Abstract
The restriction of a small molecule's motion on binding to a protein causes a loss of configurational entropy, and thus a penalty in binding affinity. Some energy models used in computer-aided ligand design neglect this entropic penalty, whereas others account for it based on an expected drop in the number of accessible rotamers upon binding. However, the validity of the physical assumptions underlying the various approaches is largely unexamined. The present study addresses this issue by using Mining Minima calculations to analyze the association of amprenavir with HIV protease. The computed loss in ligand configurational entropy is large, contributing ∼25 kcal/mol (4.184 kJ/kcal) to ΔG°. Most of this loss results from narrower energy wells in the bound state, rather than a drop in the number of accessible rotamers. Coupling among rotation/translation and internal degrees of freedom complicates the decomposition of the entropy change into additive terms. The results highlight the potential to gain affinity by designing conformationally restricted ligands and have implications for the formulation of energy models for ligand scoring.
Keywords: drug design, translation, rotamer, affinity, rotation
A drug-like molecule that binds a protein becomes less mobile, and the resulting loss in configurational entropy opposes the attractive forces that drive binding. A number of empirical energy models used in virtual ligand screening include a term to account for this entropic penalty, but the underlying physics is not well characterized and hence merits critical examination. For example, most energy models assume that the ligand's entropy change can be decomposed into additive components, although correlated motions could lead to nonadditivity (see, e.g., refs. 1 and 2). Also, energy models often account for changes in torsional entropy with a term related to the number of rotatable bonds in the ligand, based on reasoning about the number of rotamers each bond can adopt (e.g., refs. 3–8) and a computational analysis of changes in vibrational and conformational entropy on protein folding (9). However, the physical rationale and accuracy of this approach is largely unexamined, especially in the context of protein-ligand binding. Similarly, the common assumption that changes in rotational and translational entropy are constant from one ligand to another appears to be unsupported.
Recent calculations with the second-generation Mining Minima algorithm (M2) have provided insight into changes in configurational entropy upon binding for small host–guest systems (10, 11). The computed entropic penalty was found to range as high as ∼20 kcal/mol, considerably more than typically assumed in ligand–protein scoring functions. The validity of these results is supported by the fact that the computed binding free energies were accurate to within ∼1 kcal/mol. The entropy change was furthermore found to vary significantly across complexes of the same ligand with different receptors and even across different bound conformations of the same ligand and receptor. No clear correlation was observed between entropy change and the number of rotatable bonds, but the snugness of the guest's fit in the host's binding site correlated with entropy loss.
The present study uses the M2 algorithm to characterize changes in ligand entropy upon protein–ligand binding, through analysis of amprenavir's association with HIV protease. The issues examined include the magnitude of the entropy loss associated with amprenavir's motions, the validity of rotamer counting as a model for entropy loss, and the problem of decomposing the entropy change into contributions from rotation and translation, bond torsions, bond angles, and bond stretches. The results shed light on common assumptions regarding the magnitude and causes of changes in configurational entropy upon binding and may help formulate improved empirical models for ligand screening and design.
Results
Loss of Amprenavir's Configurational Entropy.
Amprenavir loses 26.4 kcal/mol worth of configurational entropy on binding HIV protease, according to the M2 calculations. This value includes both the conformational and vibrational contributions and accounts for decreases in mobility along translational, rotational, and internal coordinates. The quasiharmonic calculations yield a similar value of 25.0 kcal/mol, and a second M2 calculation with the simpler distance-dependent dielectric treatment of solvent yields an entropic penalty within 3 kcal/mol of the full calculation.
The large loss of configurational entropy results primarily from the narrowness of the energy wells of bound amprenavir relative to those of free amprenavir: the change in vibrational entropy is 24.6 kcal/mol, whereas the change in conformational entropy is only 1.8 kcal/mol. The large loss in vibrational entropy is traceable to a combination of factors, including loss of translational and rotational freedom and narrower torsional ranges, as detailed below. The small magnitude of the loss in conformational entropy results from the fact that the number of stable conformers (i.e., distinct energy wells) does not fall much upon binding. The M2 calculations generated ∼960 distinct conformations of free amprenavir within 10 kcal/mol of the most stable free conformer, but only one highly occupied conformer for bound amprenavir. Even if free amprenavir were equally stable in all 960 energy wells, the change in conformational entropy would reach only RTln 960 = 4.1 kcal/mol, much less than the vibrational contribution. The additional fact that only a few of these 960 conformations are significantly occupied causes the actual loss of conformational entropy to be even smaller, only 1.8 kcal/mol as noted above. In particular, the single most stable conformation has a probability of 0.23, the 6 most stable conformations have a combined probability of 0.51, and the 45 most stable conformations have a combined probability of 0.91.
Entropy Decomposition.
The M2 method was also used to examine the contributions of specific classes of degrees of freedom to the change in entropy. Bond stretches and angle bends are usually considered “hard” degrees of freedom, which do not contribute much to molecular flexibility and hence to changes in entropy. Not surprisingly, then, M2 calculations with bond lengths held fixed yielded an essentially unchanged entropy loss of 26.3 kcal/mol. However, fixing bond angles caused the computed entropy loss to fall by 1.7 kcal/mol to 24.7 kcal/mol, which corresponds to an average contribution of only 0.024 kcal/mol of entropy per angle or a 4% narrowing of each angle's fluctuations. Different angles contributed differently to the change in angular entropy: the largest contributions are associated with angles at sp3 carbons near the center of the molecule, but angles within the phenyl ring make no significant contribution. Repeating the M2 calculations accounting exclusively for torsional degrees of freedom yielded an entropy loss of 12.2 kcal/mol, whereas the six rotational and translational degrees of freedom alone yielded 15.7 kcal/mol. (Note that the latter varied with the standard concentration.) Interestingly, these values sum to 27.9 kcal/mol, more than the entropy loss of 26.4 kcal/mol for all degrees of freedom and the value of 24.7 kcal/mol obtained for the combination of torsions, rotations, and translations. The lack of additivity indicates coupling among the various degrees of freedom.
Table 1 presents an alternative decomposition of entropic contributions based on a covariance analysis of the entropy contributions from each type of ligand motion, for the most stable energy well of the free and the bound states, respectively. Each diagonal term is computed with only the corresponding set of degrees of freedom (e.g., all torsions). Each off-diagonal term reports on the correlation between the corresponding sets of motions and is computed as the difference in the entropy when two sets of motions are considered, versus the sum of the two entropies found when the two sets of motions are treated separately (see Methods). The off-diagonal terms are always positive because the entropy is always lower when correlations are accounted for than when they are neglected. Note that all torsional degrees of freedom are accounted for here, not just freely rotatable bonds. The entropy loss associated with rotation and translation is ∼12 kcal/mol, and the torsional losses also are substantial at 7 kcal/mol. Angle bends lead to a smaller entropy loss of ∼2 kcal/mol, but the contribution from bond stretching is again negligible. Modest but nontrivial additional entropy losses (off-diagonal elements in Table 1) result from increased correlations among the various degrees of freedom upon binding. Such correlations make it difficult to unambiguously decompose the full entropy loss into parts associated with specific degrees of freedom.
Table 1.
Decomposition of amprenavir's change in configurational entropy upon binding, calculated via submatrices of the covariance matrices of the dominant free and bound energy wells
| R/T | Tors | Angle | Stretch | |
|---|---|---|---|---|
| Free | ||||
| R/T | −7.07 | 0 | 0 | 0 |
| Tors | 54.5 | 3.6 | 0.1 | |
| Angle | 78.1 | 1.0 | ||
| Stretch | 109.52 | |||
| Bound | ||||
| R/T | 7.02 | 1.5 | 0.8 | 0.08 |
| Tors | 61.6 | 4.1 | 0.13 | |
| Angle | 80.1 | 1.2 | ||
| Stretch | 109.55 | |||
| Difference | ||||
| R/T | 12.3 | 1.5 | 0.8 | 0.08 |
| Tors | 7.1 | 0.5 | 0.03 | |
| Angle | 2.0 | 0.2 | ||
| Stretch | 0.04 |
Entropy values multiplied by −T to yield free energy contributions in kcal/mol. R/T, rotational and translation part, including RT ln(8π2/C°) for the free ligand. Tors, torsional. Ang, angle bend. Stretch, bond stretch.
Off-diagonal terms are entropy changes caused by changes in correlations between classes of motions, e.g., Stors, stretchcorr = Stors, stretch − Stors − Sstretch. Lower triangle terms are not listed because the matrix is symmetric.
The arbitrariness of such decompositions is illustrated by the fact that significantly different decompositions are obtained with the M2 and covariance analyses (above); for example, these respective methods yield values of 12 and 7 kcal/mol for the loss in torsional entropy. The differences have at least two causes. First, the M2 calculations consider multiple energy wells for the free ligand, whereas the covariance method considers only one. More importantly, removing degrees of freedom in the M2 calculations freezes them and thus reduces the fluctuations of those that remain, because of coupling; whereas removing degrees of freedom in the covariance method does not freeze them, but only removes their fluctuations and the correlations of their fluctuations with the remaining motions. In the M2 calculations, it is reasonable that torsional freedom is strongly restricted by freezing the rotations and translations of the bound ligand, because rotation, translation, and torsions are strongly coupled with each other in this case. Note that there is no coupling between torsions and rotational/translational motions for the free ligand. The covariance method, in contrast, computes the torsional entropy of the bound ligand without restraining rotation or translation; hence it yields a smaller torsional entropy loss on binding.
The quasiharmonic analysis provides an independent value for the change in rotational and translational entropy on binding, based on the fluctuations in position and orientation of the bound ligand. Because all other ligand (and protein) degrees of freedom are also mobile in the molecular dynamics simulation, the result is most suitably compared with the covariance result above, rather than the M2 result in which other degrees of freedom are frozen. The result, 11.6 kcal/mol, agrees well with the value of 12.3 kcal/mol from the covariance method. Note that both calculations use the same root atoms and protein atoms to define the position and orientation of amprenavir in the binding site. Using different root atoms would have made the two calculations difficult to compare on an equal footing.
Discussion
Importance of Configurational Entropy.
The loss of amprenavir's configurational entropy upon binding is found to be remarkably large, opposing binding by ∼25 kcal/mol. The full free energy cost of configurational entropy is almost certainly even larger, because the present calculations omit any contribution from the restriction of protein motions. The protein contribution could be especially important for HIV protease, where binding is associated with a marked reduction in the mobility of the active site flaps; significant coupling of protein and ligand motions also is expected. Interestingly, the loss of ligand configurational entropy found here is as great as the largest combined loss of host and guest configurational entropy in prior studies of host–guest systems (10, 11). It is likely that other high-affinity protein–ligand systems undergo similarly large losses of configurational entropy on binding, because there is nothing peculiar about the amprenavir–HIV protease system. However, low-affinity systems are expected to undergo much smaller entropy losses, based on entropy–energy correlation (11).
The reliability of the present results is supported by the accuracy of M2 calculations of host–guest binding free energies, for which such calculations are tractable (10, 11). It is also worth noting that the average energies, 〈U + W〉, provided by M2 for model compounds were found to agree with MD simulations to within 1 kcal/mol (12). This observation supports the accuracy of the partitioning of free energy into entropy and enthalpy by the M2 method. The marked reduction of ligand motion reflected by the present results appears to be consistent with a molecular dynamics analysis of saquinavir bound to HIV protease, which shows highly restricted torsional fluctuations of at most 10° in the course of a 1-ns simulation (13). The changes in configurational entropy reported here are robust with respect to computational methodology, based on the agreement between M2 and quasiharmonic results and the insensitivity of the M2 results to the choice of solvent model. Unfortunately, direct experimental evaluation of the present results does not appear to be possible currently: calorimetric studies provide the change in entropy upon binding, but do not yield the configurational part in isolation. It may be possible to examine these questions further with detailed MD simulations (M.K.G., B. J. Killian, and J. Y. Kravitz, unpublished work).
It has long been recognized that a more rigid ligand is likely to lose less entropy upon binding than a more flexible one, and thus should bind more tightly, all other things being equal. The large magnitude of the entropy loss observed here highlights the potential significance of this concept for the selection of candidate ligands and the optimization of lead compounds. In particular, it appears that increasing the intrinsic rigidity of even part of a ligand could readily improve affinity by several kcal/mol, i.e., by an amount more customarily associated with the formation of a new hydrogen bond or another favorable ligand–protein interaction. These results also suggest an explanation for the common observation that natural products, with their complex, rigid ring systems, are a particularly rich source of high-affinity ligands: they presumably have less entropy to lose. Thus, highly rigid natural products may be able to beat the usual entropy–energy compensation graphs (e.g., refs. 10, 11, and 14) and achieve particularly high binding efficiencies (15, 16). The challenge of a rigid ligand, of course, is that it must possess precisely the right conformation for binding, because it will not readily adjust to fit the site.
Conformational Versus Vibrational Entropy.
It is often assumed that the loss of ligand entropy upon binding results chiefly from a drop in the number of accessible rotamers. The present results contrast with this view, because only ∼2 kcal/mol (−TΔSconf) of the entropy loss is found to result from a drop in the number of stable conformers. Instead, most of the entropy loss is vibrational, resulting from narrower, rather than fewer, energy wells after binding (−TΔSvib). Given these differing physical pictures, it is worth examining the common view more closely. The loss in torsional entropy is typically estimated as 0.3–0.54 kcal/mol per torsional degree of freedom (see, e.g., ref. 17), which would imply that amprenavir, with ∼12 rotatable bonds, must possess 400–49,000 equally stable free conformers. It is not clear that these large numbers are supported by experimental or theoretical evidence. Indeed, NMR studies frequently reveal marked conformational preferences of drug-like compounds in solution, such as the “folded” conformation of tandospirone (18) and the preorganized conformation of KNI-272, despite its 15 rotatable bonds (19). Such results weigh against the suitability of ligand rotamer counting as a basis for estimating the loss of ligand entropy on binding.
There is significant literature on the contributions of conformational and vibrational entropy to changes in side-chain configurational entropy on protein folding, and it is appropriate to ask whether this literature bears on protein–ligand binding. It is often held that the loss of side-chain conformational entropy on folding is much greater than the loss of vibrational entropy (e.g., refs. 3, 9, and 20), in contrast with the present view of amprenavir binding. Perhaps the binding of amprenavir to HIV protease simply is more restrictive than the “binding” of a side chain in the interior of a protein, leading to more marked narrowing of energy wells. Alternatively, it is possible that the loss of vibrational entropy when a side chain is buried has been underestimated, as recently suggested (21). The chief evidence that changes in vibrational entropy on folding are small is found in a pioneering paper (9) comparing the mean vibrational entropy of an amino acid in folded BPTI, 34 kcal/mol per residue, with the mean vibrational entropies of short peptides modeling the unfolded state, 19–48 kcal/mol per residue depending on the type of amino acid. If the mean entropy of a residue in the unfolded protein were in the middle of this range, then the vibrational entropy would indeed change little upon folding; but if the mean were close to 48, then the loss in vibrational entropy would be substantial. It is also important to note that BPTI is a small protein and many of its side chains project into solvent rather than into the protein's interior. Solvent-exposed side chains undergo little change in entropy upon folding (21), so the average loss of entropy of BPTI's side chains may not be directly informative about the entropic consequences of side-chain burial or ligand binding.
Rotational, Translational, and Torsional Entropy.
Losses in the rotational and translational entropy of amprenavir are found here to oppose binding by 12–16 kcal/mol, depending on the choice of decomposition. These values are bracketed by the results of prior calculations (22–28) for different systems and with different methods; the range is ∼6 to 18 kcal/mol. It is physically reasonable that there should be significant variations in the translational and rotational freedom of bound ligands, depending on how snugly each ligand is held in the binding site, as noted (11). Thus, 4-hydroxy-2-butanone binds the protein FKBP with a relatively small affinity of −4.5 kcal/mol, and the computed loss in translational and rotational entropy is only several kcal/mol (23). On the other hand, small entropy losses have been reported for the high-affinity biotin–streptavidin interaction (17, 24, 29). This complicated overall picture probably is caused by the fact that the computed change in rotational and translational entropy depends on the method used to carry out the entropy decomposition and the choice of coordinate system (30, 31). For example, the position fluctuations of the ligand can be defined based on root atoms in the ligand and nearby reference atoms in the protein, as done here, or on the center of mass of the ligand and the center of mass of the protein (e.g., ref. 17), and there is no reason to expect that the results will be the same. Note, however, that the computed change in the total entropy upon binding should not depend on the choice of coordinates, even though individual components can. Other methodologic details also can affect the results, as recently highlighted (26). For example, the quasiharmonic approximation can lead to errors because it overestimates entropy when multiple energy wells are sampled (12).
The losses in torsional entropy computed here correspond to ∼0.6–1 kcal/mol per rotatable bond, depending on which decomposition is chosen. These values range higher than typical empirical values of 0.30 to 0.54 (e.g., ref. 17) and are a little higher than measured entropy changes of 0.38–0.86 kcal/mol per rotor for freezing of liquid alkanes (see, e.g., ref. 32). However, the present values are smaller than the measured entropy losses of 1.4–1.6 kcal/mol per rotor for condensation of gaseous alkanes into the solid phase (33). Arguably, the condensation data are at least as helpful in this context, because the molar entropy of a liquid alkane is lowered by intermolecular correlations (32): in effect, the alkane molecules lose entropy because they solvate each other. Such correlations are not relevant to the configurational entropy of amprenavir in water, and the condensation data avoid this problem because gas-phase alkane molecules do not interact significantly.
Implications for Scoring Functions.
The present results indicate that losses in configurational entropy affect binding free energies as strongly as better-understood energetic interactions, such as electrostatics and the hydrophobic effect. It thus seems unlikely that the energy functions used in computer-aided drug discovery will achieve high accuracy without improved treatments of entropy. A number of current models account for losses in the torsional entropy of the ligand with a penalty proportional to the number of rotatable bonds (e.g., refs. 4–6), and at least two models (7, 8) furthermore adjust the entropy loss associated with a given rotor depending on its environment, although not according to the chemical nature of the rotor, which also can affect the entropy (34). However, the present results suggest that the physical model underlying these rotamer-counting approaches overestimates the rather small number of stable conformations accessed by a drug-like ligand in the free state. Also, the common assumption that all ligands lose the same amount of rotational and translational entropy upon binding does not appear to be well supported, and both physical reasoning (32) and recent calculations suggest the opposite (11).
It is thus important to seek better ways of accounting for entropy changes within scoring functions. One suitable approach might be to use the standard rigid rotor/harmonic oscillator approximation to the free and bound system, as frequently done in quantum studies of gas-phase dimerization (e.g., ref. 35), making sure to use the 1 M standard concentration customary for solutions (36) rather than the gas-phase standard state of 1 atm (1 atm = 101.3 kPa). Indeed, the rigid rotor/harmonic oscillator approximation could be used to compute the partition functions of multiple energy wells, and these could be combined by Eq. 12. Although such an approach would omit the corrections that a full M2 calculation provides, it should capture much the same physical picture. Whatever method is used, the present study highlights the importance of accounting for couplings among the different motions of the ligand, because they produce significant nonadditivity. In fact, the increase in coupling upon binding contributes to the overall loss in entropy, as evidenced by the off-diagonal terms in Table 1. Also, the loss of angle-bending entropy may be nontrivial, the largest angular contributions here arising from angles at sp3 atoms near the center of the ligand, whereas angles in rigid rings contribute negligibly. This unexpected result, if confirmed, deserves attention in the formulation of scoring functions. Finally, the dominance of the vibrational entropy over the conformational entropy suggests that the change of ligand entropy on binding can be assessed accurately without enumerating many different energy minima.
Methods
Configurational Entropy: Definition and Decomposition.
The standard free energy of binding of a protein P and a ligand L that form a complex PL can be written as (30):
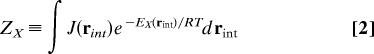 |
Here μ°X is the standard chemical potential of molecular species X, where X = PL, P, or L; R is the gas constant; T is absolute temperature; C° is the standard concentration; ZX is the configuration integral over the internal coordinates of species X; J(rint) is the Jacobian determinant for the internal coordinates; and E(rint) is the sum of the potential energy (U) and solvation free energy (W) of the molecule or complex as a function of its conformation, where the conformation is specified by the internal coordinates rint. Eq. 1 omits a pressure-volume term that is negligible for most systems of interest (30). The terms “chemical potential” and “free energy” will be used interchangeably in this article, in conformity with common usage. In addition, the term “entropy” is used loosely to mean the free energy contribution −TS associated with entropy S. For the complex, PL, the internal coordinates include 6 degrees of freedom specifying the position and orientation of the ligand relative to the protein. When the distance unit of the internal coordinates is specified in Å, C° = 1 mole/liter must be converted to 6.02 × 10−4 molecules per Å3. This concentration may be thought of as corresponding to a “standard volume” of 1/C° = 1,661 Å3 per molecule.
The standard entropy change upon binding is the temperature derivative of the standard free energy of binding at constant pressure (37):
Application of this expression to Eq. 1 allows the entropy change on binding to be partitioned, without approximation, into a configurational part, ΔS°configo, which is associated with only the degrees of freedom of the protein and the ligand, and a solvent part, ΔSsolv, which is the ensemble-averaged change in the mean solvation entropy upon binding:
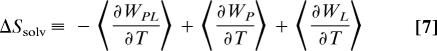 |
Here angle brackets indicate ensemble averages, Δ〈U + W〉 is the change on binding of the ensemble-averaged sum of potential energy and solvation energy, and the temperature derivative of the solvation free energy W has been identified as the solvation entropy, much as in Eq. 4. It is worth pointing out that ΔS°configo depends on the standard concentration, C°, consistent with the expectation that the loss of translational entropy upon binding depends on the standard volume allotted to each molecule in the free state. This study focuses on the configurational entropy; the implicit solvation model used here does not yield accurate temperature derivatives, so changes in solvation entropy cannot be obtained at this time.
The configurational entropy itself can be further partitioned into a “conformational” part, which reflects the number of occupied energy wells, and a “vibrational” part, which reflects the average width of the occupied wells (9):
In the special case of M equally stable energy wells, the conformational entropy equals simply −Rln M; when the energy wells are not equally stable, the conformational term generalizes to:
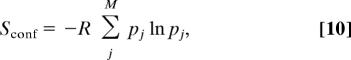 |
where pj is the probability of occupying energy well j. It is straightforward to show that the difference between this conformational entropy and the total configurational entropy is exactly the Boltzmann-averaged entropy of the individual energy wells, so that:
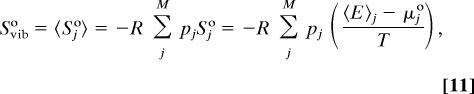 |
where 〈E〉j is the average energy of well j and μ°j is its standard chemical potential. This decomposition of configurational entropy into conformational and vibrational parts was originally provided without derivation, as recently noted (21), so a derivation is provided in Appendix. The vibrational entropy depends formally on standard concentration because each multidimensional energy well includes three translational dimensions, whose ranges are determined by the standard volume.
One can, alternatively, decompose the configurational entropy according to different classes of degrees of freedom: overall rotation and translation, bond torsion, bond angles, and bond stretches. However, the component entropies need not add up to the full entropy unless motions along the various degrees of freedom are uncorrelated with each other; not only pairwise but also higher-order correlations can occur. Here, two decompositions are applied. The first compares the entropy obtained by the M2 method (below) with all ligand degrees of freedom, to that obtained when a subset of motions, such as bond angles, are frozen; this method includes both the conformational and vibrational contributions discussed in the previous paragraph. The second approach limits attention to the single most stable energy well of the free ligand or the bound ligand. The second derivative matrix of the energy with respect to all degrees of freedom is inverted to provide the covariance matrix (38), and the entropy associated with the motions of interest (e.g., the torsional entopy Stors) is then computed from the determinant of the submatrix associated with these degrees of freedom. This calculation neglects correlations with other classes of motion (e.g., bond stretching), which can lower the net entropy, but correlations can be examined by extracting the submatrix associated with two classes of motion (e.g., torsions and stretches), computing the associated entropy (e.g., Stors, stretch), and subtracting the individual entropy terms to generate the contribution of correlation to the entropy; e.g., Stors, stretchcorrcorr ≡ Stors, stretch − Stors − Sstretch.
Calculations with Mining Minima.
Computing the change in configurational entropy upon binding with Eq. 6 requires evaluation of ΔG° and 〈U + W〉. The Mining Minima algorithm (10) is used to compute ΔG° as the difference in standard chemical potentials (Eq. 1), where the standard chemical potential of a molecule or complex is computed via a sum of contributions from M local energy wells j:
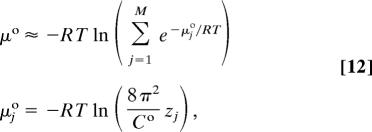 |
where μ°j and zj are, respectively, the standard chemical potential and the local configuration integral over internal coordiates of the molecule in energy well j. The probability of well j is:
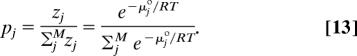 |
Each local configuration integral zj is estimated via an enhanced version of the harmonic approximation (39), in which the second derivative matrix of the energy at the energy minimum is diagonalized, and numerical integrals are evaluated along the eigenvectors with low force constants. If the numerical integral deviates from the harmonic approximation by >1 kcal/mol, then the numerical integral along that mode is substituted for the harmonic approximation. The full integral for the energy well is computed as the product of all of the single-mode integrals. In the present calculations the energy wells were so close to harmonic that the numerical integrals were never substituted for the analytic harmonic results.
The harmonicity of the wells allows the average energy of well j to be obtained via the equipartition theorem:
where Ej° is the energy at the local energy minimum and Nint is the number of internal degrees of freedom rint. The energy averaged over all energy wells is:
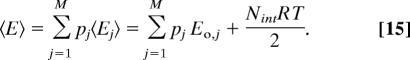 |
The calculations use a coordinate system consisting of bond lengths, bond angles, and bond torsions (36, 39). The position and orientation of the ligand are specified via the coordinates of three “root” atoms, here numbered 1, 2, and 3 for convenience, where atoms 1 and 2 are connected by a covalent bond, as are atoms 2 and 3. The position of atom 1 defines the translational position of the ligand, the orientation of the 1–2 bond defines two orientational coordinates of the ligand, and the orientation of the 2–3 bond defines the third orientational coordinate of the ligand. For the free ligand, each row (and each column) of the second derivative matrix corresponds to a bond length, bond angle, or bond torsion; the position and orientation of the free ligand do not affect the energy and hence do not appear in the matrix. For the bound ligand, six position and orientation coordinates appear in the matrix because these affect the energy when the ligand is bound.
As noted in the previous subsection, the use of bond angle–torsion coordinates permits examination of the entropic contributions of the different classes of molecular motion to the change in entropy upon binding. This is done by artificially deleting the rows and columns corresponding to undesired degrees of freedom from the second derivative matrix and repeating the M2 integrals with the reduced matrix, for the same set of energy minima. For example, the entropy change can be calculated after removing all rows and columns corresponding to bond lengths. The issue of correlation is considered in Results.
The energy minima of the free ligand were found with the Tork search algorithm (41). For the bound ligand, the protein was held rigid and candidate poses of the ligand in the protein were generated with the program Vdock (42, 43). These conformations were energy-minimized, along with the protein, to local energy minima, and duplicate conformations were eliminated with a method that accounts for chemical symmetry (44).
Quasiharmonic Approximation.
The quasiharmonic approximation (45) was used to provide a check on the M2 results for the bound complex. Although the quasiharmonic approximation has been shown to be inaccurate when it encompasses multiple energy wells (12), it should be accurate for bound amprenavir because the M2 calculations indicate that there is only one significantly occupied energy well. The method was implemented with a 300-K molecular dynamics simulation with CHARMM (46) in which both the ligand and the protein were mobile, with time steps of 2 fs, nonbonded cutoffs of 15 Å, and a generalized Born model of the solvent (47). The total simulation time was 2 ns, and snapshots were saved every 0.4 ps for the last 1.6 ns, for a total of 4,000 snapshots. The Cartesian coordinates in the saved snapshots were converted to bond angle–torsion coordinates with the same root atoms as the M2 calculations, and the covariance matrix of the bond angle–torsion coordinates was computed and used in the standard formula for entropy (45).
Structures and Parameters.
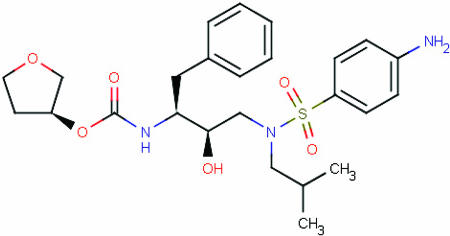
The initial structure of the complex of HIV protease and amprenavir was drawn from crystal structure 1hpv (48) in the Protein Data Bank (49, 50), and all ionizable groups were assigned protonation states normally expected at pH 7. In the M2 calculations, potential energies U(r) (Eq. 3) were computed with the CHARMM 22 parameter set (51), and the solvation energy W(r) was computed with a generalized Born model (40), using a water dielectric constant of 80. For comparison, an additional calculation was done with the distance-dependent dielectric Dij = 4rij, where rij is the distance between atoms i and j in Å. Three carbons in the tetrahydrofuran ring of amprenavir (Fig. 1) were used as root atoms. Their positions in the protein's reference frame were defined by a pseudobond, two pseudoangles, and three pseudodihedral angles rooted in atoms associated with the center of the protease flap: one hydrogen of the flap water, the main-chain nitrogen of Ile 50, and the main-chain carbon of Gly-49. (The mobility of the flap water during the molecular dynamics simulation is similar to that of nearby protein atoms.)
Fig. 1.
Amprenavir.
Acknowledgments
We thank Dr. Visvaldas Kairys (Universidade de Madeira, Funchal, Portugal) for providing setup files for the molecular systems. C.A.C. thanks Dr. J. A. McCammon for support and Drs. Tushar Jain, Jessica Swanson, and Ben Zhuo Lu for helpful discussions. M.K.G. thanks Dr. Greg Weiss for an informative discussion. This work was supported by National Institute of General Medical Sciences Grant GM61300 (to M.K.G.) and National Science Foundation Grant MCB-0506593 (to Dr. J. A. McCammon).
Appendix
This appendix derives the decomposition of the configurational entropy into a conformational and a vibrational part. Let the full configuration space of a molecule be partitioned into energy wells j, such that the configuration integrals within each well, zj, sum to the full configuration integral Z, either exactly or approximately if one neglects high energy regions between the wells as in the M2 method. The standard chemical potential of the molecule is given by Eq. 12, and the partial molar configurational entropy at standard concentration is:
The conformational entropy in Eq. 10 can be written as:
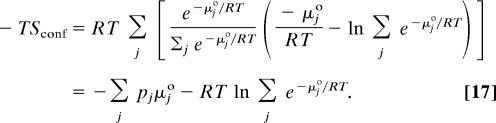 |
Eqs. 9, 12, and 15–17 yield the vibrational entropy as:
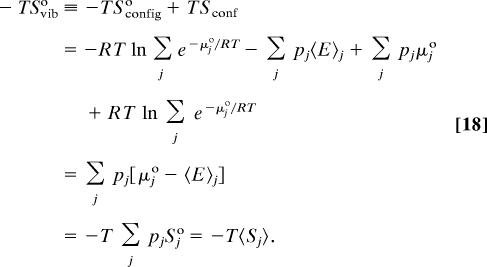 |
Thus, the difference between the total configurational entropy and the conformational entropy is the mean entropy of the individual energy wells, 〈Sj°〉, q.e.d.
Footnotes
The authors declare no conflict of interest.
References
- 1.Brady GP, Sharp KA. J Mol Biol. 1995;254:77–85. doi: 10.1006/jmbi.1995.0600. [DOI] [PubMed] [Google Scholar]
- 2.Schafer H, Daura X, Mark AE, van Gunsteren WF. Proteins Struct Funct Genet. 2001;43:45–56. doi: 10.1002/1097-0134(20010401)43:1<45::aid-prot1016>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 3.Pickett SD, Sternberg MJ. J Mol Biol. 1993;231:825–839. doi: 10.1006/jmbi.1993.1329. [DOI] [PubMed] [Google Scholar]
- 4.Böhm H-J. J Comput Aided Mol Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 5.Rarey M, Kramer B, Lengauer T, Klebe G. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 6.Wang R, Lai L, Wang S. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 7.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. J Comput Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 8.Giordanetto F, Cotesta S, Catana C, Trosset J-Y, Vulpetti A, Stouten PFW, Kroemer RT. J Chem Inf Comput Sci. 2004;44:882–893. doi: 10.1021/ci0499626. [DOI] [PubMed] [Google Scholar]
- 9.Karplus M, Ichiye T, Pettitt BM. Biophys J. 1987;52:1083–1085. doi: 10.1016/S0006-3495(87)83303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C, Gilson MK. J Am Chem Soc. 2004;126:13156–13164. doi: 10.1021/ja047115d. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Chang C, Gilson MK. Biophys J. 2004;87:3035–3049. doi: 10.1529/biophysj.104.049494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C, Chang W, Gilson MK. J Chem Theory Comput. 2005;1:1017–1028. doi: 10.1021/ct0500904. [DOI] [PubMed] [Google Scholar]
- 13.Wittayanarakul K, Aruksakunwong O, Saen-oon S, Chantratita W, Parasuk V, Sompornpisut P, Hannongbua S. Biophys J. 2005;88:867–879. doi: 10.1529/biophysj.104.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houk KN, Leach AG, Kim SP, Zhang XY. Angew Chem Int Ed. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins AL, Groom CR, Alex A. Drug Discovery Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Chang C, Gilson MK. J Am Chem Soc. 2006;128:4675–4684. doi: 10.1021/ja056600l. [DOI] [PubMed] [Google Scholar]
- 17.Lazaridis T, Masunov A, Gandolfo F. Protein Struct Funct Genet. 2002;47:194–208. doi: 10.1002/prot.10086. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura T, Igarashi J, Sunagawa M. Bioorg Med Chem Lett. 2001;11:1131–1144. doi: 10.1016/s0960-894x(01)00167-6. [DOI] [PubMed] [Google Scholar]
- 19.David L, Luo R, Head MS, Gilson MK. J Phys Chem B. 1999;103:1031–1044. [Google Scholar]
- 20.Doig AJ, Sternberg MJE. Protein Sci. 1995;4:2247–2251. doi: 10.1002/pro.5560041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schafer H, Smith LJ, Mark AE, van Gunsteren WF. Protein Struct Funct Genet. 2002;46:215–224. doi: 10.1002/prot.1166. [DOI] [PubMed] [Google Scholar]
- 22.Finkelstein AV, Janin J. Protein Eng. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Swanson JMJ, Henchman RH, McCammon JA. Biophys J. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo H, Sharp K. Proc Natl Acad Sci USA. 2002;99:10399–10404. doi: 10.1073/pnas.162365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo R, Gilson MK. J Am Chem Soc. 2000;122:2934–2937. [Google Scholar]
- 26.Lee MS, Olson MA. Biophys J. 2006;90:864–877. doi: 10.1529/biophysj.105.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu B, Wong CF. Biopolymers. 2005;79:277–285. doi: 10.1002/bip.20344. [DOI] [PubMed] [Google Scholar]
- 28.Hermans J, Wang L. J Am Chem Soc. 1997;119:2707–2714. [Google Scholar]
- 29.Dixit SB, Chipot C. J Phys Chem A. 2001;105:9795–9799. [Google Scholar]
- 30.Gilson MK, Given JA, Bush BL, McCammon JA. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boresch S, Tettinger F, Leitgeb M, Karplus M. J Phys Chem B. 2003;101:9535–9551. [Google Scholar]
- 32.Searle MS, Williams DH. J Am Chem Soc. 1992;114:10690–10697. [Google Scholar]
- 33.Matulis D. Biophys Chem. 2001;93:67–82. doi: 10.1016/s0301-4622(01)00209-5. [DOI] [PubMed] [Google Scholar]
- 34.Mammen M, Shakhnovich EI, Whitesides GM. J Org Chem. 1998;63:3168–3175. [Google Scholar]
- 35.Hobza P, Sponer J. Chem Phys Lett. 1996;261:379–384. [Google Scholar]
- 36.Potter MJ, Gilson MK. J Phys Chem A. 2002;126:563–566. [Google Scholar]
- 37.McQuarrie DA. Statistical Mechanics. New York: Harper & Row; 1973. [Google Scholar]
- 38.Levy RM, Karplus M, Kushick J, Perahia D. Macromolecules. 1984;17:1370–1374. [Google Scholar]
- 39.Chang C, Potter MJ, Gilson MK. J Phys Chem B. 2003;107:1048–1055. [Google Scholar]
- 40.Qiu D, Shenkin PS, Hollinger FP, Still WC. J Phys Chem. 1997;101:3005–3014. [Google Scholar]
- 41.Chang C, Gilson MK. J Comput Chem. 2003;24:1987–1998. doi: 10.1002/jcc.10325. [DOI] [PubMed] [Google Scholar]
- 42.David L, Luo R, Gilson MK. J Comput Aided Mol Des. 2001;15:157–171. doi: 10.1023/a:1008128723048. [DOI] [PubMed] [Google Scholar]
- 43.Kairys V, Gilson MK. J Comput Chem. 2002;23:1656–1670. doi: 10.1002/jcc.10168. [DOI] [PubMed] [Google Scholar]
- 44.Chen W, Huang J, Gilson MK. J Chem Inf Comput Sci. 2004;44:1301–1313. doi: 10.1021/ci049966a. [DOI] [PubMed] [Google Scholar]
- 45.Karplus M, Kushick J. Macromolecules. 1981;14:325–332. [Google Scholar]
- 46.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 47.Dominy BN, Brooks CL., III J Phys Chem B. 1999;103:3765–3773. [Google Scholar]
- 48.Kim EE, Baker CT, Dwyer MD, Murcko MA, Rao BG, Tung RD, Navia MA. J Am Chem Soc. 1995;117:1181. [Google Scholar]
- 49.Bernstein FC, Koetzle TF, Williams TF, Meyer GJB, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 50.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molecular Simulations Inc. CHARMM. Waltham, MA: Molecular Simulations Inc.; 1992. Version 22. [Google Scholar]