Abstract
4′,6-Diamidino-2-phenylindole (DAPI), netropsin, and pentamidine are minor groove binders that have terminal − groups. The hydration changes that accompany their binding to the minor groove of the (AATT)2 sequence have been studied using the osmotic stress technique with fluorescence spectroscopy. The affinity of DAPI for the binding site decreases with the increasing osmolality of the solution, resulting in acquisition of 35 ± 1 waters upon binding. A competition fluorescence assay was utilized to measure the binding constants and hydration changes of the other two ligands, using the DNA-DAPI complex as the fluorescence reporter. Upon their association to the (AATT)2 binding site, netropsin and pentamidine acquire 26 ± 3 and 34 ± 2 additional waters of hydration, respectively. The hydration changes are discussed in the context of the terminal functional groups of the ligands and conformational changes in the DNA.
groups. The hydration changes that accompany their binding to the minor groove of the (AATT)2 sequence have been studied using the osmotic stress technique with fluorescence spectroscopy. The affinity of DAPI for the binding site decreases with the increasing osmolality of the solution, resulting in acquisition of 35 ± 1 waters upon binding. A competition fluorescence assay was utilized to measure the binding constants and hydration changes of the other two ligands, using the DNA-DAPI complex as the fluorescence reporter. Upon their association to the (AATT)2 binding site, netropsin and pentamidine acquire 26 ± 3 and 34 ± 2 additional waters of hydration, respectively. The hydration changes are discussed in the context of the terminal functional groups of the ligands and conformational changes in the DNA.
INTRODUCTION
Water is an important structural component of DNA, and its effect on the function of DNA is also significant, especially in cellular environments where high concentrations of solutes limit the amount of water (1–3). This work considers hydration effects for small molecules that are used as drugs to target particular DNA sequences via the minor groove (4). To understand the role of water in these reactions, several approaches are used. For example, high-sensitivity calorimetry with structural data shows that the change in the heat capacity for reaction is correlated with the loss of structured water from a DNA-ligand interface (5). However, water is involved beyond the entropic contribution due to expulsion from solvent-exposed regions. To illustrate, crystallography studies demonstrate that waters make intermolecular contacts between ligands and DNA (6). Spectroscopic techniques highlight the complicated dynamics of macromolecules such as DNA, and special attention has been focused on bound water in the ligand-DNA complex (7,8). Thermodynamic studies in conjunction with theoretical calculations and x-ray crystallography demonstrate that water mediates contacts between ligands and the DNA binding site (9,10). Insight into the larger population of waters involved in the DNA-small molecule reaction is obtained from volumetric studies (11).
Another approach for measuring the total hydration changes is the osmotic stress method (12). In these experiments, water is treated as a ligand, and its involvement in the ligand-DNA association is analyzed using linkage thermodynamics (13). The activity of water in the bulk solution is affected via cosolutes that are excluded from the vicinity of the DNA (14). Via this method, water has been shown to be involved in the association of proteins with DNA, and the extent of the hydration changes is influenced by the base sequence and conformational changes (15,16). A significant result from studies of small molecules interactions with DNA is that water acquisition occurs with complex formation (17–19). To consider how the structure of the ligand influences the hydration changes, three small molecules with terminal 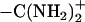 groups—4′,6-diamidino-2-phenylindole (DAPI), netropsin, and pentamidine—were reacted with the (AATT)2 binding site (Fig. 1). To measure how the binding affinities vary with osmotic pressure, the fluorescence reporter DAPI is used to measure relative equilibrium constants for the netropsin and pentamidine exchange reactions. The hydration changes are examined from the standpoint of the common terminal functional groups in the ligands and their conformational effects on DNA.
groups—4′,6-diamidino-2-phenylindole (DAPI), netropsin, and pentamidine—were reacted with the (AATT)2 binding site (Fig. 1). To measure how the binding affinities vary with osmotic pressure, the fluorescence reporter DAPI is used to measure relative equilibrium constants for the netropsin and pentamidine exchange reactions. The hydration changes are examined from the standpoint of the common terminal functional groups in the ligands and their conformational effects on DNA.
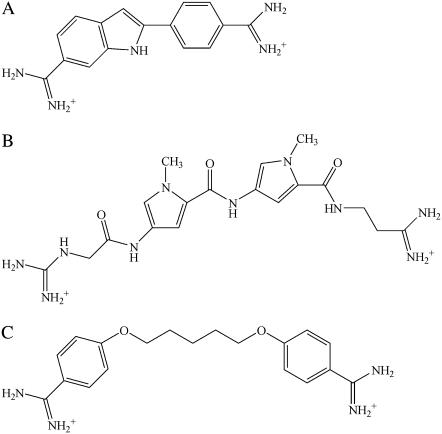
FIGURE 1.
Molecular structures of (a) DAPI, (b) netropsin, and (c) pentamidine.
MATERIALS AND METHODS
All experiments were performed in 10 mM 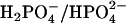 buffer with 50 mM NaCl and 1 mM Na2EDTA. Single strands of 5′-CGCGCAATTGCGCG-3′ (Integrated DNA Technologies; Coralville, IA) were annealed by slow cooling of the solution from 95°C to 10°C over 48 h. The concentration of the single stranded DNA was measured spectrophotometrically using extinction coefficient derived from the nearest neighbor approximation (125, 800 M−1 cm−1 at 260 nm) or after the complete digestion using nuclease BAL-31 (New England Biolabs; Ipswich, MA) (1). The concentrations of the drugs were determined using the following extinction coefficients: 27,000 M−1 cm−1 at 342 nm for DAPI; 21,500 M−1 cm−1 at 296 nm for netropsin; 28,900 M−1 cm−1 at 262 nm for pentamidine (20). The buffer components, netropsin, pentamidine (Sigma-Aldrich; St. Louis, MO), and DAPI (Molecular Probes; Carlsbad, CA) were used without purification. The osmolytes acetamide (Sigma-Aldrich), betaine monohydrate, trimethylamine N-oxide dihydrate (Fluka; St. Louis, MO), and triethylene glycol (TCI; Portland, OR) were also used as received. The osmolalities of the solutions were measured on a Wescor 5520 (Wescor; Logan, UT). Absorbance measurements were carried out with a Cary 50 spectrometer (Varian; Palo Alto, CA). Fluorescence data were collected on a Fluorolog spectrometer (Jobin-Yvon Horiba; Edison, NJ) at room temperature. Isothermal titration calorimetry (ITC) experiments were performed in triplicate on Microcal VP-ITC (Northhampton, MA) controlled by the Origin 7.0 software, as described earlier (19).
buffer with 50 mM NaCl and 1 mM Na2EDTA. Single strands of 5′-CGCGCAATTGCGCG-3′ (Integrated DNA Technologies; Coralville, IA) were annealed by slow cooling of the solution from 95°C to 10°C over 48 h. The concentration of the single stranded DNA was measured spectrophotometrically using extinction coefficient derived from the nearest neighbor approximation (125, 800 M−1 cm−1 at 260 nm) or after the complete digestion using nuclease BAL-31 (New England Biolabs; Ipswich, MA) (1). The concentrations of the drugs were determined using the following extinction coefficients: 27,000 M−1 cm−1 at 342 nm for DAPI; 21,500 M−1 cm−1 at 296 nm for netropsin; 28,900 M−1 cm−1 at 262 nm for pentamidine (20). The buffer components, netropsin, pentamidine (Sigma-Aldrich; St. Louis, MO), and DAPI (Molecular Probes; Carlsbad, CA) were used without purification. The osmolytes acetamide (Sigma-Aldrich), betaine monohydrate, trimethylamine N-oxide dihydrate (Fluka; St. Louis, MO), and triethylene glycol (TCI; Portland, OR) were also used as received. The osmolalities of the solutions were measured on a Wescor 5520 (Wescor; Logan, UT). Absorbance measurements were carried out with a Cary 50 spectrometer (Varian; Palo Alto, CA). Fluorescence data were collected on a Fluorolog spectrometer (Jobin-Yvon Horiba; Edison, NJ) at room temperature. Isothermal titration calorimetry (ITC) experiments were performed in triplicate on Microcal VP-ITC (Northhampton, MA) controlled by the Origin 7.0 software, as described earlier (19).
Titrations of DAPI with the oligonucleotide (DNA) were used to measure the equilibrium constant for the association reaction and to determine the numbers of waters that are exchanged (ΔNw,DAPI)
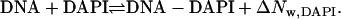 |
(1) |
The equilibrium constants were calculated as described for Hoechst 33258 using an excitation wavelength of 356 nm and the emission wavelength of 461 nm (19). Titrations of the preformed DNA-DAPI complex with the competitors netropsin or pentamidine were analyzed to determine the relative equilibrium constant for the exchange reaction
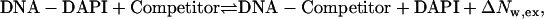 |
(2) |
where ΔNw,ex is the change in the number of waters for the reaction. ΔNw,ex and ΔNw,DAPI were used to obtain the change in the number of waters for the association of the competitors with the DNA. To maintain a reasonable fluorescence signal and to minimize drug aggregation with the DNA, a low DNA concentration (4 nM) was used. To demonstrate that the strands in the DNA-ligand complexes did not dissociate during the time period of the experiments, netropsin was titrated to the preformed DAPI-DNA complex using a hairpin-forming oligonucleotide. This oligonucleotide—5′-GCAATTGCTTTTGCAATTGC-3′—contained the (AATT)2 binding site like the duplex oligonucleotide and an additional T4 loop that covalently links the two strands. The relative equilibrium constants were similar for the hairpin and the duplex oligonucleotide. To insure that 95% of the DNA was in the preformed complex with DAPI, the equilibrium constants for the DAPI association reaction (Eq. 1) were used to calculate the DAPI concentrations needed at each osmolality. Titrations with 99% bound oligonucleotide yielded the same relative affinities, suggesting that small amounts of unbound oligonucleotide did not influence the results.
For the competition titrations, the observed emission, F, has two contributions from the bound and unbound DAPI:
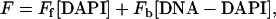 |
(3) |
where [DAPI] is the unbound DAPI concentration during the DNA titration and Ff is the intrinsic fluorescence of the unbound DAPI (for a 1 M concentration). [DNA-DAPI] and Fb are the corresponding parameters for the bound DAPI. During the titration with the competitors, the observed emission decreases as DAPI is displaced from the minor groove. The combination of mass balance with Eqs. 2 and 3 describes how the concentration of the DNA-Competitor complex changes during the titration
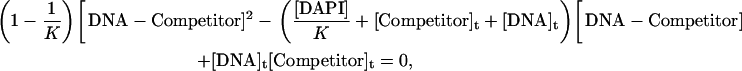 |
(4) |
where K is the equilibrium constant for the exchange reactions (Eq. 2) and the total concentrations of the competitor and oligonucleotide are [Competitor]t and [DNA]t, respectively. The values of K, Ff, and Fb were obtained from Eqs. 3 and 4 using least squares fitting of the observed emission intensity as a function of added competitor. The hydration changes (ΔNw) were measured using the change in the observed equilibrium constant (K) with the change in the osmolality (Osm) of the cosolute (12)
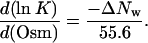 |
(5) |
RESULTS
DAPI
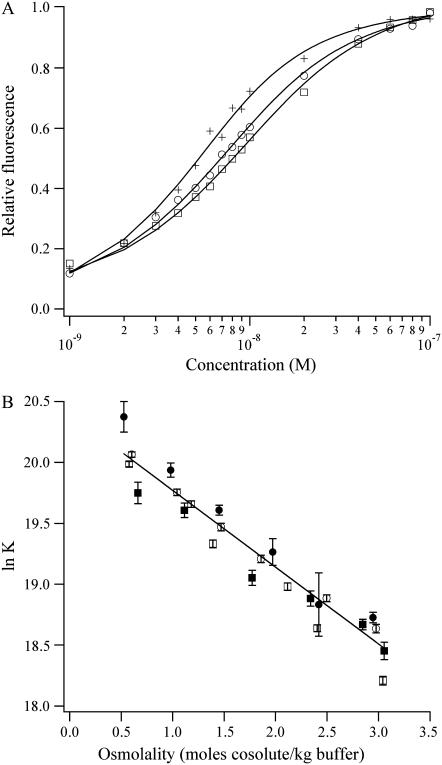
For the study of the minor groove complexes in this study, DAPI was used as the fluorescent reporter in the competitive equilibria. Thus, the reference reaction of DAPI with the (AATT)2 binding site is discussed first. The fluorescence quantum yield of DAPI associated with DNA is significantly higher relative to the free DAPI, thus allowing a distinction of bound and unbound ligand for the measurement of the equilibrium constant. DAPI has a high affinity for the minor groove of A/T-rich binding sites (21). To ascertain the hydration changes for the association reaction, DAPI was titrated with the oligonucleotide in the presence of varying concentrations of the osmolytes acetamide, betaine, triethylene glycol, and trimethylamine N-oxide. Binding isotherms (Fig. 2 A) show the decrease in the affinity with the increasing osmolyte concentration. Thus, the product is more hydrated than the reactants, with 35 ± 1 additional waters being measured using the four osmolytes (Fig. 2 B). The structures of the osmolytes differ significantly, but their influence on the equilibrium constants is similar. This observation suggests that the osmolytes are excluded from the vicinity of the DNA and thus primarily act to reduce the activity of water in the bulk solution. In further support of osmolyte exclusion, extrapolation to the buffer conditions in Fig. 2 B gives an equilibrium constant of 6.7 (± 0.1) × 108 M−1, which is consistent with the direct measurement in buffer alone (7.0 (± 0.5) × 108 M−1) (21,22). In addition, their lack of a net charge precludes electrostatic interactions with the DNA. Preferential interaction coefficients provide a direct measure of the extent of interaction of cosolutes with DNA, and the exclusion of betaine from the vicinity of the DNA has been demonstrated by vapor pressure osmometry (23). The changes in the dielectric constant due to the osmolytes are not a significant factor, as comparable results were obtained using betaine and acetamide that have opposite effects on the dielectric constant (24).
FIGURE 2.
(A) Binding isotherms for the titration of the DNA into 10 nM DAPI in buffers with 0.5 M (crosses), 1.5 m (open circles), and 2.5 m (open squares) triethylene glycol show that the affinity of DAPI for the (AATT)2 binding site decreases with increasing concentrations of triethylene glycol. (B) Plot of the natural logarithm of the observed equilibrium constant for the association of DAPI with the DNA as a function of the concentrations of acetamide (open circles), betaine (open squares), triethylene glycol (solid circles), and trimethylamine N-oxide (solid squares). A linear least-squares fit using Eq. 5 gives 35 ± 1 waters that are acquired by the complex. The extrapolation to the buffer conditions gives 6.7 (± 0.1) × 108 M−1.
Netropsin and pentamidine
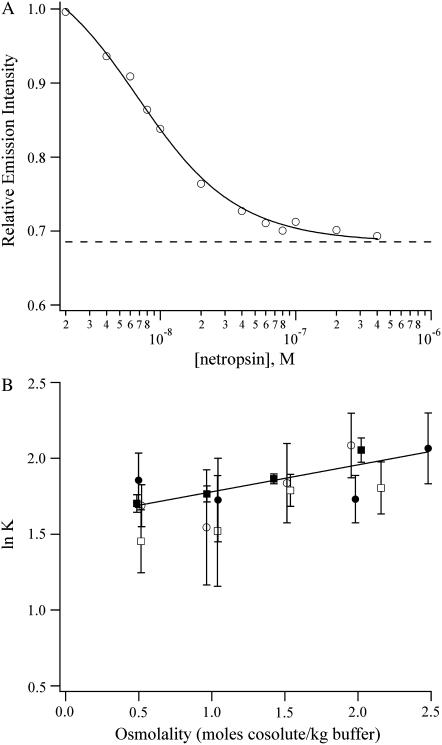
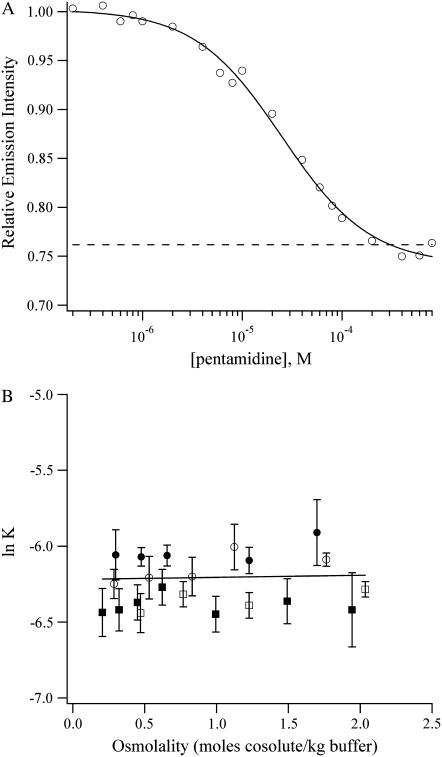
Having established the binding affinities and hydration changes that accompany the binding of DAPI with (AATT)2, netropsin and pentamidine (Fig. 1) were studied. Like DAPI, these drugs also favor binding in the minor groove of A/T-rich sequences. To measure the relative affinities of netropsin and pentamidine for the oligonucleotide, competition titrations using DAPI-(AATT)2 were used, and the progress of the titration was followed using the decrease in the fluorescence intensity (Figs. 3 A and 4 A). The netropsin and pentamidine complexes are hydrated to a comparable extent as the DAPI-(AATT)2 complex, resulting in relative binding affinities that do not change significantly the osmolality of the solution (Figs. 3 B and 4 B). Using the hydration changes for the DAPI-(AATT)2 complex, the differences in hydration between the DNA and DNA-ligand complex are 26 ± 3 waters for netropsin and 34 ± 2 waters for pentamidine.
FIGURE 3.
(A) Binding isotherm for the titration of netropsin to the DAPI-DNA complex in a buffer with 1.1 m triethylene glycol. The solid line is the fit using Eq. 4 which yields a relative equilibrium constant of 7.6 ± 0.6. The dashed line is the emission intensity of the unbound DAPI before the addition of the DNA. (B) Plot of the natural logarithm of the relative equilibrium constants for the exchange reactions (Eq. 2) with netropsin as a function of the osmolality of acetamide (open circles), betaine (open squares), triethylene glycol (solid circles), and trimethylamine N-oxide (solid squares). The change in the amount of water is −9 ± 3 for netropsin. The extrapolation to the buffer conditions gives a relative equilibrium constant of 5.0 ± 0.3, which is consistent with the direct measurement of 4.9 ± 1.1 in the buffer alone.
FIGURE 4.
(A) Binding isotherm for the titration of pentamidine to the DAPI-DNA complex in a buffer with 1.2 m triethylene glycol. The solid line is the fit using Eq. 4 which yields a relative equilibrium constant of 0.0023 ± 0.0002. The dashed line is the emission intensity of the unbound DAPI before the addition of DNA. (B) Plot of the natural logarithm of the equilibrium constants for the exchange reactions (Eq. 2) with pentamidine as a function of the osmolality of acetamide (open circles), betaine (open squares), triethylene glycol (solid circles), and trimethylamine N-oxide (solid squares). The change in the amount of water is −1 ± 2 for pentamidine. The extrapolation to the buffer conditions gives a relative equilibrium constant of 2.0 (± 0.5) × 10−3, which is consistent with the direct measurement of 1.9 (±0.2) × 10−3 in buffer alone.
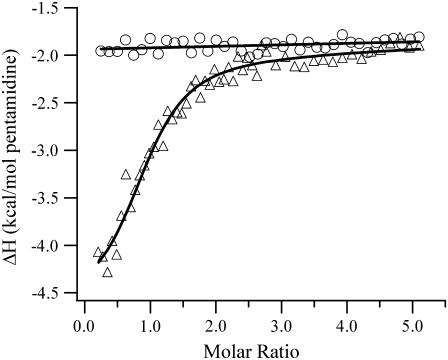
The DAPI concentration was relatively low (<90 nM at the highest osmolyte concentrations) compared to the concentrations where additional binding sites are populated, so aggregation with the DNA should not perturb the results (19,25). For the weaker binding pentamidine, higher concentrations were needed to displace the DAPI. To discern possible additional binding for pentamidine at these higher concentrations, isothermal titration calorimetry studies were used. The derived binding site size is 0.94 ± 0.06 pentamidines/oligonucleotide, which is consistent with binding in the minor groove (Fig. 5). Importantly, titration to higher stoichiometries showed no additional binding modes. To support the exchange model in Eq. 2, the DAPI emission after adding saturating amounts of netropsin or pentamidine was comparable to the emission of the unbound DAPI (Figs. 3 A and 4 A). Furthermore, the fluorescence intensity changes upon addition of netropsin or pentamidine were well fit using the model provided by Eq. 2. Relative binding constants determined from extrapolation to the buffer conditions agree with the values measured in buffer (Figs. 3 B and 4 B). This agreement was also observed for the DAPI reaction with the oligonucleotide and supports osmolyte exclusion. Using the relative binding affinities with the affinity of DAPI for the oligonucleotide, the binding constants for netropsin and pentamidine with the oligonucleotide in the buffer were determined. For netropsin, the equilibrium constant of 3.4 (± 0.7) × 109 M−1 is consistent with the high affinities measured using different experimental techniques (22,26). For pentamidine, the binding affinity of 1.3 (± 0.1) × 106 M−1 is in good agreement with the value of 1.8 (± 0.3) × 106 M−1 derived from the calorimetry studies (Fig. 5).
FIGURE 5.
Integrated heats from the calorimetric titration of 500 μM pentamidine into BPES-50 (circles) and 6 μM (CGCGCAATTGCGCG)2 (triangles) in BPES-50 at 25°C. The one-site model yielded a heat of reaction of −2.4 ± 0.2 kcal/mol from the average of three separate titrations. The heat of dilution is −1.9 ± 0.1 kcal/mol. The heat of reaction for the addition to the DNA solution approaches the heat of dilution after saturation of the minor groove binding site. The titration to the DNA was continued to 11 pentamidines:oligonucleotide with no change in the heat of reaction.
DISCUSSION
The hydration changes that accompany the interaction of three minor groove binding ligands—DAPI, netropsin, and pentamidine—with the (AATT)2 sequence have been studied. Hydration is an important feature of this binding site, as exemplified by its spine of hydration and cations (27,28). The complementary fit of these ligands into the minor groove results in displacement of most of the interfacial waters, as demonstrated by x-ray crystallography and thermodynamic studies (5,29–33). However, waters remain an important feature of these complexes. For example, the crystallography studies show that waters bridge the ligand and DNA, particularly at the ends of the ligands (29–33). The significance of terminally bound water is highlighted by recent studies of ligands whose shapes are not complementary to the minor groove binding site. For example, a diamidine compound with a linear shape was reacted with an (AATT)2 binding site (9). Based on x-ray crystallography and molecular dynamics simulations, waters mediate contacts between one of the amidine groups and the functional groups of the minor groove. The high affinity of this ligand for the binding site strongly suggests that hydration is critical for the formation of this complex. The thermodynamic significance of terminally bound waters was also demonstrated in a calorimetry study of the binding of netropsin with an (AATT)2 binding site (10,22). Two binding modes were observed, of which one was attributed to the complementary fit of the crescent shaped netropsin with the binding site. The conformational flexibility of netropsin permits the formation of alternate structures, and the second binding mode was assigned to the interaction of a more linear netropsin with the binding site. For this complex, simulations supported water-mediated contacts between the minor groove and the amidino nitrogens at the terminus of the netropsin. In addition, the enthalpy and entropy changes for this second binding mode are consistent with previous studies that considered bridging waters in the complex (34).
The above studies show that small numbers of waters link small molecule ligands and the DNA via intermolecular bonding. The involvement of a larger population of waters in the complex is supported by theoretical, spectroscopic, and thermodynamic studies. The full range of bound waters is treated in recent theoretical treatments of macromolecular hydration (35,36). Spectroscopic studies of hydration suggest that bound waters extend away from the macromolecular surface (37). Thermodynamic methods measure the total hydration changes for reactions involving macromolecules (11,12). For example, DNA binding sites gain up to 30 additional waters when associated with small molecule intercalators (18). Importantly, the amount of solvent exposure of the DNA bound ligand dictates the amount of water uptake, a trend that is supported by studies of minor groove binders. For example, Hoechst 33258 binding with the (AATT)2 minor groove enhances the hydration of the complex relative to the reactants by 60 ± 13 waters (19). Netropsin binding with mixed sequence DNA is coupled with the uptake of 50–60 waters (38). Consistent with the results from our study, higher affinity binding affinity sites such as the minor groove are associated with a smaller amount of additional waters. For both Hoechst and netropsin, volumetric techniques measure water expulsion for the binding of netropsin with A/T-rich oligonucleotides (39,40). These differences could be expected because the volumetric methods measure changes in the hydrogen bonding structure of water, whereas the osmotic stress method measures the stoichiometry of water exchange based on the exclusion of the osmolytes.
DNA conformational changes may also contribute to the water acquisition. For example, osmotic stress studies showed that oxygen binding to hemoglobin is accompanied by ∼60 additional waters (41,42). Using structural data for the two forms of hemoglobin, the additional solvation was attributed to the surface area changes due to the reaction. Other studies have also related hydration changes to alterations in the solvent exposure (43,44). For the minor groove binders used in this study, the conformational changes are small but potentially significant (45–48). To further understand the significance of this effect, enhanced solvent exposure could be probed using osmolytes with varying sizes (43,49).
CONCLUSION
Understanding the role of water is important for efforts to target specific DNA sequences using drugs such as small molecules that bind in the minor groove (10,50). The results of this study show that the association of the minor groove binders DAPI, netropsin, and pentamidine with the (AATT)2 binding site is accompanied by the acquisition of water. The ligands differ significantly with respect to their size and functional groups that bind with the DNA, but they have the common structural feature of terminal - groups. The similar amounts of water gained in the complex suggest that the termini of the ligands are important for the hydration changes. This conclusion is consistent with x-ray crystallography and thermodynamic studies which show that terminally located waters provide mediating interactions between the ligand and DNA. In addition, the ligands may produce similar conformational changes in the DNA that could result in enhanced hydration. The osmotic stress method offers a convenient method for assessing the involvement of water in DNA reactions, and our studies provide insight into structural factors that influence water acquisition.
groups. The similar amounts of water gained in the complex suggest that the termini of the ligands are important for the hydration changes. This conclusion is consistent with x-ray crystallography and thermodynamic studies which show that terminally located waters provide mediating interactions between the ligand and DNA. In addition, the ligands may produce similar conformational changes in the DNA that could result in enhanced hydration. The osmotic stress method offers a convenient method for assessing the involvement of water in DNA reactions, and our studies provide insight into structural factors that influence water acquisition.
Acknowledgments
We gratefully acknowledge the support provided by the National Institutes of Health (NIH grants R15GM071370 and P20 RR-016461 (from the National Center for Research Resource)) and the Henry Dreyfus Teacher-Scholar Awards Program. We appreciate the comments of the reviewers.
References
- 1.Bloomfield, V. A., D. M. Crothers, and I. Tinoco Jr. 2000. Nucleic Acids: Structures, Properties, and Functions. University Science Books, Sausaltio, CA.
- 2.Courtenay, E. S., M. W. Capp, C. F. Anderson, and M. T. Record, Jr. 2000. Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry. 39:4455–4471. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg, A. 2000. Ten commandments: lessons from the enzymology of DNA replication. J. Bacteriol. 182:3613–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demeunynck, M., C. Bailly, and W. D. Wilson. 2003. DNA and RNA Binders. Wiley-VCH, New York.
- 5.Haq, I. 2002. Thermodynamics of drug-DNA interactions. Arch. Biochem. Biophys. 403:1–15. [DOI] [PubMed] [Google Scholar]
- 6.Neidle, S. 2001. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 18:291–309. [DOI] [PubMed] [Google Scholar]
- 7.Andreatta, D., J. L. Perez Lustres, S. A. Kovalenko, N. P. Ernsting, C. J. Murphy, R. S. Coleman, and M. A. Berg. 2005. Power-law solvation dynamics in DNA over six decades in time. J. Am. Chem. Soc. 127:7270–7271. [DOI] [PubMed] [Google Scholar]
- 8.Pal, S. K., and A. H. Zewail. 2004. Dynamics of water in biological recognition. Chem. Rev. 104:2099–2123. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen, B., D. Hamelberg, C. Bailly, P. Colson, J. Stanek, R. Brun, S. Neidle, and W. D. Wilson. 2004. Characterization of a novel DNA minor-goove complex. Biophys. J. 86:1028–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freyer, M. W., R. Buscaglia, D. Cashman, S. Hyslop, W. D. Wilson, J. B. Chaires, and E. A. Lewis. 2006. Binding of netropsin to several DNA constructs: evidence for at least two different 1:1 complexes formed from an -AATT-containing ds-DNA construct and a single minor groove binding ligand. Biophys. Chem. In press. [DOI] [PubMed]
- 11.Chalikian, T. V., and K. J. Breslauer. 1998. Volumetric properties of nucleic acids. Biopolymers. 48:264–280. [DOI] [PubMed] [Google Scholar]
- 12.Parsegian, V. A., R. P. Rand, and D. C. Rau. 1995. Macromolecules and water: probing with osmotic stress. Methods Enzymol. 259:43–94. [DOI] [PubMed] [Google Scholar]
- 13.Wyman, J., Jr. 1964. Linked functions and reciprocal effects in hemoglobin: a second look. Adv. Protein Chem. 19:223–286. [DOI] [PubMed] [Google Scholar]
- 14.Stanley, C., and D. C. Rau. 2006. Preferential hydration of DNA: the magnitude and distance dependence of alcohol and polyol interactions. Biophys. J. 91:912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rau, D. C. 2006. Sequestered water and binding energy are coupled in complexes of λ-cro repressor with non-consensus binding sequences. J. Mol. Biol. 361:352–361. [DOI] [PubMed] [Google Scholar]
- 16.Lynch, T. W., and S. G. Sligar. 2000. Macromolecular hydration changes associated with BamHI binding and catalysis. J. Biol. Chem. 275:30561–30565. [DOI] [PubMed] [Google Scholar]
- 17.Sidorova, N. Y., and D. C. Rau. 1995. The osmotic sensitivity of netropsin analogue binding to DNA. Biopolymers. 35:377–384. [DOI] [PubMed] [Google Scholar]
- 18.Qu, X., and J. B. Chaires. 2001. Hydration changes for DNA intercalation reactions. J. Am. Chem. Soc. 123:1–7. [DOI] [PubMed] [Google Scholar]
- 19.Kiser, J. R., R. W. Monk, R. L. Smalls, and J. T. Petty. 2005. Hydration changes in the association of Hoechst 33258 with DNA. Biochemistry. 44:16988–16997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosu, F., V. Gabelica, C. Houssier, and E. De Pauw. 2002. Determination of affinity, stoichiometry and sequence selectivity of minor groove binder complexes with double-stranded oligodeoxynucleotides by electrospray ionization mass spectrometry. Nucleic Acids Res. 30:e82–e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breusegem, S. Y., R. M. Clegg, and F. G. Loontiens. 2002. Base-sequence specificity of Hoechst 33258 and DAPI binding to five (A/T)4 DNA sites with kinetic evidence for more than one high-affinity Hoechst 33258-AATT complex1. J. Mol. Biol. 315:1049–1061. [DOI] [PubMed] [Google Scholar]
- 22.Freyer, M. W., R. Buscaglia, B. Nguyen, W. David Wilson, and E. A. Lewis. 2006. Binding of netropsin and 4,6-diamidino-2-phenylindole to an A2T2 DNA hairpin: a comparison of biophysical techniques. Anal. Biochem. 355:259–266. [DOI] [PubMed] [Google Scholar]
- 23.Hong, J., M. W. Capp, C. F. Anderson, R. M. Saecker, D. J. Felitsky, M. W. Anderson, J. Record, and M. Thomas. 2004. Preferential interactions of glycine betaine and of urea with DNA: implications for DNA hydration and for effects of these solutes on DNA stability. Biochemistry. 43:14744–14758. [DOI] [PubMed] [Google Scholar]
- 24.Cohn, E. J., and J. T. Edsall. 1943. Proteins, Amino Acids, and Peptides as Ions and Dipolar Ions. Reinhold, New York.
- 25.Sovenyhazy, K. M., J. A. Bordelon, and J. T. Petty. 2003. Spectroscopic studies of the multiple binding modes of a trimethine-bridged cyanine dye with DNA. Nucleic Acids Res. 31:2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lah, J., and G. Vesnaver. 2000. Binding of distamycin A and netropsin to the 12mer DNA duplexes containing mixed AT center dot GC sequences with at most five or three successive AT base pairs. Biochemistry. 39:9317–9326. [DOI] [PubMed] [Google Scholar]
- 27.Drew, H. R., and R. E. Dickerson. 1981. Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol. 151:535–556. [DOI] [PubMed] [Google Scholar]
- 28.Shui, X., L. McFail-Isom, G. G. Hu, and L. D. Williams. 1998. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 37:8341–8355. [DOI] [PubMed] [Google Scholar]
- 29.Pjura, P. E., K. Grzeskowiak, and R. E. Dickerson. 1987. Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 197:257–271. [DOI] [PubMed] [Google Scholar]
- 30.Larsen, T. A., D. S. Goodsell, D. Cascio, K. Grzeskowiak, and R. E. Dickerson. 1989. The structure of DAPI bound to DNA. J. Biomol. Struct. Dyn. 7:477–491. [DOI] [PubMed] [Google Scholar]
- 31.Vlieghe, D., J. Sponer, and L. Van Meervelt. 1999. Crystal structure of d(GGCCAATTGG) complexed with DAPI reveals novel binding mode. Biochemistry. 38:16443–16451. [DOI] [PubMed] [Google Scholar]
- 32.Van Hecke, K., P. C. Nam, M. T. Nguyen, and L. Van Meervelt. 2005. Netropsin interactions in the minor groove of d(GGCCAATTGG) studied by a combination of resolution enhancement and ab initio calculations. FEBS J. 272:3531–3541. [DOI] [PubMed] [Google Scholar]
- 33.Edwards, K. J., T. C. Jenkins, and S. Neidle. 1992. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 31:7104–7109. [DOI] [PubMed] [Google Scholar]
- 34.Cooper, A. 2005. Heat capacity effects in protein folding and ligand binding: a re-evaluation of the role of water in biomolecular thermodynamics. Biophys. Chem. 115:89–97. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu, S. 2004. Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proc. Natl. Acad. Sci. USA. 101:1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, P. E. 2006. Chemical potential derivatives and preferential interaction parameters in biological systems from Kirkwood-Buff Theory. Biophys. J. 91:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heugen, U., G. Schwaab, E. Brundermann, M. Heyden, X. Yu, D. M. Leitner, and M. Havenith. 2006. Solute-induced retardation of water dynamics probed directly by terahertz spectroscopy. Proc. Natl. Acad. Sci. USA. 103:12301–12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidorova, N. Y., and D. C. Rau. 1995. The osmotic sensitivity of netropsin analogue binding to DNA. Biopolymers. 35:377–384. [DOI] [PubMed] [Google Scholar]
- 39.Chalikian, T. V., G. E. Plum, A. P. Sarvazyan, and K. J. Breslauer. 1994. Influence of drug-binding on DNA hydration—acoustic and densimetric characterizations of netropsin binding to the Poly(Dadt)·Poly(Dadt) and Poly(Da)·Poly(Dt) duplexes and the Poly(Dt)·Poly(Da)·Poly(Dt) triplex at 25°C. Biochemistry. 33:8629–8640. [DOI] [PubMed] [Google Scholar]
- 40.Han, F., N. Taulier, and T. V. Chalikian. 2005. Association of the minor groove binding drug Hoechst 33258 with d(CGCGAATTCGCG)(2): volumetric, calorimetric, and spectroscopic characterizations. Biochemistry. 44:9785–9794. [DOI] [PubMed] [Google Scholar]
- 41.Colombo, M. F., D. C. Rau, and V. A. Parsegian. 1992. Protein solvation in allosteric regulation: a water effect on hemoglobin. Science. 256:655–659. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu, S. 2004. Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proc. Natl. Acad. Sci. USA. 101:1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LiCata, V. J., and N. M. Allewell. 1997. Functionally linked hydration changes in Escherichia coli aspartate transcarbamylase and its catalytic subunit. Biochemistry. 36:10161–10167. [DOI] [PubMed] [Google Scholar]
- 44.Lynch, T. W., and S. G. Sligar. 2000. Macromolecular hydration changes associated with BamHI binding and catalysis. J. Biol. Chem. 275:30561–30565. [DOI] [PubMed] [Google Scholar]
- 45.Larsen, T. A., D. S. Goodsell, D. Cascio, K. Grzeskowiak, and R. E. Dickerson. 1989. The structure of DAPI bound to DNA. J. Biomol. Struct. Dyn. 7:477–491. [DOI] [PubMed] [Google Scholar]
- 46.Kopka, M. L., C. Yoon, D. Goodsell, P. Pjura, and R. E. Dickerson. 1985. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J. Mol. Biol. 183:553–563. [DOI] [PubMed] [Google Scholar]
- 47.Goodwin, K. D., E. C. Long, and M. M. Georgiadis. 2005. A host-guest approach for determining drug-DNA interactions: an example using netropsin. Nucleic Acids Res. 33:4106–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards, K. J., T. C. Jenkins, and S. Neidle. 1992. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 31:7104–7109. [DOI] [PubMed] [Google Scholar]
- 49.Zimmerberg, J., and V. A. Parsegian. 1986. Polymer inaccessible volume changes during opening and closing of a voltage-dependent ionic channel. Nature. 323:36–39. [DOI] [PubMed] [Google Scholar]
- 50.Ladbury, J. E. 1996. Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem. Biol. 3:973–980. [DOI] [PubMed] [Google Scholar]