Abstract
The assembly of simple transmembrane helix homo-oligomers is studied by combining a generalized Born implicit membrane model with replica exchange molecular dynamics simulations to sample the conformational space of various oligomerization states and the native oligomeric conformation. Our approach is applied to predict the structures of transmembrane helices of three proteins—glycophorin A, the M2 proton channel, and phospholamban—using only peptide sequence and the native oligomerization state information. In every case, the methodology reproduces native conformations that are in good agreement with available experimental structural data. Thus, our method should be useful in the prediction of native structures of transmembrane domains of other peptides. When we ignore the experimental constraint on the native oligomerization state and attempt de novo prediction of the structure and oligomerization state based only on sequence and simple energetic considerations, we identify the pentamer as the most stable oligomer for phospholamban. However, for the glycophorin A and the M2 proton channels, we tend to predict higher oligomers as more stable. Our studies demonstrate that reliable predictions of the structure of transmembrane helical oligomers can be achieved when the observed oligomerization state is imposed as a constraint, but that further efforts are needed for the de novo prediction of both structure and oligomeric state.
INTRODUCTION
Integral membrane proteins account for 30% of all proteins in the cell and play key roles in communication between the cell and its environment (1). Biological activity is clearly linked to protein folding, with misfolding leading to malfunction and disease for both membrane and non-membrane-associated proteins. However, in contrast to the wealth of available information regarding the folding of water-soluble proteins, relatively little is known about how membrane proteins fold to their native states. One idea, the two-stage model of integral membrane protein folding proposed by Popot and Engelman more than a decade ago (2,3), suggests a mechanism for helix-bundle membrane protein folding: the insertion of helix into the membrane (stage 1) and the assembly of the inserted helices in the membrane (stage 2). In earlier studies of insertion and folding, we explored aspects of stage 1 and observed a rather general mechanism governing these processes (4). Exploring the mechanism of assembly of membrane proteins is fundamentally related to our understanding of the biological functions of these proteins. Although the preponderance of transmembrane helical structure makes the prediction of membrane protein structures in one sense simpler than that of water-soluble proteins, the prediction of helix assembly in these systems remains an outstanding problem in computational biology because it requires a detailed structural and thermodynamic understanding of protein-protein and protein-lipid interactions (5).
The experimental determination of three-dimensional structures of membrane proteins is extremely difficult. Among the ∼30,000 protein structures found in the Protein Data Bank (PDB) (6), only 0.2% are of membrane proteins. Considering their biological importance and significant presence in genomes, a challenge to theory and computational biology is to assist experiment in understanding the structure and function of membrane proteins.
Several other methods have been used to explore the interfacial structures of transmembrane helices based on molecular dynamics (MD) simulation or energy minimization methods (7–11), using additional experimental information to identify the near-native structures. Engelman and co-workers developed a computational search algorithm to explore the interfacial structures of transmembrane helix homo-oligomers. They found that the van der Waals interactions alone provide sufficiently stabilizing forces to determine the specific helix association in phospholamban (7), glycophorin A (8), and synaptobrevin (9). Kukol et al. performed an exhaustive molecular dynamics global search protocol to obtain a structure of the M2 protein from the influenza A virus using the orientational data derived from site-directly infrared dichroism spectra as an unbiased refinement energy term (10). Torres et al. explored the interfacial structures of glycophorin A, the M2 protein, and phospholamban using global searching molecular dynamics simulations and helix tile as restraints (11). Ponder and co-workers performed an ab initio prediction of the glycophorin A structure using a novel potential smoothing and search algorithm (12). Helms and co-workers developed a novel scoring function for modeling structures of oligomers of transmembrane helices assuming that van der Waals interaction dominates in the packing of transmembrane helices (13). Kokubo and Okamoto used a replica-exchange Monte Carlo simulation method to study the structures of transmembrane helices of bacteriorhodopsin (14) and glycophorin A (15,16). Recently, Bowie and co-workers proposed a simple Monte Carlo method to study the association of helices using only sequence and native oligomerization state information (17,18). These approaches usually ignored the heterogeneous membrane/solvent environment and incorporated specific information from about the systems of interest from experiment, and consequently may not generalize to other cases. In this study we demonstrate that with only sequence and oligomerization state information we are able to assemble conformational ensembles that are in excellent agreement with experiment for three transmembrane assemblies, suggesting that the combination of a more physical model for the aqueous/membrane interface and enhanced sampling methods provide a more broadly applicable approach to predicting and modeling transmembrane assemblies.
An explicit membrane/solvent model provides the most detailed information to molecular modeling and represents the most accurate model (19–22). However, due to the increase in computing resources needed as the system size increases, significant efforts have been directed to the development of implicit membrane models. In general, continuum electrostatics can be used to define the electrostatic potential and the electrostatic solvation energy of a solute with arbitrary shape by solving the Poisson-Boltzmann equation using finite-difference methods (23–25). Unfortunately, the cost of solving the Poisson-Boltzmann equations has limited its application in molecular dynamics simulations (20,26). Alternatively, implicit membrane models based on generalized Born (GB) theories and dielectric screening functions have been used quite successfully to estimate the electrostatic solvation energy. Spassov et al. first extended the GB method to include an implicit membrane. They proposed an empirical approach to model the membrane within the context of a pairwise additive GB model (27). Lazaridis used an effective energy function approach to model protein solvation (28). Im et al. proposed an improved GB method based on a smooth dielectric boundary to study the structure, stability, and interactions of membrane proteins (29,30). For more information, see recent reviews by Brooks and co-workers (31,32). More recently, Feig and co-workers devised an implicit membrane model based on GB theories developed in the Brooks group (33).
Im and Brooks studied the interfacial folding and membrane insertion of designed peptides (4), using their implicit membrane GB model (29,30) and replica-exchange (REX) (34,35) molecular dynamics (MD). Their results demonstrated the mechanism of stage 1 of the two-stage model, and the success of using an implicit membrane model combined with advanced sampling methods to simulate biological membranes. In this article, we focus on the second stage, the assembly of transmembrane helices. Starting from an idealized helix, we sampled the conformational space of various oligomerization states using the implicit membrane GB model of Im et al. (29) REX/MD simulations, and the imposition of rotational symmetry to define the extent of oligomerization. We applied our method to predict the transmembrane structures of three peptides: glycophorin A (GpA), the M2 proton channel (M2-TMP), and phospholamban (PLB), which are experimentally known to form dimeric, tetrameric, and pentameric structures, respectively. We first explored the structures of each peptide in the native oligomeric state. We compared the predicted structures of GpA dimer, the M2-TMP tetramer, and PLB pentamer with experimental structures to examine our prediction with the native oligomerization state information as the structural constraint. Furthermore, we compared the potential energy between different oligomerization states for each peptide to address the challenging question of whether one can predict the native state energetically. In other words, whether one can predict the structures of helix homo-oligomers in membrane without using any experimental information.
COMPUTATIONAL MODEL AND METHODS
We began our calculations with idealized α-helices, i.e., φ = −65° and ψ = −40°, for each peptide using the sequences given in Table 1. Each structure was oriented along the membrane normal in the membrane, and then rotated by 22.5° around the Z axis to produce 16 replicas. Each replica was then translated by 20 Å from the symmetry axis in the X,Y plane, and these were taken as the initial structures of the monomers in our REX/MD simulation. We imposed m-fold rotational symmetry using the IMAGE facility in CHARMM (36) to provide putative oligomers of order m. For glycophorin A, which is a dimer in the native state, we imposed twofold, threefold, fourfold, and fivefold symmetries on the single peptide to simulate the structure of a dimeric, trimeric, tetrameric, and pentameric assembly, respectively. The M2 protein forms a tetramer and phospholamban forms a pentamer in their native states. We imposed twofold, threefold, fourfold, fivefold, and sixfold symmetries to simulate the structure of a dimeric, trimeric, tetrameric, pentameric, and hexameric peptide oligomer, for these two proteins. As a reference, a simulation of each peptide itself was also carried out.
TABLE 1.
Amino-acid sequence of glycophorin A, M2-TMP, and phospholamban peptides
| Peptides | Sequence |
|---|---|
| Glycophorin A | EITLIIFGVM AGVIGTILLI SYGI |
| M2-TMP | SSDPLVVAAS IIGILHLILW ILDRL |
| Phospholamban | LQNLFINFCL ILICLLLICI IVMLL |
The N-terminus of each peptide is blocked by an acetyl group and its C-terminus by an n-methyl amide group, except for phospholamban, for which a standard C-terminus is used.
Our studies were performed using the GBSW (a Generalized Born model with a simple SWitching function) module (29,30) in the CHARMM program (36). All MD simulations used a time-step of 2 fs and no cutoff for the nonbonded energy evaluation. The all-hydrogen parameter set PARAM22 (37) of the CHARMM force field was used. The physical parameters representing the membrane in our GB model are 0.03 kcal/(mol × Å2) for the surface tension coefficient (representing the nonpolar solvation energy), 25 Å for the thickness of the membrane hydrophobic core, and 1 Å for a membrane smoothing length over which the hydrophobic region is gradually changed to the solvent region. The planar membrane is perpendicular to the Z axis and centered at Z = 0.
The MMTSB Tool Set (38) was used to control the REX simulations. We used 16 replicas that were distributed over an exponentially spaced temperature range from 300 K to 600 K. Langevin dynamics with a friction coefficient of 5.0 ps−1 for heavy atoms was used. A cylindrical harmonic restraint with a 25 Å radius and a force constant of 1.0 kcal/(mol × Å2) was applied to prevent the peptides from drifting radially away from each other, i.e., away from the symmetry axis. (Note that this is much larger than the radius of any of the assemblies we studied.) The REX/MD simulations were carried out for 10 ns for each oligomerization state of each peptide. Every 1 ps, a replica exchange was attempted and the coordinates were saved for further analyses. The pairwise exchange ratio was ∼40% for each run.
Using the CLUSTER facility in MMTSB Tool Set (38), we clustered the sampled structures in the native state of each peptide. Due to the size limitation of the ensemble of structures used in the CLUSTER facility, we collected every other structure during the last 7 ns of the REX/MD simulation providing 3500 structures to be used in clustering stage. We chose the structure located at the center of the largest cluster as the predicted structure.
RESULTS
Transmembrane helix dimer structure of glycophorin A
Glycophorin A (GpA) is one of the most abundant proteins located on the surface of red blood cells; however, despite its ubiquitous presence, its function remains unknown. It is also one of the most well-studied model systems in the field of helix-helix interactions in membranes (39,40). The NMR structure of glycophorin A in micelles (PDB:1AFO) (41) shows that it forms a right-handed helical dimer with the packing motif LIXXGVXXGV. The two Gly residues form a flat surface to facilitate tight packing of the backbone atoms (42). The two Val residues play the key role in the van der Waals interaction between the transmembrane helices.
The 3500 sampled structures formed two clusters with group size of 1865 (53%) and 1635 (47%) structures, respectively. The representative structure from the largest cluster has a Cα root mean-square derivation (RMSD) value of 2.2 Å relative to the experimental structure, whereas the representative structure from the other cluster has a Cα RMSD value of 7.6 Å.
Fig. 1, A and B, shows the interhelical crossing angle of the simulated dimeric structures and Cα RMSD of the dimeric structures relative to the native structure (PDB:1AFO) as a function of time. For clarity, only five trajectories out of 16 during the last 7 ns are shown. We can see a few transitions between the two configurations (left-handed dimer and right-handed dimer) occurring at high temperatures, indicating the sampling efficiency of REX/MD simulation. The RMSD is well-correlated with the interhelical crossing angle. Fig. 1, C and D, shows the distribution of crossing angles and Cα RMSD of the structures sampled at the lowest temperature (300 K) during the MD simulation. Based on the distribution of crossing angles, the helices could be clustered into two distinct families of conformations: a right-handed dimer (crossing angle at −50°), and a left-handed dimer (crossing angle at 50°). The right-handed dimer has a most probable RMSD value of 2.2 Å, whereas the most probable RMSD value of the left-handed dimer is 7.8 Å. The solid line in Fig. 1, C and D, shows the integrated population. While we see both conformations occurring with some probability, the native right-handed dimer occupies >60% of the total conformations sampled. These results are relatively consistent with the clustering results using the CLUSTER facility.
FIGURE 1.
(A) Interhelical crossing angle of GpA dimeric structures during REX/MD simulation as a function of time. The crossing angle is measured by the angle between two principal axes defined by the backbone heavy atoms of each monomer. The negative sign means that it forms a right-handed dimer and the positive sign stands for a left-handed dimer. For clarity, only five trajectories are shown. (B) RMSD of the Cα atoms of GpA sampled structures relative to the native structure (1AFO) as a function of time. (C) The distribution of crossing angle of GpA dimer. The solid curve represents the integrated population. (D) The distribution of RMSD of Cα atoms of sampled structures relative to the native structure.
Fig. 2 A shows the structure of a representative dimer model of glycophorin A derived from our simulation. As shown in Fig. 2, B and C, the comparison of contact map for the Cα-Cα distances between our model and the NMR structure reveals that the interfacial residues of our model, including Leu75, Ile76, Gly79, Val80, Gly83, and Val84, are identical with those of the solution NMR structure.
FIGURE 2.
(A) The representative dimer model of GpA derived from our simulation. The stick representations show the interfacial residues. Contact map of the Cα atoms in GpA NMR structure model (B) and our predicted model (C). Color is coded by the distance between two Cα atoms.
Transmembrane helix tetramer structure of M2-TMP
The M2 protein from Influenza A contains 97 residues and is a proton selective ion channel that forms a left-handed tetrameric helical domain (43). The structure of a 25-residue (from Ser22 to Leu46) peptide, which is also called M2-TMP, was recently determined in a DMPC bilayer using rotational echo double-resonance solid-state NMR (44).
The 3500 sampled structures from the REX/MD simulation formed five clusters with group size of 1373 (39%), 919 (26%), 849 (24%), 226 (6%), and 133 (4%) structures. The representative structure from the largest cluster has a Cα RMSD value of 2.7 Å relative to the experimental structure (PDB:1NYJ). The representative structures from other clusters have a Cα RMSD value of 8.5, 5.1, 3.9, and 4.1 Å relative to the experimental structure.
Fig. 3, A and B, show the interhelical crossing angle of the sampled tetrameric structures and Cα RMSD of the sampled tetrameric structures relative to the native structure as a function of simulation time. Again, the RMSD is well-correlated with the interhelical crossing angle. The distribution of RMSD in the sampled structures at the lowest temperature in Fig. 3 D, showing the existence of five clusters, is consistent with the clustering results using the CLUSTER facility. Based on the distribution of crossing angles in Fig. 3 C, the helices could be clustered into three families of conformations: two right-handed tetramers (crossing angle at −25° and −5°), and a left-handed tetramer (crossing angle at 35°). The left-handed tetrameric state, which is also the native state, has the largest population of 50%. The population of the two right-handed tetramers is ∼30% and 20%, respectively. The left-handed tetramer has a most probable RMSD value of 2.6 Å at the lowest temperature, whereas the most probable RMSD value of the right-handed tetramer is 8.9 Å.
FIGURE 3.
(A) Interhelical crossing angle of M2-TMP tetramer structures derived from simulation as a function of time. For clarity, only five trajectories are shown. (B) RMSD of the Cα atoms of M2-TMP sampled structures relative to the native structure (1NYJ) as a function of time. (C) The distribution of crossing angle of M2-TMP tetrameric structures. (D) The distribution of RMSD of Cα atoms of sampled structures relative to the native structure.
Fig. 4 A shows the structure of a representative tetrameric model of M2-TMP derived from our simulation. Fig. 4, B and C, shows that the interfacial residues, including Val27, Ser31, Gly34, His37, Leu38, and Trp41, are identical with those of the experimental structure derived from solid-state NMR.
FIGURE 4.
(A) The representative tetramer model of M2-TMP derived from our simulation. The stick representations show the interfacial residues. Contact map of the Cα-Cα in the M2-TMP NMR structure model (B) and our predicted model (C).
Transmembrane helix pentamer structure of phospholamban
Located in the membrane of the cardiac sarcoplasmic reticulum, phospholamban (PLB) is involved in regulation of a Ca2+ pump (45). We compare our predicted model with a model structure (PDB:1PLN) (46), which was created by the direct structure modeling of mutagenesis data. Structures have been determined for the helical monomer in the solid state using rotational echo double-resonance (47), and more recently for the pentameric structure in micelles via solution NMR methods (48). The model structure differs very little from the more recent experimental structure.
The 3500 sampled structures formed four clusters with group size of 1653 (47%), 1233 (35%), 498 (14%), and 116 (4%) structures. The representative structure from the largest cluster has a Cα RMSD value of 0.62 Å relative to the model structure (PDB:1PLN). Our predicted model shows similar agreement with the pentameric NMR structure (PDB:1ZLL)(48) which, for the TM region, is in the range of 0.71–0.94 Å compared with the 20 NMR structures and has an average Cα RMSD of 0.84 Å. The representative structures from other clusters have a Cα RMSD value of 3.4, 1.6, and 4.6 Å, respectively, relative to the model structure (PDB:1PLN).
Fig. 5 A shows the distribution of crossing angle of phospholamban pentameric structures at 300 K, which suggests that the helices only form a left-handed pentamer (crossing angle at 19°). Fig. 5 B shows the distribution of Cα RMSD of phospholamban sampled structures at 300 K relative to PDB:1PLN during the MD simulation. The distribution of RMSD, which is characterized by the existence of four clusters with the most probable RMSD values of 0.62, 1.7, 3.3, and 4.6 Å, is consistent with the clustering results using the CLUSTER facility.
FIGURE 5.
(A) The distribution of crossing angle of the sampled PLB pentameric structures. (B) The distribution of RMSD of Cα atoms of the sampled structures relative to the model structure in model structure (PDB:1PLN).
Fig. 6 A shows the structure of the representative pentameric model of PLB derived from our simulation. Fig. 6, B and C, illustrates that the positions of the interfacial residues, including Leu37, Ile40, Leu44, Ile47, and Leu51, are identical to PDB:1PLN.
FIGURE 6.
(A) The representative pentamer model of PLB derived from our simulation. The stick representations show the interfacial residues. Contact map of the Cα-Cα in PLB model derived from experimental data (B) and our predicted model (C).
Predicting native oligomerization state energetically
In this section, we compare the potential energy between the different oligomerization states of each peptide. The question we would like to address is whether we can predict the native oligomerization state of each peptide energetically. We assume that the native state not only has the lowest free energy, but also the lowest potential energy.
Fig. 7 A shows the potential energy profile of each peptide for various oligomerization states at the lowest temperature (300 K), averaged over the last 7 ns of the REX/MD simulations, relative to the corresponding monomeric states. The potential energy of each oligomerization state of each peptide converged after the initial 2 ns of simulation (data not shown). In the case of GpA, the dimeric state does not have the lowest potential energy. Table 2 shows the decomposition of the potential energy. The differences in potential energy between monomeric/dimeric state and dimeric/trimeric state are −21.1 kcal/mol and −15.3 kcal/mol, respectively. The differences are dominated by the differences in van der Waals interaction between monomeric/dimeric state and dimeric/trimeric state, which are −21.8 kcal/mol and −13.3 kcal/mol, respectively. Clearly, van der Waals interactions between the interfacial residues play the key role in the packing of helices in our model.
FIGURE 7.
Potential energy (A) and free energy (B) profiles of GpA, M2-TMP, and PLB oligomers at the lowest temperature (300 K) during the REX/MD simulations, relative to the corresponding monomers.
TABLE 2.
Various average properties from the glycophorin A simulations
| Energy (kcal/mol)
|
||||
|---|---|---|---|---|
| Oligomer | W | Uvdw | ΔGnp | Welec |
| Monomer | 225.1 ± 14.2 | −37.9 ± 7.6 | 16.6 ± 3.7 | −113.1 ± 7.9 |
| Dimer | 204.0 ± 14.4 | −59.7 ± 7.5 | 17.2 ± 1.6 | −113.3 ± 7.6 |
| Trimer | 188.7 ± 14.9 | −73.0 ± 8.0 | 16.7 ± 1.3 | −114.8 ± 7.9 |
| Tetramer | 185.5 ± 14.4 | −78.9 ± 8.0 | 15.5 ± 2.2 | −112.3 ± 7.7 |
| Pentamer | 184.9 ± 14.5
|
−82.2 ± 8.6
|
17.4 ± 1.8
|
−111.4 ± 8.5
|
| ΔW
|
ΔUvdw
|
ΔΔGnp
|
ΔWelec
|
|
| Monomer/dimer | −21.1 | −21.8 | 0.6 | −0.2 |
| Dimer/trimer | −15.3 | −13.3 | −0.5 | −1.5 |
| Trimer/tetramer | −3.2 | −5.9 | −1.2 | 2.5 |
| Tetramer/pentamer | −0.6 | −3.3 | 1.9 | 0.9 |
The potential energy W is defined as the sum of the internal molecular mechanics energy, the external molecular mechanics energy (van der Waals Uvdw and Coulomb Ucoul), the electrostatic solvation energy ΔGelec, and the nonpolar solvation energy ΔGnp. Welec is the sum of Ucoul and ΔGelec.
In the case of M2-TMP, as shown in Fig. 7 A, the tetrameric state, which is the native state, does not have the lowest potential energy. Table 3 shows the decomposition of the energy. The potential energy differences between monomeric/dimeric state, dimeric/trimeric state, and tetrameric/pentameric state are all dominated by the differences in interhelical van der Waals interactions. Interestingly, the difference of interhelical van der Waals interaction in two adjacent oligomerization states becomes smaller as the oligomerization number increases. This trend is also seen in glycophorin A and phospholamban. The potential energy difference between trimeric state and tetrameric state is relatively small, compared to the other two adjacent states. The differences in interhelical van der Waals interaction between trimeric/tetrameric state are canceled by other unfavorable energy differences, such as internal energy, which is 418.6 ± 0.5 kcal/mol for monomer, dimer, and trimer, whereas it is 421.7 ± 0.2 kcal/mol for tetramer, pentamer, and hexamer. The van der Waals interaction between the helices in the pentameric state and hexameric state is identical, whereas the electrostatic interaction dominates the potential energy difference between pentameric state and hexameric state. This is perhaps due to the electrostatic interaction between the polar residues located in the interhelical interface, mainly Ser31, His37, and Trp41. As demonstrated in the solid-state NMR structure, the distance between His37 and Trp41 is <3.9 Å, which suggests that the interaction between His37 and Trp41 plays the key role in sterically closing the channel (44). We observed that the close packing of pentamer and hexamer does not have the correct handedness. As shown in Fig. 8, the sampled structures at pentameric and hexameric states are mostly right-handed, whereas the native structure should be left-handed.
TABLE 3.
Various average properties from the M2-TMP simulations
| Energy(kcal/mol)
|
||||
|---|---|---|---|---|
| Oligomer | W | Uvdw | ΔGnp | Welec |
| Monomer | −118.1 ± 14.8 | −58.6 ± 7.3 | 18.2 ± 2.0 | −497.0 ± 8.5 |
| Dimer | −147.0 ± 15.4 | −81.5 ± 8.2 | 22.5 ± 2.3 | −506.5 ± 8.7 |
| Trimer | −158.0 ± 15.7 | −89.1 ± 8.4 | 18.9 ± 1.7 | −505.9 ± 9.0 |
| Tetramer | −157.5 ± 15.0 | −95.9 ± 8.7 | 19.8 ± 1.8 | −503.3 ± 9.5 |
| Pentamer | −166.8 ± 15.2 | −102.8 ± 9.1 | 19.1 ± 1.9 | −504.5 ± 10.8 |
| Hexamer | −170.7 ± 15.1
|
−102.8 ± 7.9
|
19.0 ± 2.1
|
−508.9 ± 10.1
|
| ΔW
|
ΔUvdw
|
ΔΔGnp
|
ΔWelec
|
|
| Monomer/dimer | −28.9 | −22.9 | 4.3 | −9.5 |
| Dimer/trimer | −11.0 | −7.6 | −3.6 | 0.6 |
| Trimer/tetramer | 0.5 | −6.8 | 0.9 | 2.6 |
| Tetramer/pentamer | −9.3 | −6.9 | −0.7 | −1.2 |
| Pentamer/hexamer | −3.9 | 0 | −0.1 | −4.4 |
All the energy terms are defined in Table 2.
FIGURE 8.
The distribution of crossing angle of M2-TMP sampled structures at tetrameric, pentameric, and hexameric states.
In the case of PLB, as shown in Fig. 7 A, the pentameric state, which is the native state, has the lowest potential energy. As shown in Table 4, the difference between interhelical van der Waals interactions again dominates the potential energy difference between adjacent oligomerization states. Since the interfacial residues in PLB are all hydrophobic residues, the electrostatic interaction between the helices does not contribute significantly to the stabilization of the oligomers.
TABLE 4.
Various average properties from the PLB simulations
| Energy(kcal/mol)
|
||||
|---|---|---|---|---|
| Oligomer | W | Uvdw | ΔGnp | Welec |
| Monomer | 60.8 ± 15.1 | −59.0 ± 7.1 | 15.3 ± 2.1 | −319.2 ± 7.7 |
| Dimer | 43.5 ± 15.9 | −84.8 ± 10.6 | 20.8 ± 3.8 | −317.2 ± 8.5 |
| Trimer | 28.2 ± 15.4 | −100.6 ± 8.5 | 22.4 ± 1.9 | −318.7 ± 8.0 |
| Tetramer | 18.6 ± 15.2 | −111.2 ± 8.1 | 21.8 ± 1.5 | −316.6 ± 7.6 |
| Pentamer | 8.6 ± 15.7 | −120.2 ± 8.2 | 21.6 ± 1.4 | −317.2 ± 7.8 |
| Hexamer | 11.1 ± 15.9
|
−117.2 ± 8.9
|
20.8 ± 2.1
|
−316.9 ± 7.6
|
| ΔW
|
ΔUvdw
|
ΔΔGnp
|
ΔWelec
|
|
| Monomer/dimer | −17.3 | −25.8 | 5.5 | 2.0 |
| Dimer/trimer | −15.3 | −15.8 | 1.6 | −1.5 |
| Trimer/tetramer | −9.6 | −10.6 | −0.6 | 2.1 |
| Tetramer/pentamer | −10.0 | −9.0 | −0.2 | −0.6 |
| Pentamer/hexamer | 2.5 | 3.0 | −0.8 | 0.3 |
All the energy terms are defined in Table 2.
CONCLUDING DISCUSSION
We have investigated the membrane assembly of GpA, M2-TMP, and PLB peptide, using REX/MD and an implicit membrane GB model. Our approach is quite successful in predicting the structures of homo-oligomers, using only the native oligomerization state as a structural constraint. It is noteworthy that this property can often be gleaned from measurements utilizing analytical ultracentrifugation, equilibrium dialysis, and other biochemical approaches without the necessity of atomic level structural information (41,49–51). For our predicted models, we find the RMSD value of Cα atoms relative to the corresponding experimental and model structures are 2.2 Å (GpA), 2.7 Å (M2-TMP), and 0.62 Å (PLB), respectively. Also of interest is the observation that a distribution of conformations appear to be present in each case. Whether this is a true reflection of some level of conformational heterogeneity or a limitation of our model remains to be investigated.
Using only the peptide sequence we do not always predict the native oligomerization state as predominant based on energetic criteria. We successfully predicted the native oligomerization state for PLB, but not for GpA and M2-TMP. One explanation for this may be that we did not consider the entropy loss during helix association. Shown in Table 5 is an estimation of translational, rotational, and conformational entropy. The translational entropy and rotational entropy were calculated based on principal RMS fluctuations of the center of mass or Euler angles (52). The translational entropy can be expressed as
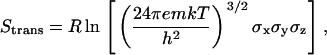 |
(1) |
where σx, σy, and σz are the principal RMS fluctuations for the center of mass of each peptide at different oligomerization states. The absolute rotational entropy can be expressed as
 |
(2) |
where σφ, σψ, and σθ are RMS fluctuations in the three Euler angles. The conformational entropy was calculated from the covariance matrices of the atomic fluctuations with quasiharmonic approximation (53,54) (using the QUASIHARMONIC facility in CHARMM (36)). The free energy is calculated as the sum of the potential energy and the entropy terms. As illustrated in Fig. 7 B, these results show that the M2-TMP pentamer and PLB tetramer have the lowest free energy, compared to other oligomerization states. We did not observe such a free energy turnover in the case of GpA, where the free energy continues to decrease with increasing oligomerization number. Including correction terms to consider the entropy loss does not provide a complete answer, and our ability to identify the native oligomerization state is still lacking.
TABLE 5.
Estimated entropy terms in each model system
| Energy(kcal/mol)
|
||||||
|---|---|---|---|---|---|---|
| Oligomer | W | −TStrans | −TSrot | −TScomform | F | |
| GpA | Monomer | 225.1 | 0 | 0 | 0 | 225.1 |
| Dimer | 204.0 | 1.3 | 5.4 | 9.0 | 219.7 | |
| Trimer | 188.7 | 1.4 | 7.3 | 20.4 | 217.8 | |
| Tetramer | 185.5 | 1.2 | 8.3 | 7.8 | 202.8 | |
| Pentamer | 184.9 | 1.1 | 9.0 | 3.9 | 198.9* | |
| M2-TMP | Monomer | −118.1 | 0 | 0 | 0 | −118.1 |
| Dimer | −147.0 | 1.4 | 5.9 | 14.7 | −125.0 | |
| Trimer | −158.0 | 1.4 | 7.4 | 12.9 | −136.3 | |
| Tetramer | −157.5 | 1.2 | 8.5 | −1.8 | −149.6 | |
| Pentamer | −166.8 | 1.2 | 9.1 | 6.6 | −149.9* | |
| Hexamer | −170.7 | 1.1 | 9.5 | 13.8 | −146.3 | |
| PLB | Monomer | 60.8 | 0 | 0 | 0 | 60.8 |
| Dimer | 43.5 | 0.8 | 5.7 | −4.8 | 45.2 | |
| Trimer | 28.2 | 1.2 | 7.9 | 1.8 | 39.1 | |
| Tetramer | 18.6 | 1.2 | 8.9 | 0.6 | 29.3* | |
| Pentamer | 8.6 | 1.2 | 9.6 | 13.5 | 32.9 | |
| Hexamer | 11.1 | 1.0 | 9.9 | 12.0 | 34.0 | |
Strans, translational entropy; Srot, rotational entropy; and Sconform, conformational entropy. –TS is defined to be 0 at monomeric state of each peptide. The free energy F is defined as the sum of potential energy W and entropy term –TS.
Lowest free energy.
More accurate evaluation of entropy loss may be needed to improve the first-principles calculation of folding and oligomerization equilibria for these peptides. However, we also find that the interhelical van der Waals interaction dominates in the packing of the GpA, M2-TMP, and PLB peptides. Thus, we anticipate that the interhelical van der Waals interaction is overestimated in our GBSW model, since we did not include peptide-lipid dispersion interactions that should compete with the peptide-peptide interactions. Currently, we are extending our implicit membrane model to include interactions between protein and lipid. The optimization of parameters in our GBSW model is ongoing.
Acknowledgments
We are thankful for the generous support of this research by the National Institute of Health (grant No. RR12255) and by the Center for Theoretical Biological Physics through funding from the National Science Foundation (grant No. PHY0216576).
References
- 1.Wallin, E., and G. von Heijne. 1998. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7:1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popot, J. L., and D. M. Engelman. 1990. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 29:4031–4037. [DOI] [PubMed] [Google Scholar]
- 3.Engelman, D. M., Y. Chen, C. N. Chin, A. R. Curran, A. M. Dixon, A. D. Dupuy, A. S. Lee, U. Lehnert, E. E. Matthews, Y. K. Reshetnyak, A. Senes, and J. L. Popot. 2003. Membrane protein folding: beyond the two-stage model. FEBS Lett. 555:122–125. [DOI] [PubMed] [Google Scholar]
- 4.Im, W., and C. L. Brooks III. 2005. Interfacial folding and membrane insertion of designed peptides studied via molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 102:6771:6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hessa, T., H. Kim, K. Bihlmaier, C. Lundin, J. Boekel, H. Andersson, I. M. Nilsson, S. H. White, and G. von Heijne. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 433:377–381. [DOI] [PubMed] [Google Scholar]
- 6.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams, P. D., I. T. Arkin, D. M. Engelman, and A. T. Brunger. 1995. Computational searching and mutagenesis suggest a structure for the pentameric transmembrane domain of phospholamban. Nat. Struct. Biol. 2:154–162. [DOI] [PubMed] [Google Scholar]
- 8.Adams, P. D., D. M. Engelman, and A. T. Brunger. 1996. Improved prediction for the structure of the dimeric transmembrane domain of glycophorin A obtained through global searching. Proteins Struct. Funct. Genet. 26:257–261. [DOI] [PubMed] [Google Scholar]
- 9.Fleming, K. G., and D. M. Engelman. 2001. Computation and mutagenesis suggest a right-handed structure for the synaptobrevin transmembrane dimer. Proteins Struct. Funct. Genet. 45:313–317. [DOI] [PubMed] [Google Scholar]
- 10.Kukol, A., P. D. Adams, L. M. Rice, A. T. Brunger, and I. T. Arkin. 1999. Experimentally based orientational refinement of membrane protein models: a structure for the influenza A M2 H+ channel. J. Mol. Biol. 286:951–962. [DOI] [PubMed] [Google Scholar]
- 11.Torres, J., J. A. G. Briggs, and I. T. Arkin. 2002. Contribution of energy values to the analysis of global searching molecular dynamics simulations of transmembrane helical bundles. Biophys. J. 82:3063–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappu, R. V., G. R. Marshall, and J. W. Ponder. 1999. A potential smoothing algorithm accurately predicts transmembrane helix packing. Nat. Struct. Biol. 6:50–55. [DOI] [PubMed] [Google Scholar]
- 13.Park, Y., M. Elsner, R. Staritzbichler, and V. Helms. 2004. Novel scoring function for modeling structures of oligomers of transmembrane α-helices. Proteins Struct. Funct. Genet. 57:577–585. [DOI] [PubMed] [Google Scholar]
- 14.Kokubo, H., and Y. Okamoto. 2004. Self-assembly of transmembrane helices of bacteriorhodopsin by a replica-exchange Monte Carlo simulation. Chem. Phys. Lett. 392:68–175. [Google Scholar]
- 15.Kokubo, H., and Y. Okamoto. 2004. Prediction of membrane protein structures by replica-exchange Monte Carlo simulations: case of two helices. J. Chem. Phys. 120:10837–10847. [DOI] [PubMed] [Google Scholar]
- 16.Kokubo, H., and Y. Okamoto. 2004. Prediction of transmembrane helix configurations by replica-exchange simulations. Chem. Phys. Lett. 383:397–402. [Google Scholar]
- 17.Kim, S., A. K. Chamberlain, and J. U. Bowie. 2003. A simple method for modeling transmembrane helix oligomers. J. Mol. Biol. 329:831–840. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S., A. K. Chamberlain, and J. U. Bowie. 2004. Membrane channel structure of Helicobacter pylori vacuolating toxin: role of multiple GXXXG motifs in cylindrical channels. Proc. Natl. Acad. Sci. USA. 101:5988–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer, W. B., and M. S. P. Sansom. 2002. Viral ion channels: structure and function. Biochim. Biophys. Acta. 1561:27–45. [DOI] [PubMed] [Google Scholar]
- 20.Murray, D., and B. Honig. 2002. Electrostatic control of the membrane targeting of C2 domains. Mol. Cell. 9:145–154. [DOI] [PubMed] [Google Scholar]
- 21.Im, W., and B. Roux. 2002. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 319:1177–1197. [DOI] [PubMed] [Google Scholar]
- 22.Petrache, H. I., A. Grossfield, K. R. MacKenzie, D. M. Engelman, and T. B. Woolf. 2000. Modulation of glycophorin A transmembrane helix interactions by lipid bilayer molecular dynamics calculations. J. Mol. Biol. 302:727–746. [DOI] [PubMed] [Google Scholar]
- 23.Warwicker, J., and H. C. Watson. 1982. Calculation of the electric potential in the active site cleft due to α-helix dipoles. J. Mol. Biol. 157:671–679. [DOI] [PubMed] [Google Scholar]
- 24.Klapper, I., R. Hagstrom, R. Fine, K. Sharp, and B. Honig. 1986. Focusing of electric fields in the active site of Cu-Zn superoxide dismutase. Proteins. 1:47–59. [DOI] [PubMed] [Google Scholar]
- 25.Nicholls, A., and B. Honig. 1991. A rapid finite difference algorithm, utilizing successive overrelaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 4:435–445. [Google Scholar]
- 26.Roux, B., S. Bernche, and W. Im. 2000. Ion channels, permeation and electrostatics: insight into the function of KcsA. Biochemistry. 39:13295–13306. [DOI] [PubMed] [Google Scholar]
- 27.Spassov, V. Z., L. Yan, and S. Szalma. 2002. Introducing an implicit membrane in Generalized Born/solvent accessibility continuum solvent models. J. Phys. Chem. B. 106:8726–8738. [Google Scholar]
- 28.Lazaridis, T. 2003. Effective energy function for proteins in lipid membranes. Proteins. 52:176–192. [DOI] [PubMed] [Google Scholar]
- 29.Im, W., M. Feig, and C. L. Brooks III. 2003. An implicit membrane generalized Born theory for the study of structure, stability, and interactions of membrane proteins. Biophys. J. 85:2900–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Im, W., M. S. Lee, and C. L. Brooks III. 2003. Generalized Born model with a simple smoothing function. J. Comput. Chem. 24:1691–1702. [DOI] [PubMed] [Google Scholar]
- 31.Feig, M., and C. L. Brooks III. 2004. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr. Opin. Struct. Biol. 14:217–224. [DOI] [PubMed] [Google Scholar]
- 32.Im, W., J. Chen, and C. L. Brooks III. 2005. Peptide and protein folding and conformational equilibria: theoretical treatment of electrostatics and hydrogen bonding with implicit solvent models. Adv. Protein Chem. In press. [DOI] [PubMed]
- 33.Tanizaki, S., and M. Feig. 2005. Molecular dynamics simulations of large integral membrane proteins with an implicit membrane model. J. Phys. Chem. B. 110:548–556. [DOI] [PubMed] [Google Scholar]
- 34.Hansmann, U. H. E. 1997. Parallel tempering algorithm for conformational studies of biological molecules. Chem. Phys. Lett. 281:140–150. [Google Scholar]
- 35.Sugita, Y., and Y. Okamoto. 1999. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314:141–151. [Google Scholar]
- 36.Brooks, B. R., R. Bruccoleri, B. Olafson, D. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J. Comput. Phys. 4:187–217. [Google Scholar]
- 37.MacKerell, A.D., Jr, D. Bashford, M. Bellott, R. L. Dunbrack Jr, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher III, B. Roux, M. Schlenkrich, J. C. Smith, T. Stote, J. E. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 38.Feig, M., J. Karanicolas, and C. L. Brooks III. 2004. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph. Model. 22:377–395. [DOI] [PubMed] [Google Scholar]
- 39.Popot, J. L., and D. M. Engelman. 2000. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69:881–922. [DOI] [PubMed] [Google Scholar]
- 40.Arkin, I. T. 2002. Structural aspects of oligomerization taking place between the transmembrane α-helices of bitopic membrane proteins. Biochim. Biophys. Acta. 1565:347–363. [DOI] [PubMed] [Google Scholar]
- 41.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science. 69:881–922. [DOI] [PubMed] [Google Scholar]
- 42.Smith, S. O., D. Song, S. Shekar, M. Groesbeek, M. Ziliox, and S. Aimoto. 2001. Structure of the transmembrane dimer interface of glycophorin A in membrane bilayers. Biochemistry. 40:6553–6558. [DOI] [PubMed] [Google Scholar]
- 43.Sakaguchi, T., Q. Tu, L. H. Pinto, and R. A. Lamb. 1997. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc. Natl. Acad. Sci. USA. 94:5000–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura, K., S. Kim, L. Zhang, and T. A. Cross. 2002. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry. 41:13170–13177. [DOI] [PubMed] [Google Scholar]
- 45.James, P., M. Inui, M. Tada, M. Chiesi, and E. Carafoli. 1989. Nature and site of phospholamban regulation of the calcium pump of sarcoplasmic reticulum. Nature. 342:90–92. [DOI] [PubMed] [Google Scholar]
- 46.Herzyk, P., and R. E. Hubbard. 1998. Using experimental information to produce a model of the transmembrane domain of the ion channel phospholamban. Biophys. J. 74:1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, S. O., T. Kawakami, W. Liu, M. Ziliox, and S. Aimoto. 2001. Helical structure of phospholamban in membrane bilayers. J. Mol. Biol. 313:1139–1148. [DOI] [PubMed] [Google Scholar]
- 48.Oxenoid, K., and J. J. Chou. 2005. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc. Natl. Acad. Sci. USA. 102:10870–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.K. P. Howard, J. D. Lear, and W. DeGrado. 2002. Sequence determinants of the energetics of folding of a transmembrane four-helix-bundle protein. Proc. Natl. Acad. Sci. USA. 99:8568–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wimley, W. C., K. Hristova, A. S. Ladokhin, L. Silvestro, P. H. Axelsen, and S. H. White. 1998. Folding of β-sheet membrane proteins: a hydrophobic hexapeptide model. J. Mol. Biol. 277:1091–1110. [DOI] [PubMed] [Google Scholar]
- 51.Cristian, L., J. D. Lear, and W. F. DeGrado. 2003. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 100:14772–14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlsson, J., and J. Åqvist. 2005. Absolute and relative entropies from computer simulation with applications to ligand binding. J. Phys. Chem. B. 109:6448–6456. [DOI] [PubMed] [Google Scholar]
- 53.Schlitter, J. 1993. Estimation of absolute entropies of macromolecules using the covariance matrix. Chem. Phys. Lett. 215:617–621. [Google Scholar]
- 54.Andricioaei, I., and M. Karplus. 2001. On the calculation of entropy from covariance matrices of the atomic fluctuations. J. Chem. Phys. 115:6289–6292. [Google Scholar]










