Abstract
The mammalian inflammatory response to infection involves the induction of several hundred genes, a process that must be carefully regulated to achieve pathogen clearance and prevent the consequences of unregulated expression, such as cancer. Recently, microRNAs (miRNAs) have emerged as a class of gene expression regulators that has also been linked to cancer. However, the relationship between inflammation, innate immunity, and miRNA expression is just beginning to be explored. In the present study, we use microarray technology to identify miRNAs induced in primary murine macrophages after exposure to polyriboinosinic:polyribocytidylic acid or the cytokine IFN-β. miR-155 was the only miRNA of those tested that was substantially up-regulated by both stimuli. It also was induced by several Toll-like receptor ligands through myeloid differentiation factor 88- or TRIF-dependent pathways, whereas up-regulation by IFNs was shown to involve TNF-α autocrine signaling. Pharmacological inhibition of the kinase JNK blocked induction of miR-155 in response to either polyriboinosinic:polyribocytidylic acid or TNF-α, suggesting that miR-155-inducing signals use the JNK pathway. Together, these findings characterize miR-155 as a common target of a broad range of inflammatory mediators. Importantly, because miR-155 is known to function as an oncogene, these observations identify a potential link between inflammation and cancer.
Keywords: cancer, inflammation, innate immunity, cytokines
The mammalian innate immune response provides a critical first line of defense against pathogens. Detection of microbial ligands is achieved through pattern recognition receptors, such as the Toll-like receptors (TLRs) that are expressed at high levels on macrophages and dendritic cells (1). TLRs have been evolutionarily conserved from Drosophila to humans, with ≈11 mammalian TLRs known that recognize a wide range of distinct chemical structures conserved in the microbial world (2). After pathogen recognition, TLRs signal through adaptor proteins of the myeloid differentiation factor 88 (MyD88) family to activate several downstream signal transduction pathways, such as NF-κB, MAPKs, and members of the IRF family (3). Upon activation, these pathways coordinate the up-regulation of several functionally distinct gene subsets (4) through both transcriptional and posttranscriptional mechanisms (5). The proteins encoded by these genes, such as the cytokines IFN-β, IFN-γ, and TNF-α, are intended to initiate microbial clearance. However, during pathological situations, these cytokines and the pathways they activate can also play a role in such conditions as cancer and autoimmunity (6–8). Although this “inflammatory” response to infection has been studied in great detail, there remain several regulatory aspects of this complex system that require further identification and understanding.
Mammalian microRNAs (miRNAs) are noncoding RNA oligonucleotides that have been highly conserved during evolution and have recently emerged as potent regulators of gene expression (9). Approximately 500 genes encoding miRNAs have been identified in mammals and have been shown to be both temporally and spatially regulated. miRNAs are transcribed by RNA polymerase II as part of a primary transcript (10, 11) that is processed by Drosha and DGCR8 into a smaller RNA molecule that is exported from the nucleus by Exportin 5 (12, 13). Upon reaching the cytoplasm, the primary miRNA undergoes further processing by Dicer and is subsequently loaded onto the RISC complex (14). Once in their mature form, miRNAs specifically bind to 3′ UTRs of target cellular mRNAs leading to either mRNA degradation or inhibition of translation (9).
Functionally speaking, mammalian miRNAs play pivotal roles in shaping cellular development and differentiation in various tissues (15). Consequently, dysregulated miRNA levels are associated with several types of malignancies including colon, breast, and lung cancers (16–18). Cells of hematopoeitic origin also express certain miRNAs that drive their development (19, 20), and altered miRNA expression has been found in leukocyte-derived tumors including pediatric Burkitt's lymphoma and chronic lymphocytic leukemia (16, 17). Despite these observations, it is presently unclear how miRNA expression in leukocytes becomes altered during the onset of cellular transformation.
Because of the profound influence of inflammatory stimuli on gene expression, the innate immune response has the potential to regulate miRNA levels. Recently, our group used microarray technology to determine whether the TLR4 ligand lipopolysaccharide (LPS) can affect miRNA expression in human THP-1 monocytes (21). Three miRNAs were up-regulated, including miR-132, miR-146, and miR-155. Further characterization of miR-146 revealed that it is regulated by NF-κB and may function as a negative regulator of IRAK1 and TRAF6 expression. Such findings demonstrate that miRNAs levels can be altered by bacterial endotoxin and may be involved in regulating innate immune responses.
To test whether virally relevant stimuli induce expression of specific miRNAs, we extended our microarray studies. miR-155 was identified as a specific target of both polyriboinosinic:polyribocytidylic acid [poly(I:C)] and IFN-β. Further investigation revealed that several TLR ligands increased miR-155 expression through either the MyD88 or TRIF signaling, whereas IFNs required TNF-α autocrine signaling to up-regulate miR-155. Inhibition of JNK blocked both poly(I:C) and TNF-α induction of miR-155, indicating a role for MAPK signaling in the regulation of miR-155 levels. Our findings reveal miR-155 as a component of the inflammatory response and suggest that this oncogenic miRNA may prove to be a link between inflammation and cancer.
Results
Our observations started from a microRNA array screen performed to identify miRNAs up-regulated by macrophages in response to stimuli with relevance to viral infections. Macrophages were matured from murine bone marrow and stimulated with either the synthetic viral intermediate, poly(I:C) (double-stranded RNA), or the host antiviral response cytokine, IFN-β. After 6 h of treatment, total RNA was extracted and used to determine the expression levels of 200 unique mouse and human miRNAs in their mature forms. miR-155 was identified as the only miRNA substantially induced both by poly(I:C) and IFN-β (Fig. 1A), and this was based on by using a P value of 0.01 as a cutoff for significance. To confirm the validity of miR-155 induction, a portion of the RNA used for the microarray was converted to cDNA and subjected to quantitative PCR (qPCR). Consistent with the microarray findings, miR-155 was strongly induced by poly(I:C) and IFN-β, a result confirmed by Northern blotting (Fig. 1B). The small nuclear RNA U6 was relatively unchanged by these stimuli (Fig. 1C), whereas the poly(I:C) and IFN-β target gene IP10 was induced by both (Fig. 1C). Some of the macrophages generated for the experiment were also stained with antigen-specific antibodies and analyzed by FACS to assay the macrophage marker CD11b and up-regulation of the poly(I:C) and IFN-β target gene CD86 after 24 h of stimulation (Fig. 1D). These data indicate that macrophages respond to viral cues by strongly up-regulating miR-155, a miRNA that is known from other studies to function as an oncogene (17, 18, 22–25).
Fig. 1.
Microarray analysis of miRNAs induced during the macrophage antiviral response. (A) WT murine macrophages were stimulated with medium (m), 2 μg/ml poly(I:C) [p(I:C)], or 1,000 units/ml IFN-β for 6 h. RNA was extracted and used to conduct a microarray analysis to determine the expression levels of 200 mammalian miRNAs. Data are presented on a scatter plot showing log10-transformed signal intensities for each probe on both channels for the Cy3-labeled media controls and samples stimulated with Cy5-labeled IFN-β (Left) or poly(I:C) (Right). (B) RNA used in A was analyzed by qPCR to assay expression of miR-155 and in a separate experiment by Northern blot analysis under the same conditions. (C) RNA used in A was assayed by qPCR to detect expression of the small nuclear RNA U6 as a loading control or IP10 mRNA to ensure equivalent stimulation by poly(I:C) and IFN-β. (D) A portion of the macrophages generated in A were stimulated with medium, poly(I:C), or IFN-β and assayed for CD11b and CD86 expression by using FACS to ensure proper macrophage development and activation, respectively.
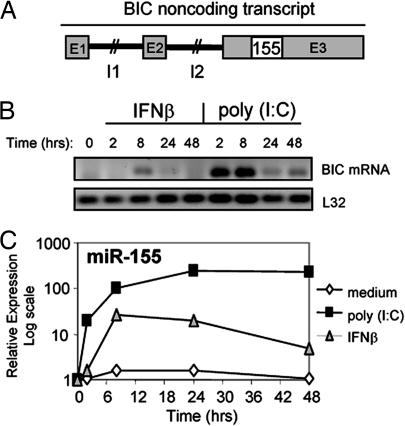
miR-155 is found within the BIC gene (17) on chromosome 21 in humans and 16 in mice (26). The genomic structure of human BIC consists of three exons, and its transcript is transcribed and processed into two differently sized mRNA molecules through alternative polyadenylation. However, BIC lacks a large ORF and therefore is unlikely to encode a protein. Rather, its sole function may be to give rise to miR-155 encoded within exon 3 (Fig. 2A). To monitor the kinetics of miR-155 induction, both BIC mRNA and mature miR-155 were assayed over a 48-h time course after poly(I:C) or IFN-β stimulation of primary macrophages. In response to poly(I:C), BIC mRNA became detectable by 2 h, remained elevated to 8 h, and was still present at reduced levels by 24 and 48 h after stimulation (Fig. 2B). miR-155 induction by poly(I:C) followed a similar pattern of expression as BIC, with the exception of remaining at its highest levels at the 24- and 48-h time points (Fig. 2C). IFN-β did not induce BIC mRNA by 2 h, but it was detected by 8 h and was nearly undetectable by 24 and 48 h (Fig. 2B). IFN-β induction of miR-155 followed the same delayed pattern of induction as BIC, reaching its highest levels by 8 h and slowly decreasing by 24 and 48 h after stimulation (Fig. 2C). These findings provide evidence that the regulation of miR-155 levels involves BIC mRNA up-regulation by poly(I:C) or IFN-β. Furthermore, miR-155 is an immediate early target gene of poly(I:C)-induced signaling, whereas its induction is relatively delayed downstream from IFN-β stimulation.
Fig. 2.
Kinetics of poly(I:C) and IFN-β induction of BIC mRNA and mature miR-155. (A) Unscaled depiction of the genomic structure of the human BIC noncoding RNA gene and location of miR-155 (155) in exon 3. E, exon; I, intron. (B) After stimulation with 2 μg/ml poly(I:C) or 1,000 units/ml IFN-β, macrophage expression of BIC mRNA was analyzed over a 48-h time course by reverse transcription with an oligonucleotide dT primer followed by detection using PCR and agarose gel electrophoresis. Primers were designed to target BIC sequences extending outside of miR-155. L32 mRNA detection is included as a control. (C) RNA from B was also used to assay mature miR-155 by qPCR over a 48-h time course.
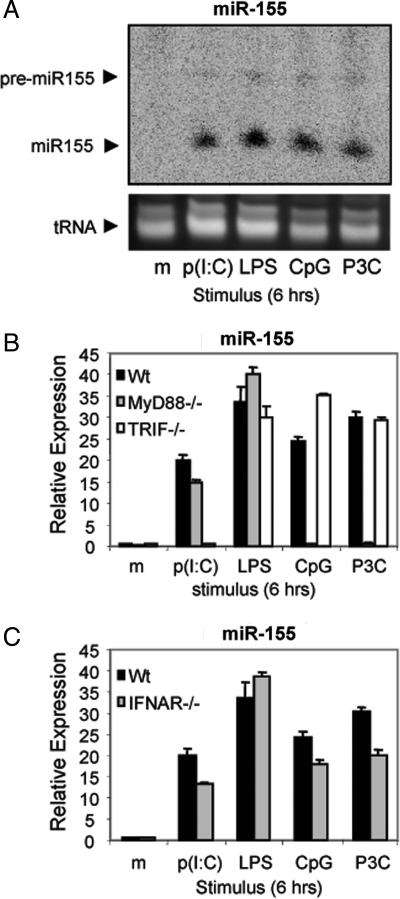
TLR3 is a receptor for poly(I:C) (27). Therefore, other TLR ligands were tested to determine whether they also could induce miR-155. Macrophages were stimulated with either poly(I:C); LPS, which signals through TLR4 (28, 29); hypomethylated DNA (CpG), a TLR9 ligand (30); or Pam3CSK4, a synthetic lipoprotein that activates TLR2 (31). After 6 h, RNA was harvested from these cells and analyzed by Northern blotting (Fig. 3A), or the same RNA was converted to cDNA and assayed by qPCR (data not shown) to determine miR-155 levels. All four TLR ligands tested induced strong expression of miR-155, whereas miR-155 was not detected by Northern blotting in cells treated with medium alone (Fig. 3A). Therefore, TLRs that are known to recognize pathogen-associated molecular patterns from both viruses and other pathogens can strongly induce miR-155.
Fig. 3.
TLRs induce miR-155 expression through MyD88- or TRIF-dependent signaling pathways. (A) WT (Wt) murine macrophages were stimulated with medium (m), 2 μg/ml poly(I:C) [p(I:C)], 5 ng/ml LPS, 2 μg/ml Pam3CSK4 (P3C), or 100 nM CpG for 6 h and assayed by Northern blot analysis with a miR-155-specific probe. (B) WT, MyD88−/−, or TRIF−/− macrophages were stimulated with medium, poly(I:C), LPS, CpG, or Pam3CSK4 for 6 h, and miR-155 expression was assayed by qPCR. (C) WT or IFNAR−/− macrophages were stimulated with medium, poly(I:C), LPS, CpG, or Pam3CSK4 for 6 h, and miR-155 expression was assayed by qPCR.
TLRs signal through the MyD88 family of adaptor proteins (3). Of these adaptors, TLR2 and TLR9 signaling is known to require MyD88, whereas TLR3 utilizes TRIF. Adaptors can also play partially redundant roles; for instance, TLR4 signals through either MyD88 or TRIF (3). To test the requirement of these adaptors for TLR induction of miR-155, macrophages deficient in either TRIF or MyD88 were stimulated with different TLR ligands and miR-155 levels were assayed by qPCR after 6 h. CpG (TLR9)- or Pam3CSK4 (TLR2)-treated macrophages required MyD88, but not TRIF, to induce miR-155, whereas poly(I:C) (TLR3) required TRIF but not MyD88 (Fig. 3B). TLR4 up-regulated miR-155 in the absence of either single adaptor (Fig. 3B). Such results confirm the specificity of the TLR ligands used and demonstrate that either MyD88- or TRIF-dependent signaling pathways are sufficient to induce miR-155.
A subset of TLR-responsive genes require IFN-β autocrine/paracrine signaling for their induction (4). Because miR-155 is up-regulated by both TLRs and IFN-β, we tested whether TLR induction of miR-155 required IFN-β autocrine/paracrine signaling. However, both WT and IFNAR−/− macrophages increased miR-155 expression in response to either poly(I:C), LPS, CpG, or Pam3CSK4 as assayed by qPCR (Fig. 3C). These data indicate that TLRs do not require IFN-β production for early up-regulation of miR-155.
Similar to IFN-β, IFN-γ is produced in response to viral and bacterial infections and plays an important role in macrophage activation (32). IFN-γ also induced miR-155 in macrophages after 6 h of stimulation (Fig. 4A Left). Because IFN induction of BIC mRNA and mature miR-155 was delayed compared with that of poly(I:C), it appeared that IFNs might use a protein intermediate to up-regulate miR-155. We noted a recent study identifying a role for TNF-α autocrine/paracrine signaling after IFN-γ stimulation of macrophages (33). Using TNFR1−/− macrophages, we found that IFN-β and IFN-γ failed to up-regulate miR-155 in the absence of TNFR1 signaling as compared with the induction observed in WT cells (Fig. 4A Left). Furthermore, TNF-α was sufficient to induce miR-155 expression in a TNFR1-dependent manner (Fig. 4A Right) and induced miR-155 with faster kinetics than did IFN-β [supporting information (SI) Fig. 6]. Furthermore, both IFN-β and IFN-γ induced TNF-α mRNA expression (Fig. 4B Left), whereas IFN-β induction of IP10 remained intact in the TNFR1−/− macrophage (Fig. 4B Right), demonstrating that these cells can still respond to IFN treatment. Finally, whereas poly(I:C) induced TNF-α expression (Fig. 4C Left), this TLR3 ligand did not require TNF-α autocrine signaling to induce miR-155 by 6 h after stimulation (Fig. 4C Right). Together, these findings identify TNF-α as an inducer of miR-155 and indicate that IFNs require TNF-α autocrine/paracrine signaling to up-regulate miR-155 in macrophages.
Fig. 4.
IFNs induce miR-155 expression through TNF-α autocrine/paracrine signaling. (A) WT (Wt) and TNFR1−/− murine macrophages were stimulated with medium (m), 1,000 units/ml IFN-β, 50 ng/ml IFN-γ, or 10 ng/ml TNF-α for 6 h, and miR-155 was assayed by qPCR. (B) (Left) WT macrophages were stimulated with medium, IFN-β, or IFN-γ for 6 h, and TNF-α mRNA was analyzed by qPCR. (Right) WT and TNFR1−/− macrophages were stimulated with IFN-β for 6 h and assayed for IP10 mRNA expression by qPCR. (C) (Left) WT macrophages were stimulated with medium or 2 μg/ml poly(I:C) for 6 h and assayed for TNF-α expression by qPCR. (Right) WT and TNFR1−/− macrophages were stimulated with medium or poly(I:C) for 6 h, and miR-155 was assayed by qPCR.
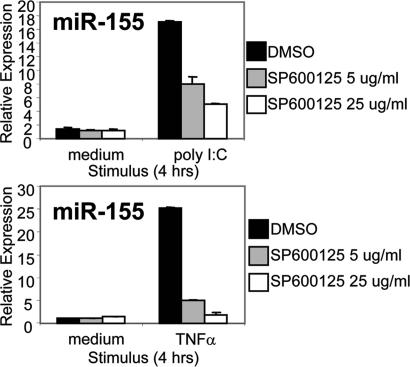
To identify downstream signaling pathways involved in miR-155 induction, we conducted a promoter sequence analysis to identify transcription factor binding sites within the promoter region of the BIC gene that are conserved between mouse and human. A region extending ≈75 bp upstream from the transcriptional start site exhibited ≈85% sequence homology between mouse and human and contained two consensus binding sequences for AP-1 (data not shown). This transcriptional complex is known to be activated by inflammatory stimuli, such as TLR ligands and TNF-α, and requires signaling by JNK (1). To test whether the JNK pathway is involved in miR-155 induction, we stimulated macrophages that had been treated with an inhibitor of JNK (sp600125) with poly(I:C) or TNF-α for 4 h. Vehicle-treated cells up-regulated miR-155 levels by 4 h after stimulation, whereas the JNK inhibitor blocked miR-155 induction by both stimuli in a dose-dependent manner (Fig. 5). As a control, the ERK inhibitor uo126 did not reduce poly(I:C) induction of miR-155 (data not shown). These findings suggest that the JNK pathway is involved in the up-regulation of miR-155 expression in response to poly(I:C) or TNF-α.
Fig. 5.
Pharmacological inhibition of JNK blocks poly(I:C) and TNF-α induction of miR-155. WT murine macrophages were pretreated for 30 min with DMSO or sp600125 at 5 or 25 μg/ml and subsequently stimulated with medium, 2 μg/ml poly(I:C), or 10 ng/ml TNF-α in the presence of the vehicle or inhibitor. After 4 h, RNA was collected, and miR-155 expression was assayed by qPCR.
Discussion
Dysregulation of miRNA levels has been associated with several types of cancers, including those involving leukocytes. However, the ability of environmental factors, such as inflammatory ligands, to alter miRNA expression is just beginning to be explored. Our microarray screen identified miR-155 as the only miRNA of the 200 we tested that was substantially induced by either poly(I:C) and IFN-β. In addition to poly(I:C), miR-155 was induced by other TLR ligands through either MyD88- or TRIF-dependent signaling pathways, and by the cytokines IFN-β and IFN-γ through TNF-α autocrine/paracrine signaling. Therefore, our study identifies and characterizes miR-155 as a component of the primary macrophage response to different types of inflammatory mediators, expanding our initial observation that miR-155 is induced by LPS in human THP-1 monocytes (21). Because miR-155 is known to function as an oncogene (17, 18, 22–25), such findings have implications for both innate immunity and cancer biology.
Although TLRs induced miR-155 as an immediate early gene, IFN induction of miR-155 was weaker and exhibited delayed kinetics because of the need for TNF-α autocrine/paracrine signaling. The existence of such a signaling loop was recently described, when it was shown that TNFR1 was required for IFN-γ induction of nitric oxide in macrophages (33). Such a system enables IFNs to induce TNF-α target genes, including miR-155, in addition to the well established IFN-responsive genes (34). Although it is presently unknown whether this pathway also is relevant in other cell types besides macrophages (35), it highlights the complexity of the cytokine networks that respond after exposure to infectious pathogens.
The IFN system has long been known to provide all nucleated cells with antiviral capabilities after infection (34). Although the protein-encoding genes induced during this response have been well studied, the involvement of miRNAs has not been investigated. Interestingly, our microRNA array screen identified only miR-155 as a substantial target of IFN-β signaling in macrophages. As discussed earlier, the induction required TNF-α autocrine/paracrine signaling. These observations would suggest that miRNAs may not be direct targets of the canonical JAK/STAT pathway known to transactivate several hundred antiviral genes in response to IFN signaling. However, whereas our screen included 200 unique miRNAs, there are presently close to 500 in the miRNA registry, leaving many to be tested. It is also possible that a portion of the miRNAs analyzed are regulated at the RNA processing level, and, therefore, their mature forms were not detected. This phenomenon has recently been observed for miR-138 in the brain (36). Such a scenario may require additional signals to activate processing and might only occur during infection with a live, whole pathogen.
It will be of importance to determine which host mRNAs with relevance to immunity are being regulated by miR-155, or whether miR-155 can directly target viral genomes or transcripts, as miR-32 has been shown to do (37). To date, the only characterized target of miR-155 is the angiotensin II type I receptor in fibroblasts (38). In addition to miR-155, we have previously found miR-146 and miR-132 at higher levels in human THP-1 monocytes after exposure to LPS (21). Further characterization of miR-146 revealed that it can attenuate expression of the innate signaling proteins IRAK1 and TRAF6, identifying a possible regulatory role for miR-146 during TLR signaling. Although clearly defining the physiological role of miR-155 as well as other miRNAs expressed in myeloid cells remains an unfinished task, collectively, these findings provide evidence that miRNAs are part of the antimicrobial gene program induced by infectious pathogens.
Our data suggest that the JNK pathway, which activates the AP-1 complex, is involved in miR-155 induction by both TLRs and TNF-α, presumably through transcriptional activation of the miR-155 encoding BIC gene. Although both the human and mouse promoter regions of BIC contain binding sites for AP-1 near the transcriptional start site, there are a few putative binding sites for NF-κB family members further upstream (ref. 39 and our unpublished data). However, the involvement of NF-κB in up-regulating miR-155 levels remains unclear. One group reported that overexpression of an IκBα dominant active protein does not block BCR mediated induction of BIC in Ramos B cells (39), whereas another gene chip study identified a reduction in BIC transcripts in response to inhibitors of IKK administered to lymphoma cells with constitutively active NF-κB (40). As both JNK and NF-κB have been implicated in mediating oncogenesis (41, 42), miR-155 may prove to be an important target of these transcription factors during this process.
Both miRNAs and microbially induced inflammation are associated with cancer (8, 16). Our current findings suggest the possibility that miR-155 could be a link between the two. The miR-155-encoding gene BIC was originally identified as a common retroviral integration site of avian leukosis virus during induction of B cell lymphomas in chickens, which exhibit high BIC expression (22, 25). miR-155 also has been found at high levels in human B cell lymphomas and other tumors (17, 18, 24, 39, 43), whereas enforced overexpression of miR-155 in mouse B cells is sufficient to trigger murine B cell lymphoma (23). These data demonstrate that inappropriate expression of miR-155 can promote cancer, and it is therefore important to understand how miR-155 levels are regulated. Although we show that a broad range of inflammatory mediators can induce miR-155 in macrophages and monocytes, B and T lymphocytes are known to up-regulate miR-155 after BCR or TCR activation, respectively (39, 44). Like macrophages, B and T cells also express a variety of TLRs that could induce miR-155 expression and contribute to both physiological and pathological responses. Furthermore, it will be of interest to determine whether miR-155 is also involved in cancers derived from myeloid cells, such as acute myeloid leukemias.
The present findings identify up-regulation of miR-155 as a consequence of exposure to a broad range of inflammatory mediators, emphasizing that the avoidance of oncogenic transformation is a key rationale for the need to rapidly resolve inflammatory responses.
Materials and Methods
Cell Culture and Reagents.
Bone marrow cells were isolated from the tibias and femurs of mice as described (4). In short, RBCs were lysed by using a RBC lysis buffer (Invitrogen, San Diego, CA), and the remaining bone marrow cells were plated out in DMEM containing 10% FBS, 100 units/ml penicillin, and 100 units/ml streptomycin and supplemented with macrophage colony-stimulating factor-conditioned medium at a previously established concentration. Cells were cultured in a humidified incubator with 5% CO2 at 37°C. After 7 days of culture, a portion of the macrophages were stained with specific antibodies and analyzed by FACS to ensure proper differentiation (CD11b+F4/80+CD11c−) and subsequently used for experiments. Primary macrophages were stimulated by using fresh DMEM containing one of the following: 5 ng/ml 055-B5 LPS (Sigma, St. Louis, MO), 100 nM CpG 1668 oligonucleotides (Invitrogen), 2 μg/ml Pam3CSK4 (Invitrogen), 2 μg/ml poly(I:C) (Amersham Pharmacia, Piscataway, NJ), 1,000 units/ml mIFN-β (R&D Systems, Minneapolis, MN), 50 ng/ml mIFN-γ (Ebioscience, San Diego, CA), or 10 ng/ml mTNF-α (Roche, Basel, Switzerland). The chemical inhibitors sp600125 and uo126 were dissolved in DMSO and used at various concentrations (Calbiochem, La Jolla, CA).
Mice.
WT, MyD88−/−, TRIF−/−, IFNAR−/−, and TNFR1−/− mice, all of which are on a C57BL/6 genetic background, were bred and housed in the University of California Division of Laboratory Animal Medicine facility and killed according to established protocols approved by the Animal Research Committee.
Microarray Analysis.
The microarray screening procedure was the same as described (21). RNA from stimulated macrophages was collected by using the mirVana RNA isolation kit (Ambion, Austin, TX); 30 μg was enriched for small RNAs, tailed by using the mirVana miRNA labeling kit (Ambion), and labeled with either Cy3 (control samples) or Cy5 (stimulated samples) fluorescent dyes (Amersham Pharmacia). The stimulated and control samples were next mixed and incubated for 14 h with miRNA array slides. The epoxy-coated slides (Schott–Nexterion, Louisville, KY) were prepared in quadruplicate by using robotics for the spotting of 200 mouse and human sequences complimentary to different mammalian miRNAs (mirVana miRNA Probe Set; Ambion). After hybridization, microarrays were scanned with a GenePix 4200A scanner (Axon Instruments, Foster City, CA) by using Gene Pix 5.0 software (Axon Instruments). Raw data were imported into the Resolver gene expression data analysis system version 4.0 (Rosetta Biosoftware, Seattle, WA) for further processing.
Messenger RNA Detection.
Total RNA was harvested from bone-marrow-derived macrophages by using the TRIzol Reagent (Invitrogen), and 1 μg of total RNA was converted to cDNA by using iScript (Invitrogen) both according to the manufacturer's protocol. Sybergreen-based real-time qPCR was performed by using the 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA) and gene-specific primers for TNF-α, IP10, and L32 as described (4, 45). All qPCR data has been normalized to L32 values. To detect the expression of BIC and L32 mRNA, cDNA was subjected to PCR and run out on a 2% agarose gel containing ethidium bromide at 1 μg/ml. The primer sequences used to detect BIC were 5′-ttggcctctgactgactcct-3′ (forward) and 5′-gcagggtgactcttggactt-3′ (reverse).
MicroRNA Detection.
For detection of miR-155 by Northern blotting, RNA was extracted by using the TRIzol reagent according to the manufacturer's protocol (Invitrogen). Fifteen micrograms of total RNA was electrophoretically separated on a 12% polyacrylamide denaturing gel, and tRNA was visualized by using ethidium bromide staining to ensure the quality and relative amount of the RNA. Total RNA was next transferred to a GeneScreenPlus membrane (PerkinElmer, Boston, MA) by using a semidry Transblot electrophoresis apparatus (Bio-Rad, Hercules, CA). The RNA was crosslinked to the membrane by using UV radiation. Hybridization was carried out by using ULTRAHybOligo solution according to the manufacturer's instructions (Ambion). The probe sequence was complementary to the mature form of miR-155, and was labeled with γ-32P. After being washed, the membranes were imaged by using a STORM phosphorimager. Detection of miR-155 and U6 was also performed by using the mirVana qRT-PCR miRNA detection kit according to the manufacturer's instructions (Ambion).
Flow Cytometry.
To detect expression of CD86 or CD11b, RBC-depleted splenocytes were stained in FACS buffer (1× PBS/0.1% BSA/2% FBS/0.1% normal mouse serum) by using phycoerythrin-conjugated anti-CD11b or FITC-conjugated anti-CD86 (Ebiosciences, San Diego, CA) and fixed with paraformaldehyde (1% final concentration). Surface expression was assayed by using a FACScan flow cytometer (Becton Dickenson, Franklin Lakes, NJ).
Promoter Analysis.
Promoter analysis software from Genomatix (Munich, Germany) was used to identify putative binding sites within the BIC promoter.
Supplementary Material
Acknowledgments
We thank Bruce Beutler (The Scripps Institute, La Jolla, CA) for providing TRIF mutant mice, Shizuo Akira (Osaka University, Osaka, Japan) for the MyD88-deficient mice; Arash Shahangian for help with macrophage culture and RNA preparation; and Jose Luis Riechmann, Vijaya Rao, Jaclyn Shingara, and David Brown for help with microarray work. This work was supported by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology and by grants from the National Institutes of Health.
Abbreviations
- miRNA
microRNA
- LPS
lipopolysaccharide
- MyD88
myeloid differentiation factor 88
- poly(I:C)
polyriboinosinic polyribocytidylic acid
- qPCR
quantitative PCR
- TLR
Toll-like receptor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610731104/DC1.
References
- 1.Akira S, Takeda K. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Modlin RL, Cheng G. Nat Med. 2004;10:1173–1174. doi: 10.1038/nm1104-1173. [DOI] [PubMed] [Google Scholar]
- 3.Dunne A, O'Neill LA. FEBS Lett. 2005;579:3330–3335. doi: 10.1016/j.febslet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 5.Kracht M, Saklatvala J. Cytokine. 2002;20:91–106. doi: 10.1006/cyto.2002.0895. [DOI] [PubMed] [Google Scholar]
- 6.Anders HJ, Zecher D, Pawar RD, Patole PS. Arthritis Res Ther. 2005;7:215–224. doi: 10.1186/ar1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balkwill F. Cancer Metastasis Rev. 2006;25(3):409–416. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 8.Karin M, Lawrence T, Nizet V. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 10.Cai X, Hagedorn CH, Cullen BR. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, Croce CM. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 17.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CZ, Li L, Lodish HF, Bartel DP. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 20.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taganov KD, Boldin MP, Chang KJ, Baltimore D. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clurman BE, Hayward WS. Mol Cell Biol. 1989;9:2657–2664. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, Lee EJ, Schmittgen TD. Genes Chromosomes Cancer. 2006;45:103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- 25.Tam W, Ben-Yehuda D, Hayward WS. Mol Cell Biol. 1997;17:1490–1502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam W. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 27.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 28.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 29.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 30.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 31.Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, Maitland M, Norgard MV, Plevy SE, Smale ST, et al. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 32.Schroder K, Sweet MJ, Hume DA. Immunobiology. 2006;211:511–524. doi: 10.1016/j.imbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Calder CJ, Nicholson LB, Dick AD. J Immunol. 2005;175:6286–6293. doi: 10.4049/jimmunol.175.10.6286. [DOI] [PubMed] [Google Scholar]
- 34.Hertzog PJ, O'Neill LA, Hamilton JA. Trends Immunol. 2003;24:534–539. doi: 10.1016/j.it.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 35.van Boxel-Dezaire AH, Rani MR, Stark GR. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 38.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. J Biol Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 39.van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S. Genes Chromosomes Cancer. 2003;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 40.Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, Nong Y, Wen D, Adams J, Dang L, Staudt LM. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- 41.Karin M. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 42.Shaulian E, Karin M. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 43.Tam W, Dahlberg JE. Genes Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 44.Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, et al. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 45.Covert MW, Leung TH, Gaston JE, Baltimore D. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.