Abasic sites are a family of DNA lesions that do not have a nucleobase. Formal hydrolysis of a nucleotide's glycosidic bond to produce a typical abasic site (AP) occurs 10000 times per day per cell.[1] Several oxidized abasic sites (e.g. L, C4-AP), whose chemical properties and biological effects are distinct from those of AP, have also been identified.[2,3] For
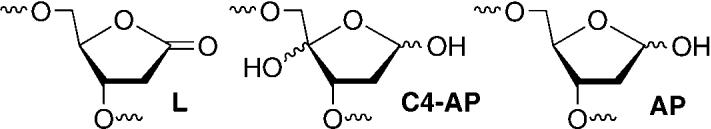
instance, 2-deoxyribonolactone (L) forms DNA–protein cross-links with repair enzymes and the outcome of its bypass during replication in E. coli is distinct from that of an AP site.[4-6] The lactone lesion is often associated with DNA damage induced by antitumor agents, such as neocarzinostatin and the bis-phenanthroline complex of copper, but was believed to form in low yields when diffusible species (e.g. hydroxyl radical) react with DNA due to the inaccessibility of the hydrogen atom at C1′.[7-10] However, 2-deoxyribonolactone was reported to be formed to the exclusion of AP sites when DNA was exposed to tert-butylperoxy radical.[11] Furthermore, recent reports indicate that ionizing radiation produces this biologically interesting lesion in larger amounts than previously thought.[12-15] Herein we report a new reagent for quantitating the formation of 2-deoxyribonolactone and its use in studies directed towards elucidating how the lesion is produced by the most common means of oxidative damage, γ radiolysis.
We recently exploited the selective reactivity of L to develop a biotinylated probe, which covalently tags the lesion with a label that can be quantitated spectroscopically when used in conjunction with avidin and horseradish peroxidase.[16] Building on this basis, we synthesized 1, which offers the
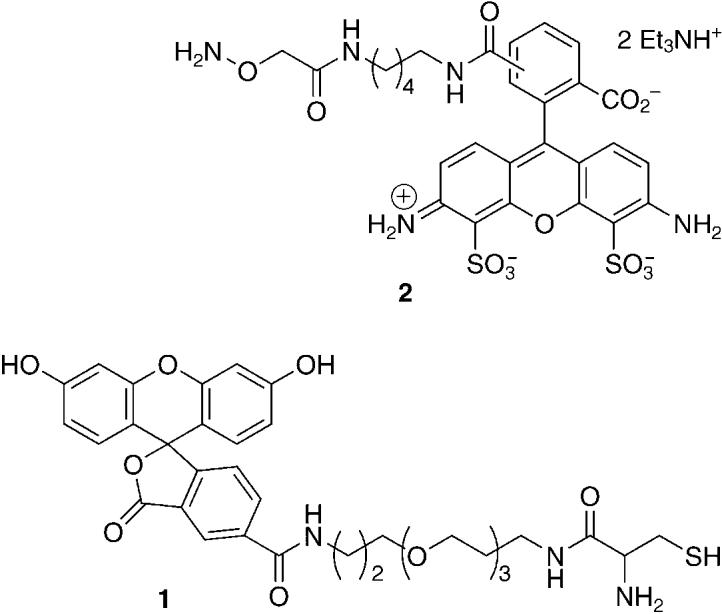
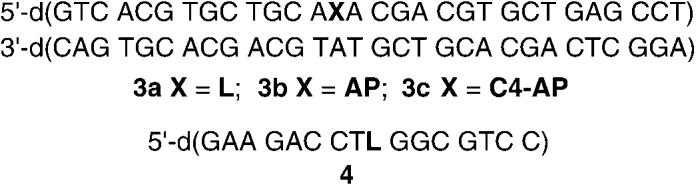
convenience of direct fluorescence detection.[17] The ability of 1 to selectively tag the lactone and not AP or C4-AP sites was determined using duplexes 3a–c that contain the independently synthesized lesions, as previously described (Figure 1).[16] DNA containing the abasic sites was cleaved using N,N′-dimethylethylenediamine (DMEDA), and the butenolide was trapped by 1. Selective 2-deoxyribonolactone detection was achieved by taking advantage of the heat lability of the adducts formed between 1 and aldehyde-containing lesions (AP, C4-AP). Adduct formation was confirmed by MALDI-TOF mass spectrometric analysis of the reaction of 1 with 4.[17] Multiple replications of the
reaction between 1 and L in duplex DNA 3a established the yield for adduct formation to be (17.5 ± 2.2)%.[17] This yield was used in subsequent experiments involving damaged calf-thymus DNA in order to transform the fluorescence signal into a value for the absolute amount of L. Similar studies carried out using 2 established that AP and C4-AP, but not L, react with this probe. Commercial reagent 2 (Alexa Fluor 488 hydroxylamine; Molecular Probes) is a fluorescent version of an aldehyde-reactive probe, which was originally designed for AP site detection.[18] However, alkoxyamine reagents have been found to be useful for labeling a variety of aldehydes in DNA, including C4-AP.[17-20]
Figure 1.
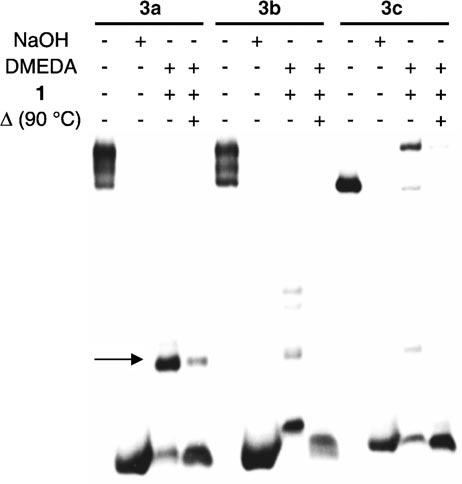
Autoradiogram demonstrating selective detection of 2-deoxyribonolactone (L) in duplex DNA by 1 (50 mm) in the presence of DMEDA (50 mm).
Bernhard and co-workers utilized the release of 5-methylene-2-furanone (5, Scheme 1) upon heating to quantitate L produced in DNA irradiated with X-rays.[14,15] They concluded that 2-deoxyribonolactone accounted for at least 30 % of the sugar damage and postulated that the lesion was formed through an O2-independent mechanism. The fraction of sugar damage attributable to L was based on a comparison of the amounts of 5 and unaltered free base released. We directly compared the yields of respective sugar lesions by using 1 to tag the lactone and 2 to trap aldehyde-containing lesions. Calf-thymus DNA was pretreated with NaOH (0.1m, 37 °C, 20 min) to remove any abasic sites and rehybridized prior to its exposure to either 137Cs or [FeII(EDTA)](EDTA = ethylenediaminetetraacetic acid). Individual samples were split into equal parts and treated with 1 and 2. The yields of L and aldehyde-containing lesions varied linearly over the dose range (25–500 Gy) of 137Cs that the DNA was exposed to (Figure 2). The yield of 2-deoxyribonolactone formation was more than threefold greater than for AP (and all other lesions that react with 2). These data indicate that L accounts for 76 % of the sugar lesions produced in calf-thymus DNA upon exposure to γ radiolysis that are detectable by these reagents. Irradiating the DNA under anaerobic conditions (Figure 2) resulted in a 3.2-fold decrease in 2-deoxyribonolactone formation and 3.4-fold decrease in AP production, indicating that over 75 % of AP and L lesions form through O2-dependent mechanisms.
Figure 2.
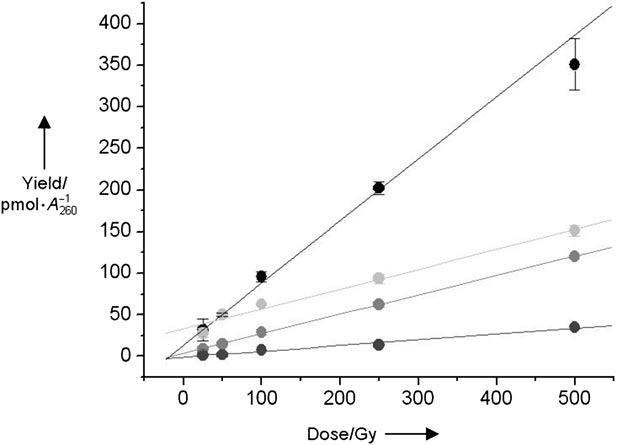
Yield of 2-deoxyribonolactone and AP lesions obtained from g radiolysis of calf-thymus DNA (100 μg/100 μL), normalized for the amount of recovered DNA under aerobic and anaerobic conditions as a function of the dose (Gy = Gray). L aerobic (black); L anaerobic (pale gray); AP aerobic (mid-gray); AP anaerobic (dark gray).
Scheme 1.
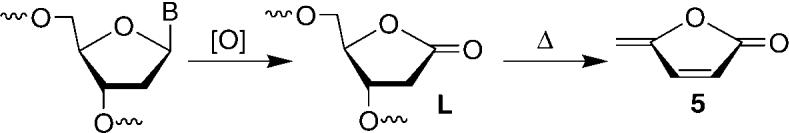
The release of 5-methylene-2-furanone (5) upon heating L.[14,15]
Although these experiments clearly show that 2-deoxyribonolactone is produced in far greater amounts than other abasic lesions (e.g. AP, C4-AP), how L is formed is less certain as a result of the complexities of ionizing radiation. γ Radiolysis damages DNA by two general pathways that produce common products.[21] The “direct” effect of γ radiolysis is the ionization of DNA and the production of cation radicals, including nucleobase holes that are implicated in electron transfer. The cation radicals may react with water and/or deprotonate to form the same radicals generated by hydroxyl radicals, which result from the ionization of water (the “indirect” effect of g radiolysis). Mannitol is often used to probe for the involvement of hydroxyl radicals.[21] Irradiation of calf-thymus DNA containing 1 or 10 mm mannitol (Figure 3) significantly depressed the yields of L and AP, but did not alter their ratio to one another. The role of the hydroxyl radical in the formation of L and AP was probed more directly by analyzing calf-thymus DNA that was treated with [FeII(EDTA)] and H2O2 (8.8 mm).[22] [FeII(EDTA)] generates the hydroxyl radical, which produces a DNA cleavage pattern that is remarkably similar to that of γ radiolysis, indicating that direct strand breaks produced by these agents arise through common reactive species.[23] However, the spectrum of abasic lesions produced by [FeII-(EDTA)] is unknown, and no AP sites were reported in a previous report using tBuOOH.[24] Quantitation of 2-deoxyribonolactone and lesions that react with 2 in DNA treated with [FeII(EDTA)] and H2O2 reveals a linear dependence on the concentration of the metal complex (Figure 4). The ratio of products produced (L:AP = 3.8) is only slightly larger than when DNA is exposed toγradiolysis, and suggests that L accounts for at least 79 % of the detected sugar damage.
Figure 3.
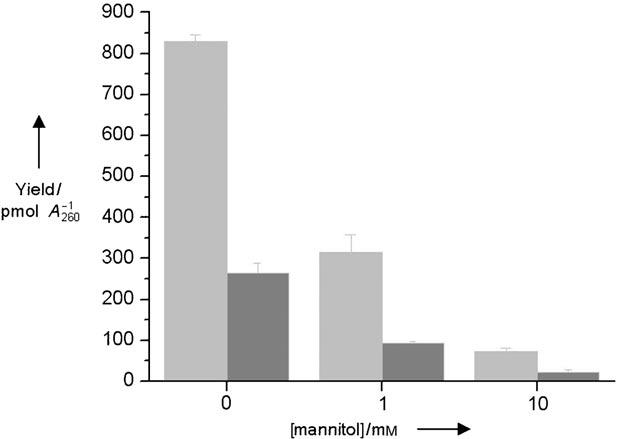
Effect of mannitol concentration on the yield of 2-deoxyribonolactone (light gray) and AP (dark gray) produced in calf-thymus DNA (1.5 μg/10 μL) exposed to γ radiolysis (500 Gy).
Figure 4.
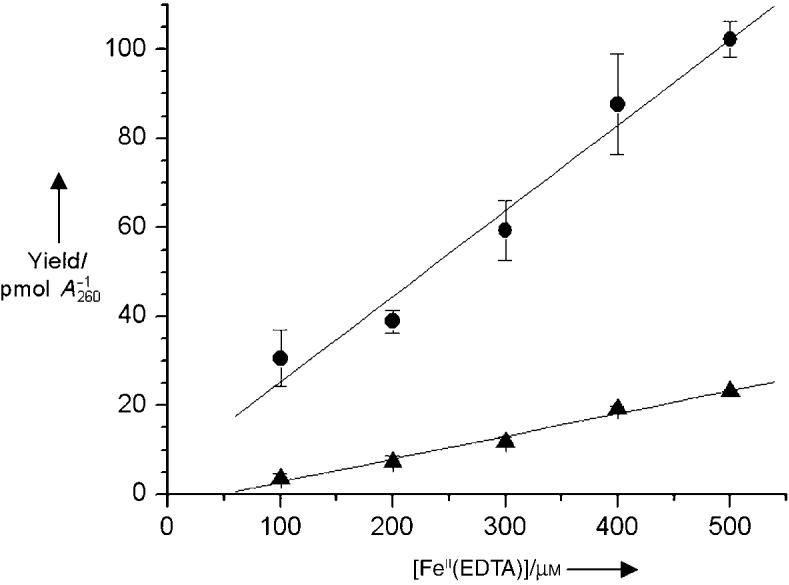
Yield of 2-deoxyribonolactone (●) and AP (▲) sites produced in calf-thymus DNA (50 μg/50 μL) exposed to H2O2 (8.8 mm) and varying concentrations of [FeII(EDTA)].
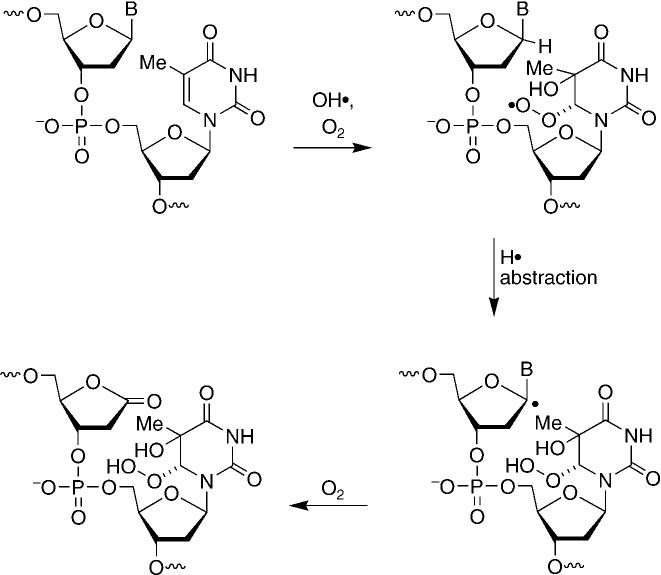
The similar distribution of L and other sugar oxidation products produced byγradiolysis and [FeII(EDTA)] suggest that the agents form these lesions by a common pathway. As [FeII(EDTA)]damages DNA by generating hydroxyl radicals, we propose that the majority of these lesions are produced under these conditions through the indirect effect of γ radiolysis. AP sites may arise from scission of a weakened glycosidic bond resulting from radical addition to the nucleobases. However, formation of 2-deoxyribonolactone requires formal hydrogen atom abstraction of a well-concealed carbon–hydrogen bond and further oxidation of the C1′ radical. Consequently, direct C1′ hydrogen atom abstraction by the hydroxyl radical is expected to be a minor contributor to formation of L.[9] In addition, the large reduction in the yield of L upon removal of O2 suggests that oxidation of a C1′ radical by a proximal purine reactive intermediate is also not a major contributor to the formation of the lesion by γ radiolysis because this mechanism is independent of O2.[14] A recently proposed mechanism for the formation of L is consistent with the predominant generation of nucleobase radicals by γ radiolysis and the large effect of O2 on the production of the lesion (Scheme 2).[12,21] Formation of 2-deoxyribonolactone as part of a tandem lesion enables γ radiolysis to utilize the major family of reactive intermediates it produces and overcomes the inaccessibility of the C1′ hydrogen atom. The biological effects of such tandem lesions are unknown. However, these experiments suggest that they warrant investigation.
Scheme 2.
A recently proposed mechanism for the formation of L.[12,21]
Supplementary Material
Footnotes
We are grateful for support of this research by the National Institute of General Medical Sciences (GM-063028). We thank Professor William Bernhard (University of Rochester) for helpful comments and Dr. Mark Fisher (Varian) for assistance with the fluorimeter.
Supporting information for this article (synthesis and spectral characterization of 1; experimental procedure for the reaction and analysis of calf-thymus DNA; MALDI-TOF MS analyses) is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Kroeger KM, Jiang YL, Kow YW, Goodman MF, Greenberg MM. Biochemistry. 2004;43:6723–6733. doi: 10.1021/bi049813g. [DOI] [PubMed] [Google Scholar]
- 3.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto M, Greenberg MM, Kow YW, Hwang J-T, Cunningham RP. J. Am. Chem. Soc. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- 5.Kroeger KM, Hashimoto M, Kow YW, Greenberg MM. Biochemistry. 2003;42:2449–2455. doi: 10.1021/bi027168c. [DOI] [PubMed] [Google Scholar]
- 6.DeMott MS, Beyret E, Wong D, Bales BC, Hwang J-T, Greenberg MM, Demple B. J. Biol. Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 7.Kappen LS, Goldberg IH. Biochemistry. 1989;28:1027–1032. doi: 10.1021/bi00429a016. [DOI] [PubMed] [Google Scholar]
- 8.Bales BC, PitiK M, Meunier B, Greenberg MM. J. Am. Chem. Soc. 2002;124:9062–9063. doi: 10.1021/ja026970z. [DOI] [PubMed] [Google Scholar]
- 9.Miaskiewicz K, Osman R. J. Am. Chem. Soc. 1994;116:232–238. [Google Scholar]
- 10.Dizdaroglu M, Neuwald K, von Sonntag C. Z. Naturforsch. B. 1976;31:227–233. [Google Scholar]
- 11.Harkin LA, Burcham PC. Biochem. Biophys. Res. Commun. 1997;237:1–5. doi: 10.1006/bbrc.1997.7065. [DOI] [PubMed] [Google Scholar]
- 12.Carter KN, Greenberg MM. J. Am. Chem. Soc. 2003;125:13376–13378. doi: 10.1021/ja036629u. [DOI] [PubMed] [Google Scholar]
- 13.Hong IS, Carter KN, Greenberg MM. J. Org. Chem. 2004;69:6974–6978. doi: 10.1021/jo0492158. [DOI] [PubMed] [Google Scholar]
- 14.Roginskaya M, Bernhard WA, Marion RT, Razskazovskiy Y. Radiat. Res. 2005;163:85–89. doi: 10.1667/rr3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roginskaya M, Razskazovskiy Y, Bernhard WA. Angew. Chem. 2005;117:6366–6369. doi: 10.1002/anie.200501956. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:6210–6213. doi: 10.1002/anie.200501956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato K, Greenberg MM. J. Am. Chem. Soc. 2005;127:2806–2807. doi: 10.1021/ja0426185. [DOI] [PubMed] [Google Scholar]
- 17. See Supporting Information.
- 18.Ide H, Akamatsu K, Kimura Y, Michiue K, Makino K, Asaeda A, Takamori Y, Kubo K. Biochemistry. 1993;32:8276–8283. doi: 10.1021/bi00083a031. [DOI] [PubMed] [Google Scholar]
- 19.Byrns MC, Vu CC, Neidigh JW, Abad J-L, Jones RA, Peterson LA. Chem. Res. Toxicol. 2006;19:414–420. doi: 10.1021/tx050302k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong YC, Sangaiah R, Nakamura J, Pachkowski BF, Ranasinghe A, Gold A, Ball LM, Swenberg JA. Chem. Res. Toxicol. 2005;18:51–60. doi: 10.1021/tx049853l. [DOI] [PubMed] [Google Scholar]
- 21.Von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair. Springer; Berlin: 2006. [Google Scholar]
- 22.Price MA, Tullius TD. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- 23.Pogozelski WK, McNeese TJ, Tullius TD. J. Am. Chem. Soc. 1995;117:6428–6433. [Google Scholar]
- 24.Kanazawa A, Sawa T, Akaik T, Maeda H. Cancer Lett. 2000;156:51–55. doi: 10.1016/s0304-3835(00)00439-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.