A, Horsfield JA, et al. In situ hybridization screen in zebrafish for the selection of genes encoding secreted proteins. Dev Dyn. 2001;222:637–44. doi: 10.1002/dvdy.1218. [DOI] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Kieran MW, Chan FY, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–7. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Durbin L, Brennan C, Shiomi K, et al. Eph signaling is required for segmentation and differentiation of the somites. Genes Dev. 1998;12:3096–109. doi: 10.1101/gad.12.19.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furthauer M, Lin W, Ang SL, et al. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4:170–4. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- Gates MA, Kim L, Egan ES, et al. A genetic linkage map for zebrafish: comparative analysis and localization of genes and expressed sequences. Genome Res. 1999;9:334–47. [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–9. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hauptmann G. Two-color detection of mRNA transcript localizations in fish and fly embryos using alkaline phosphatase and beta-galactosidase conjugated antibodies. Dev Genes Evol. 1999;209:317–21. doi: 10.1007/s004270050258. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–45. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci U S A. 1999;96:9745–50. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Kelly PD, Chu F, Woods IG, et al. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 2000;10:558–67. doi: 10.1101/gr.10.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapik EW, Goodman A, Ekker M, et al. A microsatellite genetic linkage map for zebrafish (Danio rerio) Nat Genet. 1998;18:338–43. doi: 10.1038/ng0498-338. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–87. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Mugford JW, Diamond BA, et al. phospholipase C gamma-1 is required downstream of vascular endothelial growth factor during arterial development. Genes Dev. 2003;17:1346–51. doi: 10.1101/gad.1072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–83. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–36. doi: 10.1016/s1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Liao W, Bisgrove BW, Sawyer H, et al. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124:381–9. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2:956–66. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Johnson SL, Midson CN, et al. A genetic linkage map for the zebrafish. Science. 1994;264:699–703. doi: 10.1126/science.8171321. [DOI] [PubMed] [Google Scholar]
- Qian F, Zhen F, Ong C, et al. Microarray analysis of zebrafish cloche mutant using amplified cDNA and identification of potential downstream target genes. Dev Dyn. 2005;233:1163–72. doi: 10.1002/dvdy.20444. [DOI] [PubMed] [Google Scholar]
- Sieger D, Tautz D, Gajewski M. The role of Suppressor of Hairless in Notch mediated signalling during zebrafish somitogenesis. Mech Dev. 2003;120:1083–94. doi: 10.1016/s0925-4773(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Stan RV, Tkachenko E, Niesman IR. PV1 is a key structural component for the formation of the stomatal and fenestral diaphragms. Mol Biol Cell. 2004;15:3615–30. doi: 10.1091/mbc.E03-08-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–41. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, et al. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–19. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–69. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Tsang M, Friesel R, Kudoh T, et al. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4:165–9. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- Tsang M, Maegawa S, Kiang A, et al. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131:2769–79. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- Weber GJ, Choe SE, Dooley KA, et al. Mutant-specific gene programs in the zebrafish. Blood. 2005;106:521–30. doi: 10.1182/blood-2004-11-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin J, Lardelli M. Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Development, Genes, and Evolution. 1997;207:51–63. doi: 10.1007/s004270050091. [DOI] [PubMed] [Google Scholar]
- Yoda H, Momoi A, Esguerra CV, et al. An expression pattern screen for genes involved in the induction of the posterior nervous system of zebrafish. Differentiation. 2003;71:152–62. doi: 10.1046/j.1432-0436.2003.710206.x. [DOI] [PubMed] [Google Scholar]
- Zhang QH, Ye M, Wu XY, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–60. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


