Abstract
Networks of transcriptional regulatory proteins dictate specification of neural lineages from multipotent retinal progenitors. Rod photoreceptor differentiation requires the basic motif–leucine zipper (bZIP) transcription factor NRL, because loss of Nrl in mice (Nrl−/−) results in complete transformation of rods to functional cones. To examine the role of NRL in cell fate determination, we generated transgenic mice that express Nrl under the control of Crx promoter in postmitotic photoreceptor precursors of WT and Nrl−/− retina. We show that NRL expression, in both genetic backgrounds, leads to a functional retina with only rod photoreceptors. The absence of cones does not alter retinal lamination, although cone synaptic circuitry is now recruited by rods. Ectopic expression of NRL in developing cones can also induce rod-like characteristics and partially suppress cone-specific gene expression. We show that NRL is associated with specific promoter sequences in Thrb (encoding TRβ2 transcription factor required for M-cone differentiation) and S-opsin and may, therefore, directly participate in transcriptional suppression of cone development. Our studies establish that NRL is not only essential but is sufficient for rod differentiation and that postmitotic photoreceptor precursors are competent to make binary decisions during early retinogenesis.
Keywords: cell fate determination, development, gene regulation, retina, synaptic organization
Neuronal cell fate is determined by a hierarchical, stepwise process of binary decisions, commencing with multipotent progenitors that give rise to distinct cell lineages (1–3). The neural retina is an attractive model to investigate cell-fate determination; it contains seven major cell types that derive from common pool(s) of multipotent progenitor cells (4, 5). These retinal progenitors pass through sequential waves of competence, during which postmitotic cells can be specified to only a subset of neuronal fates (1, 6). Birth-dating studies in rodents indicate that ganglion cells, horizontal cells, cone photoreceptors, and amacrine cells are born prenatally, whereas most rod photoreceptors, bipolar cells, and Müller glia are generated postnatally (7–9). The orderly sequence of cell birth and a considerable overlap in their generation suggest a sequential program of cell intrinsic mechanisms and extrinsic signals that control cell fate decisions (10–16).
Each neuronal lineage is meticulously established by highly coordinated transcription factor network(s) in response to local microenvironmental cues (17). Although extrinsic factors can promote differentiation (18, 19), heterochronic mixing experiments demonstrate that progenitor cells at a particular time in development cannot be induced to generate temporally inappropriate cell types (1, 20). Additionally, intrinsic priming of retinal progenitors appears to supersede the influence of environmental signals in specifying cell fate (21). Whether commitment of lineage-restricted precursors to a specific differentiation pathway is unidirectional has not been clearly elucidated.
Postmitotic plasticity was first revealed in low-density retinal cell cultures derived from embryonic chicks; cells isolated on embryonic day (E)6 became photoreceptors shortly after terminal mitosis, whereas those from E8 embryos gave rise to nonphotoreceptors, suggesting that the fate of a cell could be changed in response to the microenvironment (22). In another demonstration of cellular plasticity, treatment of retinal explants with ciliary neurotrophic factor (CNTF) was sufficient to block rhodopsin expression in postmitotic rod precursors and resulted in the expression of bipolar interneuron markers (23). Interestingly, ETS transcription factors are selectively expressed during motor and sensory neuron development but only after their axons reach the periphery, suggesting that these proteins confer postmitotic subtype identity during the establishment of selective connections (24, 25). These findings indicate that neural identity can be specified even after the terminal cell cycle exit; however, direct in vivo evidence of postmitotic plasticity has not been reported.
The Maf-family transcription factor Nrl is expressed specifically in postmitotic rod photoreceptors of the retina and in the pineal gland (26, 27). NRL interacts with the homeodomain protein CRX (28), orphan nuclear receptor NR2E3 (29), and other retinal proteins (30–32) to regulate the expression of rod-specific genes (26, 33). NRL is essential for rod differentiation, because rods are transformed to functional S-cones in the Nrl−/− mouse retina (26, 34, 35). The apparent switch from rod to S-cone fate in the Nrl−/− mouse suggests that postmitotic photoreceptor precursors retain some degree of plasticity. In this report, we generated a series of transgenic mice that express Nrl during early and late stages of photoreceptor differentiation in WT or Nrl−/− background. We demonstrate that NRL is sufficient to guide postmitotic photoreceptors toward rod lineage and that photoreceptor precursors are competent to make binary decisions of acquiring rod versus cone identity.
Results
Overexpression of Nrl in Photoreceptor Precursors Drives Rod Differentiation at the Expense of Cones.
We hypothesized that if cones develop from a unique pool of competent cells, early cone precursors would not be responsive to NRL. On the other hand, transformation of cone precursors to rods by NRL would indicate an intrinsic capacity to give rise to both rods and cones. To directly test this, we generated transgenic mouse lines (Crxp-Nrl/WT) expressing Nrl under the control of a previously characterized 2.5 kb proximal promoter of the Crx gene (Crxp-Nrl), which is specifically expressed in postmitotic cells that can develop into either cone or rod photoreceptors (36, 37).
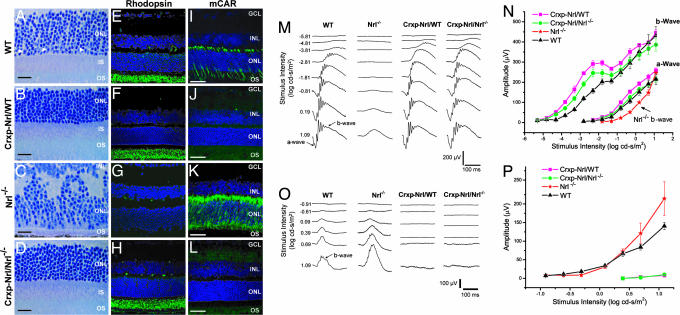
Light micrographs of semithin (plastic) sections of Crxp-Nrl/WT mouse retina showed normal laminar organization (Fig. 1A and B). Immunofluorescence studies demonstrated comparable rhodopsin expression relative to WT and Nrl−/− mice (Fig. 1 E–G); however, staining of cone-specific markers [cone arrestin, peanut agglutinin (PNA), S-opsin, and M-opsin] was undetectable in cryosections and flat-mount preparations from transgenic retinas (Fig. 1 I–K, and data not shown). Confocal examination of the outer nuclear layer revealed only the photoreceptor nuclei with dense chromatin [supporting information (SI) Fig. 6 A and B] that are characteristics of rods in the WT retina (38). Dark-adapted corneal flash electroretinograms (ERGs) from Crxp-Nrl/WT mice revealed normal rod function even at 6 mo (Fig. 1 M and N and data not shown), whereas the cone-derived photopic ERG response was absent at all ages (Fig. 1 O and P, and data not shown). These studies suggested a complete absence of cone functional pathway in the Crxp-Nrl/WT mice. Consistent with these results, quantitative RT-PCR analysis demonstrated no expression of cone phototransduction genes in the Crxp-Nrl/WT retina, with little or no change in rod-specific genes (SI Fig. 6C).
Fig. 1.
Expression of Nrl in cone precursors. (A–L) Toluidine blue stainings of WT (A), Crxp-Nrl/WT (B), Nrl−/− (C), and Crxp-Nrl/Nrl−/− (D) retinal sections demonstrate unique chromatin pattern in the photoreceptor layer for cones (indicated by arrowhead) and rods. Normal laminar structure is observed in both Crxp-Nrl/WT (B) and Crxp-Nrl/Nrl−/− (D) plastic sections. Immunohistochemical markers for rod photoreceptors (rhodopsin) can be detected in WT (E), Crxp-Nrl/WT (F), and Crxp-Nrl/Nrl−/− (H) retina but not in Nrl−/− (G). The pan cone photoreceptor marker, cone arrestin, is present only in WT (I) and Nrl−/− (K) retina, but is largely absent in the Crxp-Nrl/WT (J) and Crxp-Nrl/Nrl−/− (L). (M–P) ERG intensity series and responses were recorded from 2-mo-old WT, Nrl−/−, Crxp-Nrl/WT, and Crxp-Nrl/Nrl−/− mice under dark- (scotopic ERG; M and N) and light-adapted (photopic ERG; O and P) conditions. The x axes for M and O indicate time lapsed after flash. Stimulus energy is indicated (log cd-s/m2). OS, outer segments; IS, inner segments, ONL, outer nuclear layer; INL, inner nuclear layer. (Scale bars: A–D, 25 μm; E–L, 50 μm.)
We then bred the Crxp-Nrl transgenic mice into the Nrl−/− background (Crxp-Nrl/Nrl−/−) to test whether Nrl expression in a cone-only retina could convert a retina composed solely of cones to rods as seen in the Crxp-Nrl/WT mice. Analysis of retinal morphology uncovered a remarkable transformation of a dysmorphic retina with whorls and rosettes in the Nrl−/− mice (34) to a WT-like appearance (Fig. 1 C and D). Images from toluidine blue-stained retinal sections revealed clear extended outer segments and a highly organized laminar structure (Fig. 1D). Similar to the WT (38), and unlike the all-cone retina in Nrl−/− mice (34), the outer nuclear layer of Crxp-Nrl/Nrl−/− retina had rod-like nuclei with dense chromatin. Immunolabeling of adult Crxp-Nrl/Nrl−/− retinal sections demonstrated a complete absence of cone proteins (cone arrestin data are shown in Fig. 1L). In contrast to the Nrl−/− retinas (Fig. 1G), Crxp-Nrl/Nrl−/− mice displayed normal levels of rhodopsin (Fig. 1H). No photoreceptor degeneration was evident by histology or ERG at least up to 6 mo (Fig. 1, data not shown).
Retinal Synaptic Architecture Is Modified in the Absence of Cones.
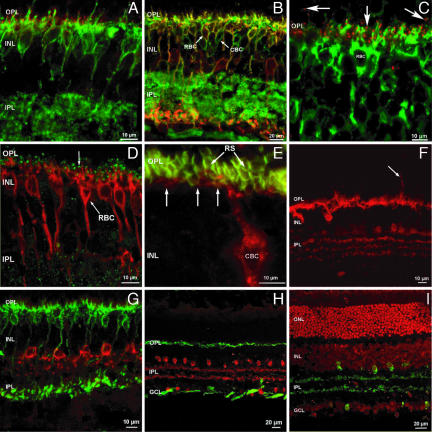
Given that a complete rod-only retina did not reveal gross changes in retinal morphology, we contemplated whether cones are essential for proper development and lamination of cone-connected neurons. Cones are presynaptic to dendrites originating from the cell bodies of horizontal cells and to at least nine different types of cone bipolar neurons (39, 40). Immunostaining of Crxp-Nrl/WT retinas with a panel of cell-type-specific antibodies (41) did not reveal any major difference in the distribution of the marker proteins for horizontal, bipolar, amacrine, and glial cells (Fig. 2). Despite the absence of cones, it was apparent that both the ON and OFF subtypes of cone bipolar cells were retained (Fig. 2 A, B, and E). All ON bipolar neurons (both rod and cone bipolar cells) carried metabotropic glutamate receptors on their dendritic tips (mGluR6), and thus, presumably, they were postsynaptic to rod spherules. It was unclear from these studies whether cone bipolar cells belonging to the OFF functional type received synapses from rod photoreceptors. The dendrites of one type of OFF cone bipolar cells, marked with Neurokinin receptor 3 (NK3-R), form basal (or flat) junctions with cone pedicles in the outer plexiform layer (SI Fig. 7). Although confocal microscopy does not reach the necessary resolution to detect such putative contacts, it is apparent from the preparations that not all of the dendrites of NK3-R-positive cone bipolar cells come in close apposition to the rod spherules and that basal junctions are therefore unlikely (Fig. 2E). It remains to be established whether and how rod spherules make connections to OFF cone bipolar cells and whether the OFF channel gains access to the scotopic pathway.
Fig. 2.
Synaptic organization of the inner retina in the absence of cones. (A) The glutamatergic receptor mGluR6 is clustered selectively at puncta (red) in the OPL, on the dendritic tips of ON bipolar cells, labeled by G0α antibodies (green). (B) G0α antibody labels the whole population of ON bipolar cells (green signal), whereas PKCα labels rod bipolar cells only (RBC; red). Rod bipolar neurons are therefore double-labeled by both antibodies and appear yellow. Green cells are ON cone bipolar cells (indicated as CBC). (C) mGluR6 receptors are labeled as red puncta located at the dendritic tips of rod bipolar cells, labeled green by PKCα antibodies. In addition, clusters of mGluR6 are visible in the OPL, but not in association with rod biolar cell dendrites. These clusters are likely to be associated to the dendrites of ON cone bipolar cells. (D) Rod bipolar cells (RBC), labeled by PKCα (red), are postsynaptic to photoreceptors in the OPL at ribbon synapses (indicated by R), as indicated by antibodies against kinesin, a synaptic ribbon marker (green). (E) High magnification of one type of cone bipolar cell (CBC), labeled with NK3-R antibody (red). Rod spherules (RS) are labeled with anti-PSD95 antibody (green). Few dendrites of cone bipolar cells reach the basal aspect of some spherules (arrows); however, many spherules do not appear apposed to CBC dendrites, although these belong to one of the most abundant types of retinal cone bipolar cell. (F) Calbindin staining (red) of the Crxp-Nrl/WT retina shows a normal distribution of intensely labeled horizontal cells and weakly fluorescent amacrine cells with their processes in the IPL. Occasionally, horizontal cell sprouts are observed (arrow). (G) AII amacrine cells (the most abundant population of mammalian amacrines) are specifically stained with DB3 antibodies (red). They exhibit a typical, bistratified morphology. The innermost dendrites terminate in apposition to the axonal endings of rod bipolar cells, stained green by PKCα antibodies. (H) Cholinergic amacrine cells are stained in the transgenic retina by ChAT antibodies (red). The cells form two mirror symmetric populations of neurons. Axonal complexes of horizontal cells are labeled with neurofilament antibodies (green). Axonal fascicles of ganglion cells are also intensely stained in the optic fiber layer. (I) Ethidium bromide nuclear staining (red) and ChAT immunostaining (green) demonstrate the normal layering and lamination of the transgenic retina. OS, outer segments; ONL, outer nuclear layer; INL, inner nuclear layer; OPL, outer plexiform layer; IPL, inner plexiform layer.
To study the morphology of horizontal cells, we stained Crxp-Nrl/WT retinas with a calbindin antibody (Fig. 2F). Although no gross changes were observed, we noticed rare ectopic sprouts emerging from the outer plexiform layer and extending into the outer nuclear layer. Other examined markers also revealed a normal distribution throughout the retina (see Fig. 2 G–I). AII amacrine neurons exhibited their peculiar bistratified morphology (Fig. 2G). Cholinergic amacrine cells (Fig. 2 H and I) showed a typical distribution in two mirror-symmetric populations. Dopaminergic amacrines and Müller glial cells also showed normal organization (data not shown). Thus, besides the likely reconnections of ON cone bipolar and horizontal cells to rods, the retina from Crxp-Nrl/WT mice was indistinguishable from WT.
Ectopic Expression of NRL Can Suppress Cone Function and Induce Rod Characteristics in a Subset of Photoreceptors Expressing S-opsin.
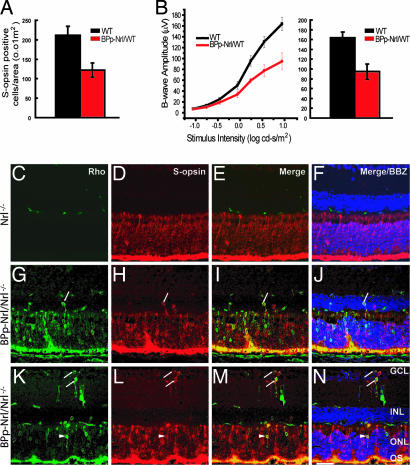
The onset of S-opsin expression begins at E16–E18 in rodents (42, 43). To further delineate NRL's role in cell fate determination, we generated transgenic mouse lines (BPp-Nrl/WT) expressing NRL under the control of a previously characterized S-opsin promoter (44). Immunostaining revealed a significant decrease of S-opsin-positive cells in the inferior region of flat-mounted retinas (Fig. 3A). Consistent with histological and immunohistochemical analysis, ERGs from the BPp-Nrl/WT mice showed a 50% reduction in the photopic b-wave amplitude compared with the WT (Fig. 3B); however, scotopic ERG a- and b-wave amplitudes were largely unaffected (data not shown).
Fig. 3.
Ectopic expression of Nrl in S-opsin-expressing cone photoreceptors. (A and B) Quantification of S-cones in the inferior domain of flat-mounted retinas from WT and BPp-Nrl/WT mice with anti-S-opsin antibody (A) revealed a 40% decrease in S-cones. Light-adapted ERG photoresponses from WT and BPp-Nrl/WT mice are shown in B [photopic b-wave (Left) and photopic b-wave at maximum intensity (Right)]. In BPp-Nrl/WT mice, ≈50% reduction in the photopic b-wave amplitude is observed compared with the WT mice. (C–N) Immunostaining of cryosections from Nrl−/− retina show the lack of rhodopsin expression and higher S-opsin expression in the ONL (C–F). In the BPp-Nrl/Nrl−/− retina rhodopsin expression can be detected in the ONL and the OS (G and K). Hybrid photoreceptors expressing both S-opsin (H and L) and rhodopsin can be observed in the ONL, INL, and the GCL (G–N). OS, outer segments; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; BBZ, bisbenzamide. (Scale bar: C–N 50 μm.) Arrows refer to colocalization of S-opsin and rhodopsin in the INL and GCL; arrowheads refer to colocalization of S-opsin and rhodopsin in the ONL.
We then transferred the BPp-Nrl transgene to the Nrl−/− background (BPp-Nrl/Nrl−/−) mice. Ectopic expression of Nrl in the all-cone Nrl−/− retina, even at this stage (i.e., under the control of S-opsin promoter), resulted in rhodopsin staining in the ONL; however, as in the Nrl−/− mice (Fig. 3 C–F) the outer and inner segments remained stunted (Fig. 3 G–N). The BPp-Nrl/Nrl−/− retina also revealed hybrid cells that expressed both S-opsin and rhodopsin in ONL, INL, and ganglion cell layer (Fig. 3 G–N; SI Fig. 8A). ERG data showed that, although the phototopic b-wave (cone-derived) was somewhat reduced, the scotopic b-wave amplitude was still undetectable in BPp-Nrl/Nrl−/− mice (data not shown).
To examine the fate of S-opsin-expressing cells, we mated the BP-Cre transgenic mice (that expresses Cre-recombinase under the control of the same S-opsin promoter) (44) with the R26R reporter line and the BPp-Nrl/WT line (SI Fig. 8 B–K). A large number of Cre-negative cells were labeled with β-galactosidase in the BP-Cre; R26R; BPp-Nrl/WT background (SI Fig. 8 B–K). Approximately 40% of β-galactosidase-positive cells did not colocalize with S-opsin. Their position in the ONL and the lack of S-opsin staining indicate that these are rod photoreceptors, suggesting a possible fate switch in response to ectopic NRL expression. However, we could not validate their identity as rods because the rod marker, rhodopsin, does not clearly label the nuclear layer. TUNEL staining of sections from E18 retina did not detect obvious differences between WT and BPp-Nrl/WT mice (data not shown).
NRL Can Associate with Cone-Specific Promoter Elements.
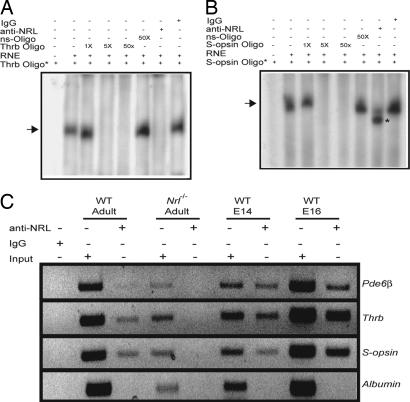
NRL is established as a positive transcriptional regulator of rod-specific genes (28, 31, 33, 45–47). To examine whether NRL can directly modulate cone-specific promoters, we screened 3 kb of 5′ upstream promoter regions of the two cone-expressed genes, Thrb [encoding Trβ2 that is involved in M-cone differentiation, (14)] and S-opsin, for the presence of Nrl or Maf response element (NRE/MARE) (46). Oligonucleotides spanning the single putative NRE sites, identified within the Thrb and S-opsin promoters, were used for EMSA with bovine retinal nuclear extracts. We detected a shifted band that could be specifically competed by the addition of 50-fold molar excess of unlabeled NRE-oligonucleotide but not a random oligonucleotide (Fig. 4A and B). The addition of anti-NRL antibody abolished the shifted band for the Trβ2 oligonucleotide (Fig. 4A), whereas S-opsin promoter–protein complex demonstrated an increased mobility in the native polyacrylamide gel (Fig. 4B). Notably, disappearance of the shifted band may occur because of the dynamic nature of some DNA–protein interactions, whereas the net charge-to-mass (e/m) ratio of the ternary complex determines their rate of mobility in a native polyacrylamide gel (48). Similar results were obtained when the radiolabeled oligonucleotides were incubated with anti-NRL antibody simultaneously with the retinal nuclear extract or with the nuclear extract preincubated with the anti-NRL antibody for 15 min. (data not shown). No effect on the gel-shift was observed in the presence of control rabbit IgG.
Fig. 4.
Association of Nrl to cone-specific promoters. (A and B) EMSA. Radiolabeled double-stranded oligonucleotides from Thrb and S-opsin promoters were incubated with RNE, followed by nondenaturing PAGE. Lanes are as indicated. Arrows represent specific shifted bands. Competition experiments were performed with increasing concentration (1-, 5-, or 50-fold molar excess, respectively) of unlabeled specific oligonucleotide or 50-fold higher concentration of nonspecific (ns) oligonucleotide, to validate the specificity of band shift. Anti-NRL or normal rabbit IgG was added in some of the reactions, as indicated. Disappearance (see A) or increased mobility of the shifted band (B; shown by asterisk) was detected with anti-NRL antibody but not IgG. These experiments were performed three times, and similar results were obtained. (C) ChIP assay. WT or Nrl−/− mouse retina was used for ChIP with anti-NRL or rabbit IgG antibody. The positive and negative controls for ChIP assays are Pde6b and albumin, respectively. Lanes are as indicated. Input DNA served as positive control for PCR.
To further evaluate the association of NRL with Thrb and S-opsin promoter elements in vivo, we performed ChIP assays using WT embryonic and adult mouse retinas. PCR primer sets spanning the Thrb and S-opsin NRE-amplified specific products with DNA immunoprecipitated with the anti-NRL antibody but not with the rabbit IgG (Fig. 4C). ChIP experiments using the Nrl−/− mouse retina (negative control) did not reveal specific amplified products (Fig. 4C).
Discussion
Specification of neuronal cell fate involves changes in progenitor competence over time (1). In the developing retina, progenitor cells show heterogeneity in their developmental competence and have the potential to produce many or all fates. Genesis of rod photoreceptors overlaps with the birth of all other retinal cell types. How regulatory factors orchestrate this decision-making has not been clearly delineated. Because CRX is expressed in all postmitotic photoreceptor precursors (36), we hypothesized that CRX-expressing cells are not committed to a specific fate and are plastic, and that NRL dictates the rod fate over a developmental time window. Our transgenic data (see Fig. 1) strongly supports the hypothesis of postmitotic plasticity (see ref. 22) in mammalian retina, because expression of NRL in even CRX-expressing cone precursors produces functional rods. It is therefore the timing of expression, availability, and amount/activity of NRL that determine whether the postmitotic precursor will acquire a rod or a cone fate (Fig. 5; SI Fig. 9). Surprisingly, ectopic expression of NRL can still drive a subset of presumptive S-opsin-expressing cone photoreceptors toward the rod lineage, although not to a fully functional phenotype.
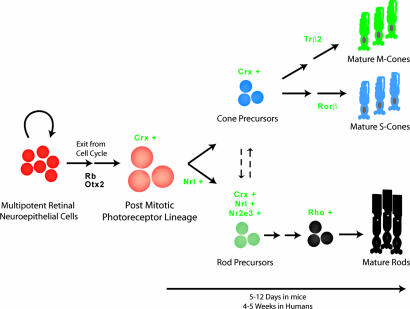
Fig. 5.
A model of photoreceptor specification. Otx2 and Rb influence multipotent retinal neuroepithelial cells to exit cell cycle. We hypothesize that Crx is the competence factor in postmitotic photoreceptor precursors. The cells that express Nrl are committed to rod photoreceptor fate, with subsequent expression of Nr2e3. The cells expressing only Crx are cone precursors. We propose that a degree of plasticity exists in early retinal development, such that changes in Nrl and/or Nr2e3 expression can lead to alterations in final ratio of rod and cone photoreceptors. Additional transcription factors (14, 62) are required to guide the development to mature photoreceptors.
NR2E3 is a photoreceptor-specific orphan nuclear receptor that is shown to suppress cone-specific gene expression in cultured cells (49, 50) and in transgenic mice (37). NRL is upstream of NR2E3 in regulatory hierarchy of photoreceptor differentiation (29, 34) and is an established activator of rod-specific genes (33, 51). It was therefore of interest to examine whether NRL can also directly suppress cone genes or all of its effects on cone differentiation are mediated by NR2E3. The finding of NRL binding to Thrb and S-opsin promoter sequences suggests that NRL functions, probably together with NR2E3, in suppressing cone differentiation in vivo. NRL's role as a transcriptional repressor is not surprising, because key regulatory proteins can control transcription through context-dependent combinatorial mechanisms (52). Notably, distinct phosphorylated isoforms of NRL are expressed during retinal development (27), and phosphorylation differences are suggested to modulate transcriptional activity of NRL by altering nuclear translocation, DNA binding, or protein interactions (63). Additional studies are required to evaluate whether different NRL isoforms participate in transcriptional activation versus repression during early photoreceptor development.
The loss of cones produces an alteration in retinal synaptic connectivity such that ON cone bipolar cells are now connected to rods in the absence of their natural synaptic partner (i.e., cones). Our results complement the previous findings (53), demonstrating that neurons of the rod pathway are recruited by newly generated cones in the Nrl−/− retina. Our data provide support to the hypothesis that, although intrinsic mechanisms may guide the formation of synaptic connections, the strength(s) of afferent input determine the final establishment of synaptic circuitry (54–56). In the cone-only Nrl−/− retina and the rod-only transgenic mice reported here, a lack of input from the other competing afferent neurons leads to functional synaptic connections that are not usually observed in the WT mouse retina.
Our studies establish that NRL is not only essential (34) but is also sufficient for rod genesis. To what extent NRL can dictate rod specification in proliferating cells or other subsets of retinal precursors will require further investigations using transgenic mice expressing NRL under the control of early cell-type promoters. In utero infection of mouse embryonic retinas (57) with Nrl may also demonstrate whether we can stretch the developmental potential of retinal progenitors. Our studies give rise to the prospect of exploiting the plastic nature of retinal precursors to replenish dying rods in degenerative retinal diseases by ectopic NRL expression in retinal stem cells (58, 59) or by transplantation of NRL-expressing progenitors (60).
Materials and Methods
Plasmid Constructs and Generation of Transgenic Mice.
A 2.3-kb mouse Crx promoter DNA (from −2286 to +72, GenBank accession nos. AF335248 and AF301006) and the Nrl coding region (GenBank accession no. NM008736) with an additional Kozak sequence were amplified and cloned into a modified promoterless pCl (pCIpl) vector (44). The 3.7-kb Crxp-Nrl insert was purified and injected into fertilized Nrl−/− (mixed background of 129 × 1/SvJ and C57BL/6J) mouse oocytes (University of Michigan transgenic core facility). Transgenic founders were bred to the Nrl−/− mice to generate F1 progeny. The progeny was also mated to C57BL/6J to generate Crxp-Nrl/WT mice. The BPp-Nrl transgenic mice were generated in a similar manner, except that a 520-bp mouse S-opsin promoter DNA (44) was used. All studies involving mice were performed in accordance with institutional and federal guidelines and approved by the University Committee on Use and Care of Animals at the University of Michigan.
Immunohistochemistry and Confocal Analysis.
Retinal sections and dissociated cells were prepared as described (29, 41) and probed with specific antibodies (listed in SI Methods). Sections were visualized under an Olympus FluoView 500 laser scanning confocal microscope (Olympus, Melville, NY) or a Leica TSC NT confocal microscope (Leica Microsystems, Milan, Italy), equipped with an argon–krypton laser. Images were digitized by using FluoView software version 5.0 or Metamorph 3.2 software.
ChIP.
Mouse retinas from different developmental stages were subjected to ChIP analysis using a ChIP-IT kit (Active Motif, Carlsbad, CA). IP was performed by using anti-NRL or normal rabbit Ig (IgG). PCR primers, derived from the Thrb and S-opsin promoter region (GenBank accession nos. NT_039340.6 and NT_039595.6, respectively) spanning the putative NRE, were used for amplification (from nucleotides 26331250 to 26331458 and 13773280 to 13773502, respectively) by using immunoprecipitated DNA as template. The albumin PCR primers were 5′-GGACACAAGACTTCTGAAAGTCCTC-3′ and 5′-TTCCTACCCCATTACAAAATCATA-3′.
EMSA.
Oligonucleotides spanning the putative NRE were radiolabeled by using [γ-]32P-ATP (Amersham Biosciences, Piscataway, NJ) and incubated in binding buffer (20 mM Hepes, pH 7.5/60 mM KCl/0.5 mM DTT/1 mM MgCl2/12% glycerol) with bovine retinal nuclear extract [RNE; (30)] (20 μg) and 50 μg/ml poly(dI-dC) for 30 min at room temperature, as described (61) For competition experiments, nonradiolabeled oligonucleotides were used in molar excess of the labeled oligonucleotides. In some experiments, antibodies were added after the incubation of 32P-labeled oligonucleotides with RNE. Samples were analyzed by 7.5% nondenaturing PAGE, followed by autoradiography.
Electroretinography.
ERGs were recorded as described (34).
Supplementary Material
Acknowledgments
We thank R. Masland, P. Raymond, P. F. Hitchcock, J. Brzezinski, T. Glaser, P. Gage, H. Cheng, and T. Saunders for stimulating discussions and/or comments on the manuscript; S. Lentz, M. Gillett, and M. Van Keuren for technical assistance; and S. Ferrara for administrative support. This work was supported by National Institutes of Health Grants EY011115, EY007003, and EY012654; The Foundation Fighting Blindness; and Research to Prevent Blindness. The core facilities of the Michigan Diabetes Research and Training Center (NIH5P60 DK20572) were also used for this work.
Abbreviations
- En
embryonic day n
- ERG
electroretinogram
- NRE
Nrl response element.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605934104/DC1.
References
- 1.Livesey FJ, Cepko CL. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 2.Malicki J. Curr Opin Neurobiol. 2004;14:15–21. doi: 10.1016/j.conb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 4.Turner DL, Snyder EY, Cepko CL. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- 5.Wetts R, Fraser SE. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 6.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Proc Natl Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young RW. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 8.Carter-Dawson LD, LaVail MM. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- 9.Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- 10.Dyer MA, Livesey FJ, Cepko CL, Oliver G. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- 11.Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Proc Natl Acad Sci USA. 2006;103:6218–6223. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 13.Hatakeyama J, Kageyama R. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 15.Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 17.Edlund T, Jessell TM. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 18.Levine EM, Fuhrmann S, Reh TA. Cell Mol Life Sci. 2000;57:224–234. doi: 10.1007/PL00000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe T, Raff MC. Development (Cambridge, UK) 1992;114:899–906. doi: 10.1242/dev.114.4.899. [DOI] [PubMed] [Google Scholar]
- 20.Belliveau MJ, Young TL, Cepko CL. J Neurosci. 2000;20:2247–2254. doi: 10.1523/JNEUROSCI.20-06-02247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cayouette M, Barres BA, Raff M. Neuron. 2003;40:897–904. doi: 10.1016/s0896-6273(03)00756-6. [DOI] [PubMed] [Google Scholar]
- 22.Adler R, Hatlee M. Science. 1989;243:391–393. doi: 10.1126/science.2911751. [DOI] [PubMed] [Google Scholar]
- 23.Ezzeddine ZD, Yang X, DeChiara T, Yancopoulos G, Cepko CL. Development (Cambridge, UK) 1997;124:1055–1067. doi: 10.1242/dev.124.5.1055. [DOI] [PubMed] [Google Scholar]
- 24.Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 25.Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- 26.Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, et al. Proc Natl Acad Sci USA. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swain PK, Hicks D, Mears AJ, Apel IJ, Smith JE, John SK, Hendrickson A, Milam AH, Swaroop A. J Biol Chem. 2001;276:36824–36830. doi: 10.1074/jbc.M105855200. [DOI] [PubMed] [Google Scholar]
- 28.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 29.Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 30.Mitton KP, Swain PK, Khanna H, Dowd M, Apel IJ, Swaroop A. Hum Mol Genet. 2003;12:365–373. doi: 10.1093/hmg/ddg035. [DOI] [PubMed] [Google Scholar]
- 31.Lerner LE, Gribanova YE, Ji M, Knox BE, Farber DB. J Biol Chem. 2001;276:34999–35007. doi: 10.1074/jbc.M103301200. [DOI] [PubMed] [Google Scholar]
- 32.Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, et al. Hum Mol Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, et al. Hum Mol Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 34.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 35.Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN., Jr Invest Ophthalmol Vis Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furukawa A, Koike C, Lippincott P, Cepko CL, Furukawa T. J Neurosci. 2002;22:1640–1647. doi: 10.1523/JNEUROSCI.22-05-01640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. Hum Mol Genet. 2006;15:2588–2602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter-Dawson LD, LaVail MM. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wassle H. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- 40.Pignatelli V, Strettoi E. J Comp Neurol. 2004;476:254–266. doi: 10.1002/cne.20207. [DOI] [PubMed] [Google Scholar]
- 41.Strettoi E, Porciatti V, Falsini B, Pignatelli V, Rossi C. J Neurosci. 2002;22:5492–5504. doi: 10.1523/JNEUROSCI.22-13-05492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szel A, Rohlich P, Mieziewska K, Aguirre G, van Veen T. J Comp Neurol. 1993;331:564–577. doi: 10.1002/cne.903310411. [DOI] [PubMed] [Google Scholar]
- 43.Chiu MI, Nathans J. Vis Neurosci. 1994;11:773–780. doi: 10.1017/s0952523800003072. [DOI] [PubMed] [Google Scholar]
- 44.Akimoto M, Filippova E, Gage PJ, Zhu X, Craft CM, Swaroop A. Invest Ophthalmol Vis Sci. 2004;45:42–47. doi: 10.1167/iovs.03-0804. [DOI] [PubMed] [Google Scholar]
- 45.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 46.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pittler SJ, Zhang Y, Chen S, Mears AJ, Zack DJ, Ren Z, Swain PK, Yao S, Swaroop A, White JB. J Biol Chem. 2004;279:19800–19807. doi: 10.1074/jbc.M401864200. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. Molecular Cloning. 3rd Ed. Vol 3. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 49.Chen J, Rattner A, Nathans J. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 51.Yu J, He S, Friedman JS, Akimoto M, Ghosh D, Mears AJ, Hicks D, Swaroop A. J Biol Chem. 2004;279:42211–42220. doi: 10.1074/jbc.M408223200. [DOI] [PubMed] [Google Scholar]
- 52.Levine M, Tjian R. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 53.Strettoi E, Mears AJ, Swaroop A. J Neurosci. 2004;24:7576–7582. doi: 10.1523/JNEUROSCI.2245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buffelli M, Burgess RW, Feng G, Lobe CG, Lichtman JW, Sanes JR. Nature. 2003;424:430–434. doi: 10.1038/nature01844. [DOI] [PubMed] [Google Scholar]
- 55.Kasthuri N, Lichtman JW. Nature. 2003;424:426–430. doi: 10.1038/nature01836. [DOI] [PubMed] [Google Scholar]
- 56.Katz LC, Shatz CJ. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 57.Dejneka NS, Surace EM, Aleman TS, Cideciyan AV, Lyubarsky A, Savchenko A, Redmond TM, Tang W, Wei Z, Rex TS, et al. Mol Ther. 2004;9:182–188. doi: 10.1016/j.ymthe.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 58.Coles BL, Angenieux B, Inoue T, Del Rio-Tsonis K, Spence JR, McInnes RR, Arsenijevic Y, van der Kooy D. Proc Natl Acad Sci USA. 2004;101:15772–15777. doi: 10.1073/pnas.0401596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banin E, Obolensky A, Idelson M, Hemo I, Reinhardtz E, Pikarsky E, Ben-Hur T, Reubinoff B. Stem Cells. 2006;24:246–257. doi: 10.1634/stemcells.2005-0009. [DOI] [PubMed] [Google Scholar]
- 60.MacLaren RE, Pearson RA, MacNeil A, Douglas RH, Salt TE, Akimoto M, Swaroop A, Sowden JC, Ali RR. Nature. 2006;444:203–207. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- 61.Khanna H, Akimoto M, Siffroi-Fernandez S, Friedman JS, Hicks D, Swaroop A. J Biol Chem. 2006;281:27327–27334. doi: 10.1074/jbc.M605500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Mol Endocrinol. 2006;20:1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 63.Kanda A, Friedman JF, Nishiguish KM, Swaroop A. Hum Mutat. 2007 doi: 10.1002/humu.20488. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.