Abstract
The Wnt pathways are evolutionarily well-conserved signal transduction pathways that are known to play important roles in all Metazoans investigated to date. Here, we examine the Wnt pathway genes and target genes present in the genome of the echinoderm Strongylocentrotus purpuratus. Analysis of the Wnt genes revealed that eleven of the thirteen reported Wnt subfamilies are represented in sea urchin, with the intriguing identification of a Wnt-A ortholog thought to be absent in deuterostomes. A phylogenetic study of the Frizzled proteins, the Wnt receptors, performed throughout the animal kingdom showed that not all Frizzled subfamilies were present in the metazoan common ancestor, e.g. Fz3/6 emerged later during evolution. Using sequence analysis, orthologs of the vast majority of the cellular machinery involved in transducing the three types of Wnt pathways were found in the sea urchin genome. Further, of about one hundred target genes identified in other organisms, more than half have clear echinoderm orthologs. Thus, these analyses produce new inputs in the evolutionary history of the Wnt genes in an animal occupying a position that offers great insights into the basal properties of deuterostomes.
Keywords: Sea urchin, genome survey, Wnt, Frizzled, canonical, planar cell polarity (PCP), Wnt/calcium
Introduction
The Wnt pathways are evolutionarily conserved signaling pathways that regulate multiple aspects of metazoan development. They are required throughout development, from early axis specification to organogenesis (e.g. Logan and Nusse, 2004; Cadigan and Nusse, 1997; Katoh, 2005; Veeman et al., 2003a). The Wnt pathways operate by three distinct mechanisms referred to as the canonical pathway, the planar cell polarity (PCP) pathway and the calcium/Wnt pathway (Fig.S1). Of these three signaling the canonical Wnt pathway is the best characterized. Its activation results in entry of β-catenin into the nucleus of the cell where in concert with a TCF/Lef family member it activates transcription of target genes (Nusse, 1999). The PCP pathway operates through small G proteins such as RhoA and Rac to activate target genes, usually through the inducement of an AP-1 transcriptional complex (Hwang et al., 2005; Veeman et al., 2003b). The third Wnt pathway, the calcium/Wnt pathway, utilizes other small GTPases to control calcium-dependent molecules, including CamKII and PKC, leading to the transcriptional activation of its target genes (Kohn and Moon, 2005). Each of the three Wnt pathways is activated in the same way by the binding of a Wnt ligand to a Frizzled receptor. Transduction of the ligand/receptor interaction subsequently involves more than a hundred proteins that relay and modify the signal on the way to target actuation in a Wnt-pathway specific manner. Proteins in the canonical Wnt pathway regulate the stability and movement of the key transcriptional activator β-catenin (Logan and Nusse, 2004). This pathway is principally required in many organisms for axis specification and endoderm differentiation (Croce and McClay, 2006; Imai et al., 2000; Wikramanayake et al., 2004; Zorn et al., 1999). The PCP pathway is not commonly associated with tissue specification but is known instead to control cytoskeletal rearrangements and cell movements such as convergent extension (CE) occurring during gastrulation in deuterostomes (Croce et al., 2006; Heisenberg et al., 2000; Kilian et al., 2003; Wallingford et al., 2002). Finally, the calcium/Wnt pathway also acts on cellular behavior regulating cell adhesion and movement. However, this pathway functions through a different set of proteins that modulate the intracellular concentrations of free calcium and cyclic guanosine monophosphate (cGMP) (Wang and Malbon, 2003). To date, in addition to the main Wnt pathways components, more than eighty other molecules have been reported as modifiers that amplify, degrade, stabilize, or alter the trajectory of the signals (Klein and Mlodzik, 2005; Logan and Nusse, 2004; Wang and Malbon, 2003). Most of these proteins have been identified in fully sequenced genomes, including human, ascidian and flies, though some of them have also been reported from individual studies in other more basal organisms such as cnidarians. Here, we present the first exhaustive report of the Wnt pathways genes in the phylum Echinodermata.
Emerging just after the protostome/deuterostome divergence, the Echinodermata offer insights into the earliest and the most basic of the deuterostome animals (Fig.1A). This phylum contains diverse non-chordate marine organisms including sea urchins. Due to its phylogenetic position, the newly available genomic sequence of Strongylocentrotus purpuratus provides, therefore, key material for evolutionary history analyses. In this study, using bioinformatics techniques, we report the identification of the signaling components and the target genes of the three Wnt pathways in the S. purpuratus genome. The results clearly show that the entire three Wnt pathways are highly conserved with more than 95% of the more than 100 genes in the pathway represented in the sea urchin genome. Further, in cases where there are multiple members within the same gene family, such as the Wnt family, the sea urchin genome reveals many deuterostome-like properties but also contains, as might be expected from its basal position, genes that are absent in more derived deuterostome groups but that are present in cnidarians and/or protostomes.
Fig. 1.
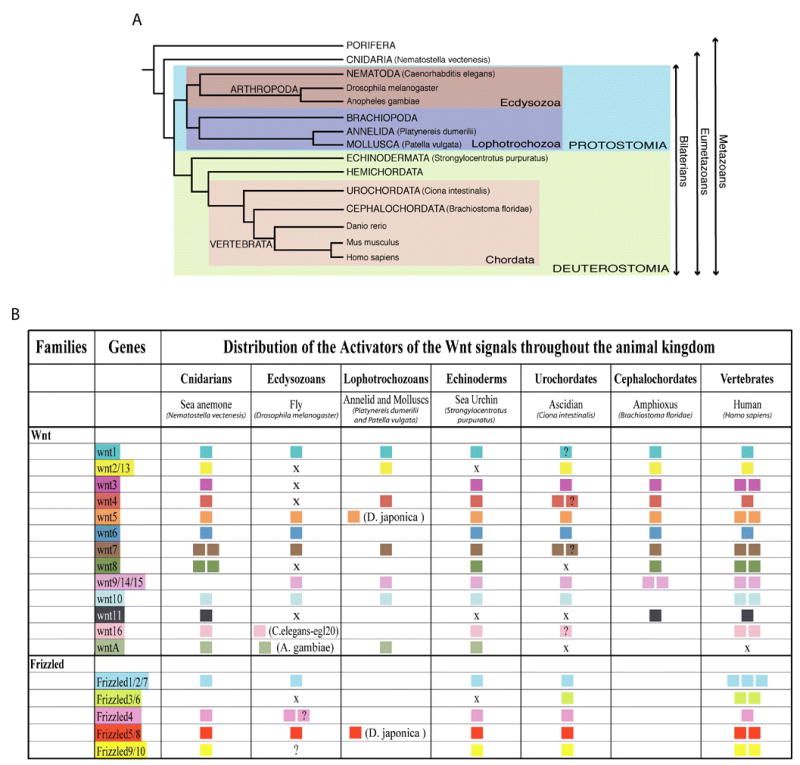
A. A phylogeny of metazoan evolution. This tree depicts likely relationships of the species used in this study within the metazoan subkingdom. B. Distribution of Wnt and Frizzled genes throughout the metazoan sub-kingdom. An X designates the absence of a member of that subfamily in the corresponding annotated genome. Question marks underline uncertainty of the orthology. The absence of information indicates that no corresponding orthologs have been reported in these species for which genomic databases are not yet available. For lineages other than the Echinodermata, data derive from Hino et al., 2003; Holland, 2002; Hotta et al., 2003; Jockusch and Ober, 2000; Katoh and Katoh, 2005; Kusserow et al., 2005; Lee et al., 2006; Leveugle et al., 2004; Maloof et al., 1999; Marsal et al., 2003; Nusse, 2001; Prud’homme et al., 2002; and Schubert et al., 2000).
Material and methods
Resources of genes sequences
The genomic sequence of Strongylocentrotus purpuratus was deciphered and provided by BCM-HGSC [Baylor College of Medicine- Human Genome Sequencing Center] (http://www.hgsc.bcm.tmc.edu/blast/blast.cgi?organism=Spurpuratus). Additional genomic resources were avalaible from several ESTs databases all accessible online at http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=7668. Proteins sequences for Wnt pathways related genes and targets from other animals were obtained from Genbank (http://www.ncbi.nlm.nih.gov/Genbank/index.html) (Benson et al., 2005), Ensembl (http://www.ensembl.org/index.html) (Hubbard et al., 2005), Pfam (http://www.sanger.ac.uk/Software/Pfam/) (Bateman et al., 2004), StellaBase (http://www.stellabase.org/) (Sullivan et al., 2006), Aniseed (Ascidian Network for In Situ Expression and Embryological Data) (http://crfb.univ-mrs.fr/aniseed/index.php) and by BLAST searches (Altschul et al., 1997) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/). Sea urchin sequences were all identified by blastp and blastn, using as guidance against genomic databases confirmed genes identified from a range of diverse animal species. All predicted sequences were reciprocally blasted against non-redundant NCBI databases leading to the identification of the best human hit, which was reblasted against the S. purpuratus genomic sequences for bi-directional best-hit analysis.
Phylogenetic Analysis
Proteins alignments were carried out using ClustalX (Thompson et al., 1997) and saved as nexus files. Phylogenetic trees were generated using four complementary methods when needed. First, neighbor-joining trees (Saitou and Nei, 1987) were run with the PAUP 4.0 program (Swofford, 1998) and with 5000 bootstrap replicates. Second, unweighted maximum parsimony reconstructions were performed for each tree with a heuristic search of 1000 replicates. Third, Maximum likelihood trees were computed using RAxML VI-1.0 (Stamatakis et al., 2005) and employing the Jones-Taylor-Thornton model of amino acid substitution, with otherwise default settings. Finally, Bayesian trees were performed with Mr. Bayes v.3.1.1 (Huelsenbeck et al., 2001; Ronquist and Huelsenbeck, 2003) and analyses were run for 500000 generations and node probabilities were calculated after a burn-in of 50000 generations. All trees were displayed with TreeView X 0.5.0, saved as svg files, and colored and converted into JPG with Adobe Illustrator. Accession numbers of all sequences used for those trees are available in tables S1 and S2 in supplementary materials.
Embryonic expression and RT-PCR
Embryonic expression of annotated sequences was confirmed using the tiling array database (Samanta et al., 2006). For the Wnt genes additional RT-PCR was performed from total RNA from various developmental stages. After extraction using the method of Cathala et al. (1983), cDNA were synthesized with a TaqMan kit from Clontech. RT-PCRs were carried out using standard protocol with specific primers designed against S. purpuratus Wnt predicted sequences.
Results
1. The activators of the Wnt pathways: Wnt and Frizzled proteins
1.a. Wnt family
The Wnt signaling molecules are a large family of secreted glycoproteins characterized by an invariant pattern of 22 to 24 highly conserved cysteine residues usually present in the last 70 C-terminal amino-acids (Van Ooyen et al., 1985). In humans, nineteen Wnt proteins have been identified that define twelve distinct subfamilies named from Wnt-1 to Wnt-11 and Wnt-16 (Miller, 2002) (Fig.1B). The Cnidaria, a basal non-bilaterian phylum thought to more closely reflect the eumetazoan ancestor (Fig.1A), possesses 14 Wnt orthologs that sort into twelve distinct subfamilies, one of which, WntA, does not have a human representative (Kusserow et al., 2005) (Fig.1B). In sea urchin, prior to the availability of the genome four members of the Wnt family (Wnt-1, -4, -5 and -8) had been identified. Each of these genes is expressed in embryos and of these, Wnt-8 has been shown to be required for endoderm specification during embryogenesis (Ferkowicz et al., 1998; Wikramanayake et al., 2004).
In silico analysis of the Strongylocentrotus purpuratus genomic sequence revealed the presence of eleven Wnt genes, including Wnt-1, -4, -5 and -8 (Table S1). To determine whether these genes were expressed, EST and tiling data were evaluated (Samanta et al., 2006) and RT-PCR analysis was performed on S. purpuratus cDNA at different developmental stages (Table S1). Based on these three independent methods, we conclude that the eleven Wnt genes identified are expressed during development. Moreover, the RT-PCR analysis at different developmental stages indicated that more than one Wnt is expressed at each stage. At the gastrula stage, for example, nine of the eleven Wnts are transcribed (Fig.2A), indicating a highly complex utilization of the Wnt pathways during embryonic development. To identify the orthology of these Wnt genes, molecular phylogenetic analyses were performed using Wnt sequences from representatives of the cnidarian, protostome and vertebrate lineages (Table S2). This analysis showed that, all sea urchin Wnt genes appear as unique members of the Wnt subfamilies to which they belong, indicating that eleven of the thirteen Wnt subfamilies recorded are represented (Fig.2B). Orthologs of Wnt-1, and Wnt-3 to -10 were found, supporting the maintenance of these subfamilies in the Echinodermata from the eumetazoan common ancestor. Exhaustive searches failed to detect S. purpuratus orthologs for Wnt-2 or -11 proteins. Based on the distribution of Wnt subfamilies throughout the animal kingdom (Fig.1), loss of the Wnt-2 ortholog appears uncommon in the Metazoa, whereas the absence of Wnt-11 is frequently encountered, at least among the groups of animals investigated.
Fig. 2.
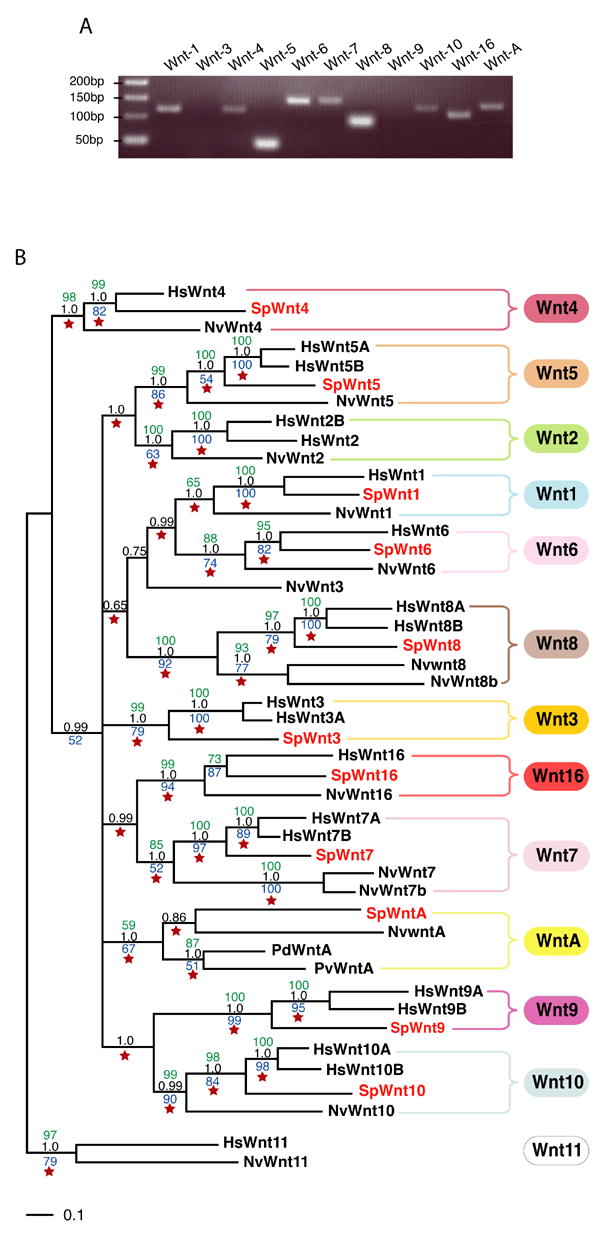
A. RT-PCR carried out on Strongylocentrotus purpuratus cDNA prepared from gastrula stage embryos. B. Phylogenetic analysis of the sea urchin Wnt family. Confidence values were calculated using neighbor joining (green), maximum parsimony (blue), maximum likelihood (red star) and Bayesian (black) methods. The outgroup is defined by Wnt11 proteins. Accession numbers of all the proteins sequences used to elaborate the tree are indicated in Tables S1 and S2. Hs, Homo sapiens; Nv, Nematostella vectensis; Hv, Hydra vulgaris; Pd, Platynereis dumerilii; Pv, Patella vulgata; Sp, Strongylocentrotus purpuratus.
The discovery of a Wnt-A ortholog in the sea urchin genome provides the most intriguing result. Wnt-A proteins are presents in cnidarians, ecdysozoans and lophotrochozoans but have not been reported in any chordate lineage to date (Fig.1). Prior to this study, the Wnt-A genes were therefore presented as defining a subfamily restricted to non-deuterostome animals (Kusserow et al., 2005). However, the presence of SpWntA modifies this point of view. The accuracy of that sequence as being a WntA ortholog has been assessed by four independent and complementary phylogenetic analyses and they all support the nature of that sequence with great confidence (bootstrap values and node probabilities ranging between 59 to 100 depending on the analysis – Fig.2B). Thus, based on the discovery of SpWntA in the sea urchin, the WntA subfamily is present in the deuterostomes, though how broadly Wnt-A genes occur in the deuterostomata remains to be established. Nevertheless, this result underscores the value of increasing the number of phyla containing at least one fully sequenced genome to provide valuable insights allowing more reliable evolutionary conclusions.
Thus, our analysis supports the notion that of the thirteen Wnt subfamilies, twelve were present in the eumetazoan common ancestor; the Wnt-9 subfamily having apparently emerged only in the Bilateria. Further, it appears that the Wnt-A subfamily was present in the common ancestor of deuterostomes, but apparently was lost during chordate evolution. Finally, since only one sea urchin ortholog has been found in each represented Wnt subfamily, it is likely that the common ancestor of the deuterostomes possessed only one Wnt gene per subfamily.
1.b. Frizzled family
The Wnt ligands activate each of the three pathways by binding to their cognate cell surface receptors of the Frizzled family (Bhanot et al., 1996). All Frizzled family members possess an extracellular region including a signal peptide and a cysteine-rich Wnt binding domain (CRD), seven transmembrane domains, a characteristic of the G protein coupled receptors, and a cytoplasmic tail displaying the conserved KTXXXW motif (Huang and Klein, 2004). In humans, ten Frizzled genes have been reported that group into five distinct subfamilies. By comparison, only four Frizzled proteins are present in the sea anemone genome and five in the ascidian (Wnt homepage website, Hotta et al., 2003; and our analysis) (Fig.1). Thus, as for the Wnt subfamilies the Frizzled subfamilies appear to be well conserved through the animal kingdom. Only one subfamily, Fz3/6, which has no orthologs identified yet in cnidarian lineages seems to have emerged later after the divergence of the Bilateria. To date, only one sea urchin Frizzled receptor has been reported and characterized. This protein is a member of the Fz5/8 subfamily and signals through the PCP pathway within the secondary mesenchyme cells to control initiation of archenteron invagination during gastrulation (Croce et al., 2006).
Via a tBlastn search on the S. purpuratus genome using human Frizzled sequences, four Frizzled candidates emerged including the previously identified Fz5/8 homolog (Table S1 and Fig.1). Transcription of all these predicted genes was supported by tiling array and in some cases by EST data as well (Table S1). Phylogenetic analysis of this family is presented in Figure 3. At first sight, each sea urchin Frizzled sequence identified affiliates to a different subfamily and not all Frizzled subfamilies are therefore represented, suggesting that the deuterostome ancestor might have possessed only four Frizzled genes, or alternatively, that the Echinoderms lost Frizzled genes. Intriguingly, as with cnidarians and protostomes, no Fz3/6 ortholog was found in the S. purpuratus genome. The Fz3/6 subfamily seems to have appeared with the emergence of the chordates. In addition, the position of this subfamily within the Fz1/2/7 subfamily suggests that Fz3/6 genes may diverge form of the chordate Fz1/2/7 molecules (Fig.3). DmFz3 and DmFz4 are quite divergent compared to the other Frizzleds. Analyzed with the neighbor-joining method and by maximum parsimony they appeared as orphan molecules defining their own subfamily. Further analysis by maximum likelihood and Bayesian analyses, however, revealed with some confidence that DmFz3 and DmFz4 belong to the Fz4 subfamily (Fig.3). Thus, it seems that the protostome Drosophila melanogaster has four Frizzled genes, one Fz5/8 ortholog (DmFz2), one Fz1/2/7 ortholog (DmFz), and two Fz4 homologs, though it is possible that one of these genes might functionally be a homolog of the Fz9/10 proteins.
Fig. 3.
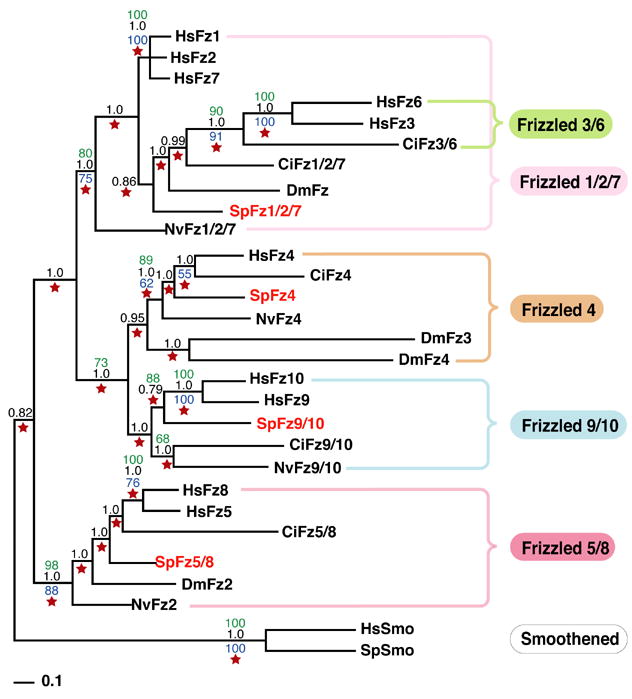
Phylogenetic analysis of the sea urchin Frizzled family. Confidence values were calculated using neighbor joining (green), maximum parsimony (blue), maximum likelihood (red star) and Bayesian (black) methods. Smoothened proteins, which are related to Frizzled, were used as an outgroup. Accession numbers of all the Frizzled sequences used to generate the tree are indicated in Tables S1 and S2. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ci, Ciona intestinalis; Nv, Nematostella vectensis; Sp, Strongylocentrotus purpuratus.
In summary, including information from the cnidaria, ecdysozoa, and protostomia in our analysis allowed us to model the evolutionary history of the Frizzled genes. Of the five known Frizzled subfamilies four (Fz1/2/7, Fz4, Fz5/8, and Fz9/10) were already present in the eumetazoan common ancestor, while the Fz3/6 subfamily appears to have arisen later during the emergence of the chordate line.
2. Signaling components of the Wnt pathways
Based on a comprehensive search including very helpful information from the Wnt homepage website (http://www.stanford.edu/~rnusse/wntwindow.html) and from many studies throughout the animal kingdom, an exhaustive analysis of proteins reported to be involved in the transduction of the three Wnt signaling pathways from the extracellular compartment to the nucleus (excluding Wnt and Frizzled proteins) is presented in Table S3. It contains 96 genes with additional molecules corresponding to paralogs or subunits of the same proteins. For each protein, the S. purpuratus genome was queried for orthologs and each discovered was examined using the tiling database for expression in the embryo. Overall, orthologs of 88.5% of the Wnt pathways components present in the table were identified in the sea urchin genome. Only eleven Wnt pathway genes could not be identified. Nine of these are associated with canonical Wnt signaling and two are involved in the PCP pathway. Although all reported components of the calcium/Wnt pathway were found, this result does not necessarily mean that this pathway is more conserved than the other two since only eight of 96 proteins in this study were related to the calcium/Wnt pathway. By contrast, two-thirds of the identified proteins play a role in the canonical Wnt pathway, indicating not only that this pathway is stringently controlled by modifiers, but also that this pathway has been better characterized and described than the two non-canonical pathways. Further, among the different cellular compartments where Wnt pathways components act (extracellular, membrane, cytoplasmic and nuclear), each compartment looks to be roughly represented by 90% of all possible components, suggesting that the level of conservation is the same along the cascade of the signal from ligand to target. Thus, the transducing apparatus of the Wnt pathways appears well conserved during evolution. Below we discuss the identification of Dickkopf orthologs and the composition of the SFRP family in the sea urchin genome.
2.a. The Dickkopf proteins
The Dickkopf (Dkk) proteins are powerful inhibitors of the canonical Wnt pathway (Glinka et al., 1998; Wu et al., 2000). They have their own cognate receptor, Kremen, for which a sea urchin ortholog has been found (TableS3). To down-regulate Wnt signaling Dkks, in binding to Kremen, also bind to the same Wnt co-receptor LRP as that required by Frizzleds (Mao et al., 2001; Mao et al., 2002; Semenov et al., 2001). Dkk proteins are cysteine-rich secreted proteins that are characterized by the presence of two cysteine-rich domains (CRD1 and CRD2), each of which has a specific highly conserved cysteine distribution (Krupnik et al., 1999). In vertebrates, four Dkk proteins have been reported (Dkk1, Dkk2, Dkk3 and Dkk4) that group into two subfamilies, the Dkk124 and the Dkk3. As yet, no Dkk homologs have been identified in protostomes, including Drosophila or C. elegans, while four Dkk homologs have been reported in the cnidarians Hydra vulgata and Hydra magnipapillata (three Dkk124 a, b and c and one Dkk3) (Augustin et al., 2006; Fedders et al., 2004; Guder et al., 2006). Intriguingly, whereas all vertebrate Dkks and the cnidarian Dkk3 possess the two common CRD motifs, the cnidarian Dkk1/2/4 possesses only one CRD domain that is most homologous to the vertebrate CRD2(Augustin et al., 2006; Guder et al., 2006) (Fig.4C).
Fig. 4.
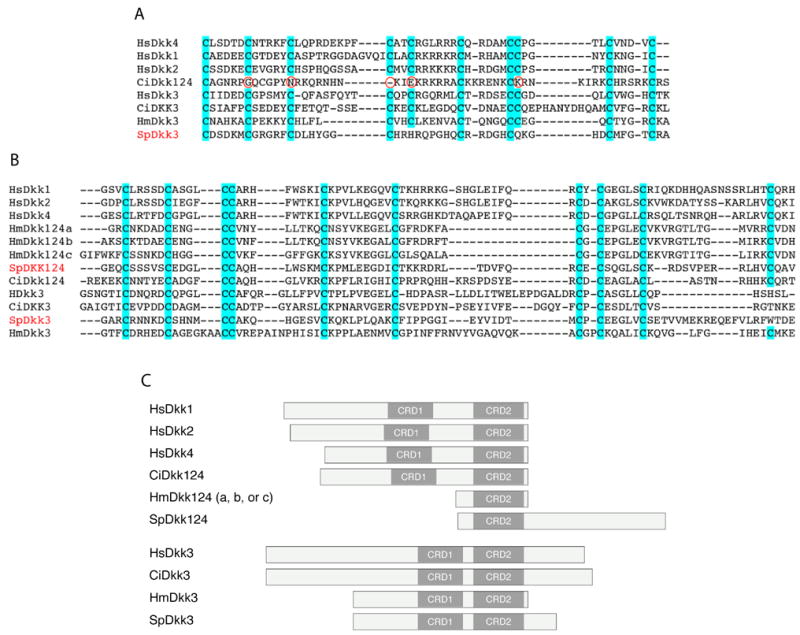
Protein structural analysis of Dickkopf (Dkk) proteins. A,B. Amino acid sequence comparison of cysteine-rich domain 1 (A) and 2 (B) of vertebrate (human, Hs), urochordate (ascidian, Ci), echinoderm (sea urchin, Sp) and cnidarian (Hydra, Hm) Dkk124 and Dkk3 related proteins. C. Schematic representation of the domain structure of Dkk proteins throughout the animal kingdom. Accession numbers of all the Dkk sequences used are indicated in Tables S2 and S3. Hs, Homo sapiens; Ci, Ciona intestinalis; Hm, Hydra magnipapillata; Sp, Strongylocentrotus purpuratus.
Based on these results we looked for Dkk orthologs in the S. purpuratus genome. Two sequences with homology with Dkk genes, particularly within the CRD domains (Fig.4A, B), were found and their embryonic expression was confirmed by the tiling assay (Table S3). Structural analysis revealed that the protein identified as SpDkk3, like other Dkk3s identified throughout the animal kingdom, contains two CRD domains (Fig.4C), both of which have a similar cysteine distribution compared to the vertebrate, the urochordate and the cnidarian Dkk3s (Fig.4A, B). By contrast, although vertebrate Dkk1, Dkk2, and Dkk4 molecules also have two CRD domains, SpDkk124 has a single CRD domain that is structurally highly similar to the vertebrate CRD2 (Fig.4B, C). This structure is reminiscent of the Hydra Dkk124 that also has only one CRD domain (Fig.4B, C). Furthermore, we found that the urochordate Ciona intestinalis also has only two Dkk genes, CiDkk124 and CiDkk3, both of which contain, like vertebrate Dkks, two CRD domains (Fig.4A-C). In the case of CiDkk124, however, sequence alignment revealed that out of the 10 cysteines present in the vertebrate CRD1 motif only five are present in the ascidian sequence (Fig.4A). Thus, the Dkk124 proteins appear to have expanded from one to two CRD motifs, relative to cnidarians, and then the Dkk genes were duplicated within the vertebrate lineages.
2.b. The SFRP family
Many extracellular proteins have been reported for their role as regulators of Wnt activity. Among them, the SFRPs (secreted Frizzled-related proteins) represent a family of genes characterized by their similarities to Frizzled receptors. As in Frizzleds, the SFRPs contain a peptide signal sequence and a single Cysteine-Rich Domain (CRD) that is very similar to the CRD of Frizzled. However, SFRPs lack transmembrane domains and instead terminate with a netrin-like domain, thus making them secreted proteins. In the extracellular space, SFRP proteins bind to Wnt proteins thereby sequestering the ligands before they reach their cognate receptors. Several SFRPs have been reported and although their preference for Wnt ligands has not yet been established, it appears that they antagonize both canonical and non-canonical Wnt pathways (for reviews see Kawano and Kypta, 2003; Jones and Jomary, 2002).
In humans, there are five SFRPs comprising two groups: the SFRP1/2/5 and the SFRP3/4 (Fig.4). In comparison, four SFRPs are present in the urochordate C. intestinalis genome (Hino et al., 2003; Hotta et al., 2003) and only one, suSFRP1, has been previously reported in the sea urchin (Illies et al., 2002). Using both the human and the ascidian SFRP sequences as queries two additional SFRP genes were found in the S. purpuratus genome. A phylogenetic analysis of the three sea urchin SFRPs sequences along with the human and the ascidian SFRPs is presented in figure 4. While one of the sea urchin SFRPs, SpSFRP1/5, appears related to the human SFRP1 and 5 proteins the other two sequences, suSFRP1 and SpSFRP3/4, branch within the SFRP3/4 group providing new information about this group. Previously the SFRP3/4 group was modeled with the two ascidian SFRP3/4 a and b as duplicate orthologs of the human SFRP3 and SFRP4 (Hotta et al., 2003). However, the introduction of the sea urchin sequences to the analysis changes this interpretation revealing a different evolutionary history of the SFRPs in the deuterostome clade. The sea urchin sequences suSFRP1 and SpSFRP3/4 appear to be independently related to human SFRP3 and 4 and to urochordate SFRP3/4 a and b respectively, each relation being supported by high bootstrap values (100 for each node). Thus, it is likely that the deuterostome common ancestor had two distinct SFRP3/4 proteins that then evolved separately within the chordate lineage. The analysis suggests the urochordates kept one gene, while vertebrates kept the other gene, both of them independently duplicating to give rise to SFRP3 and SFRP4 in human and SFRP3/4a and SFRP3/4b in ascidian. Based on tiling and EST data, SpSFRP3/4 is not transcribed in embryos, suggesting therefore that this gene is unlikely to function during embryonic development in the sea urchin.
3. Target genes of the Wnt pathways
Because Wnt signaling plays a key role in development and cancer, many studies have been conducted to determine target genes that are activated by these pathways. Studies, performed mostly on human cell lines, vertebrate embryos, and Drosophila, have led to the identification of approximately one hundred Wnt target genes (listed in table S4). Examination of this table reveals once again that much more information is available regarding the activities and roles of the canonical Wnt pathway versus the non-canonical pathways. To date, more than 90% of identified target genes of the Wnt pathways are activated by the canonical pathway, although a few of them can also be activated by the PCP pathway (Pongracz and Stockley, 2006).
Genes targeted by the Wnt pathways were grouped into three sets of molecules: signaling components (either secreted, transmembrane receptors or cytoplasmic proteins), transcription factors and other nuclear proteins, and cell adhesion molecules (Table S4). Some Wnt pathway components were presented as targets in some reports, offering feedback regulation of pathway function but these were excluded from this section. An attempt to identify orthologs of each of the reported target genes yielded recognizable proteins for 57% of the targets (Table S4). Of the three groups of targets, cell adhesion molecules are the least well represented (only 22%), whereas transcription factors were identified as the most recognizable (79%, against 49% for the signaling molecules). In agreement with our results on cell adhesion molecules, many vertebrate adhesion molecules are missing from the sea urchin genome according to the analysis presented in the Whittaker et al., paper (this issue).
Among the signaling molecules listed, paralogs or precursors for some of these proteins have been annotated, though it was difficult to conclude in a number of cases that an annotated sea urchin gene was a direct ortholog. This is the case for autotaxin, an ectonucleotide pyrophosphatase / phosphodiesterase (ENPP) protein, osteocalcin, PPAR delta and RAR gamma (see Table S4). Nevertheless, in the future additional functional data may add these genes and others as targets of the Wnt pathways. Alternatively, since these molecules have only been established as Wnt targets in vertebrates, it may be that their regulation has specifically emerged as a new role only in the vertebrate lineages.
Finally, some Wnt targets identified in Drosophila melanogaster, such as the transcription factor Stripe, have sea urchin orthologs, but do not have identified orthologs in vertebrates. In contrast, proteins exclusive (to date) to zebrafish or to Xenopus (tagged in yellow and pink respectively) have no recognizable orthologs in S. purpuratus (e.g. transcription factors Dharma/Bozozok, Siamois and Twin). Thus, although preliminary, these data suggest that additional Wnt targets may be found in the vertebrates and that vertebrates have acquired Wnt targets that were not present in the deuterostome common ancestor.
Discussion
The Wnt pathways, particularly the canonical and the PCP pathways, have been reported throughout the animal kingdom to be functionally well conserved (Cadigan and Nusse, 1997; Croce and McClay, 2006; Klein and Mlodzik, 2005; Mlodzik, 2002). Wnt ligands, Frizzled receptors and many other main components of these signaling pathways have been characterized from cnidarians to vertebrates, indicating that in all metazoan lineages the pathways play similar roles. The canonical pathway is heavily used in metazoans to establish axial asymmetries and drive endoderm formation during embryogenesis (Croce and McClay, 2006; Holland, 2002; Imai et al., 2000; Schier and Talbot, 2005; Wikramanayake et al., 2004; Zorn et al., 1999). In the same way, the PCP pathway in vertebrates, urochordates, protostomes and as recently shown in echinoderms, controls convergent extension movements involved in many morphogenetic events including gastrulation (Croce et al., 2006; Heisenberg et al., 2000; Kilian et al., 2003; Sasakura and Makabe, 2001; Shulman et al., 1998; Wallingford et al., 2002). Thus, to have a similar, widespread impact on the development of organisms as different as cnidarians, echinoderms, and vertebrates, the proteins responsible for the transduction of the signals, their relationships to one another, and the gene targets they control should be well conserved. In agreement with that prediction, among the signaling components that compose the Wnt pathways from the extracellular compartment to the nucleus, more than 85% are conserved at least between sea urchin and human (Fig.6). In addition, more than 55% of the proteins reported as Wnt target genes in vertebrates also have a closely corresponding echinoderm ortholog (Fig.6). Thus, in addition to being functionally conserved our study also supports that the Wnt pathways are also structurally well preserved.
Fig. 6.
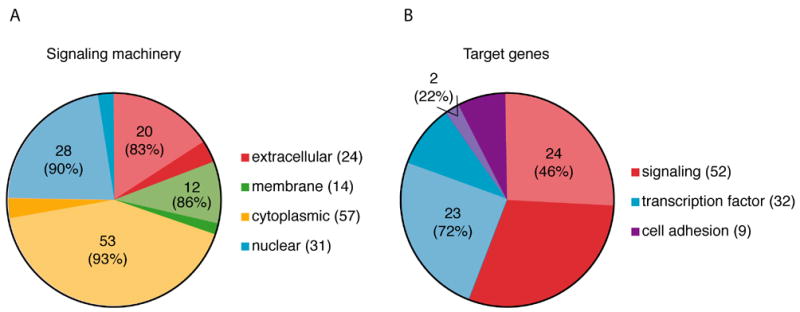
Pie charts representative of (A) Wnt pathway components and (B) target genes found in the sea urchin genome compared to vertebrates. Each color defines a distinct set of molecules with the lighter color corresponding to the proportion of these molecules that have clear orthologs in the sea urchin genome. Numbers in parentheses indicate the total number of genes used for each group of molecules. Numbers on the pie chart illustrate the number of corresponding sea urchin orthologs found and the percentage of sea urchin orthologs compared to the total of genes identified in vertebrates for this particular set of molecules.
The activators of Wnt signaling
Deciphering the S. purpuratus genome led to identification of eleven Wnt ligands, each belonging to a different Wnt subfamily (Figs.1 and 2). This analysis provides new information on Wnts throughout the animal kingdom. One of the most important findings is the presence of a Wnt-A protein in the sea urchin, modifying the previous conclusion that the Wnt-A family was only represented in non-deuterostome organisms (Kusserow et al., 2005). Further, no Wnt-11 subfamily members were found in S. purpuratus. Wnt-11 has recently been established in Xenopus as being the maternal Wnt ligand activating the canonical Wnt signaling in order to specify the dorso-ventral axis (Tao et al, 2005). However, intriguingly, although a Wnt-11 gene has been reported in sea anemones (Kusserow et al., 2005), no Wnt-11 orthologs have either been detected in protostomes, lophotrochozoans or urochordates. This absence of Wnt-11 genes in animals other than cnidarians and vertebrates and the lack of information concerning the role of Wnt-11 in sea anemone leave the interpretation of that result open. One possibility is that the function of Wnt-11 as an activator of the maternal Wnt pathway is specific to the vertebrate lineages, while in other phyla this function is conducted by another maternal Wnt ligand not yet identified; i.e. in sea urchins RT-PCR analyses indicate that among the eleven Wnts isolated five are present maternally in the egg.
Whereas an exhaustive literature about the evolutionary history of Wnts is available, this is not the case for the Frizzled receptors for which also fewer functional analyses have been conducted. Our study presents the first evolutionary analysis of Frizzled receptors including Frizzled data from cnidarians to humans. This analysis indicates that the Eumetazoan common ancestor most likely possessed four distinct Frizzled receptors (orthologs to the Fz1/2/7, Fz4, Fz5/8, and Fz9/10) whereas the other Frizzled subfamily Fz3/6 appears to have emerged later after the divergence of the Chordata from the common deuterostome ancestor.
The Wnt/Frizzled interactions represent one of the most complex relationships between extracellular ligands and receptors, and the specificity of these interactions remains unclear. As in humans, ascidians, or flies, sea urchins have at least twice as many Wnts as Frizzleds, indicating that each receptor must have more than one ligand or that the mechanism underlying signal specificity is not yet well understood. Studies performed mostly in cell culture and in vertebrates have shown that several Wnts can interact with a given Frizzled (e.g. Wang et al., 2005; Wu and Nusse, 2002), but whether this promiscuity of ligand-receptor interaction occurs in vivo has not been resolved. In the sea urchin each of Wnt or Frizzled subfamily is represented by a single ortholog, offering an opportunity that may simplify analyses of Wnt-receptor specificity. This analysis has additional importance since nine of the eleven Wnts identified are simultaneously expressed at the gastrula stage, making it imperative to sort specificity of the responses. An obvious way to control Wnt/Frizzled interactions could be to regulate the temporal and spatial distribution of the components. However, with nine Wnts expressed in the gastrula stage embryo, careful analysis of the Frizzled receptors is crucial. In agreement, in situ analysis, performed in the cnidarian Nemanostella vectensis for all Wnt ligands, revealed that some are expressed at the same time and in the same tissue during embryogenesis (Kusserow et al., 2005). Alternatively, this ligand-receptor specificity may be controlled by the extracellular regulators and/or by Frizzled co-receptors. Unfortunately, to date little is known about the expression or the roles of these molecules in echinoderms and urochordates.
Signaling components and target genes of the Wnt pathways
Throughout the animal kingdom roughly one hundred Wnt signal transduction components and one hundred target genes have been identified (Tables S3 and S4). Using these molecules, orthologs of more than 88% of the Wnt pathways components and 57% of the target genes were identified with confidence in the S. purpuratus genome (Fig.6). Given the preservation of the roles of the Wnt pathways through evolution, the conservation of the signaling machinery was not surprising. Further, while the percentage of the targets found may seem low, most of the described target genes were originally identified in vertebrates, and genome duplications may have increased the number of targets and diversified them to the extent that orthologs can no longer readily be identified in the sea urchin genome.
The number of identified signal transduction genes in the canonical Wnt pathway has recently been elevated beyond that reported here by an RNAi study in Drosophila. This study identified 238 potential regulators that include most of the known core canonical pathway members plus many others (DasGupta et al., 2005). Reciprocal-best-blast analyses have already shown that 50% of these genes have human orthologs. However these data were not included in our table. The number of signal transduction components and target genes related to the Wnt pathways are therefore likely to be expanded in the future, in sea urchins as in other organisms, by novel systematic investigations performed as the one conducted by DasGupta and collaborators.
In summary, the study presented here provides an exhaustive overview of the signaling components and target genes of the crucial developmental Wnt pathways in the sea urchin genome. The comprehensive analysis covers a large number of genes and should provide a resource for further detailed investigations of the function of individual Wnt pathway genes or for evaluating the evolutionary history of these gene families.
Supplementary Material
Fig. 5.
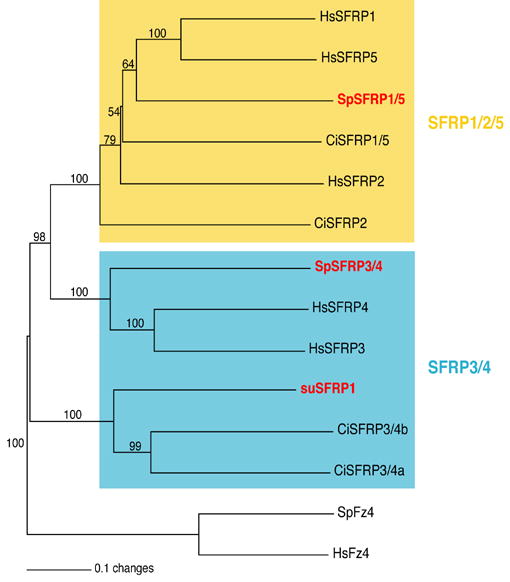
Neighbor-joining tree of SFRP family. Statistical bootstrap values appear above the branches. The two SFRP subfamilies are delimited by colored rectangles and the CRD domains of Frizzled 4 proteins were used as an outgroup. Accession numbers of all sequences used for that tree are indicated in Tables S2 and S3. Hs, Homo sapiens; Ci, Ciona intestinalis; su or Sp, sea urchin Strongylocentrotus purpuratus.
Acknowledgments
We thank the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) team who generated the S. purpuratus genome and coordinated the project. We also thank James Balhoff for his helpful contribution in the generation of the trees and Tsuyoshi Momose and Evelyn Houliston for their help on the cnidarians Frizzled sequences. This work was supported by AHA Postdoctoral Fellowship 0420074Z (C.A Byrum), by NIH grants GM61464 and HD14483 (DRM), by the National Science Foundation, the Ingeborg v. F. McKee Fund, and the George F. Straub Trust of the Hawaii Community Foundation (AHW), and by grants from the CNRS, the University Pierre et Marie Curie (ParisVI), the Association pour la Recherche sur le Cancer and Marine Genomics Europe (CG).
Footnotes
Supplementary material is accessible alongside the online version of the paper at doi:…
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin R, Franke A, Khalturin K, Kiko R, Siebert S, Hemmrich G, Bosch TC. Dickkopf related genes are components of the positional value gradient in Hydra. Dev Biol. 2006;296:62–70. doi: 10.1016/j.ydbio.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–8. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–30. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cathala G, Savouret JF, Mendez B, West BL, Karin M, Martial JA, Baxter JD. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2:327–333. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Croce J, Duloquin L, Lhomond G, McClay DR, Gache C. Frizzled5/8 is required in secondary mesenchyme cells to initiate archenteron invagination during sea urchin development. Development. 2006;133:547–57. doi: 10.1242/dev.02218. [DOI] [PubMed] [Google Scholar]
- Croce J, McClay DR. The Canonical Wnt Pathway in Embryonic Axis Polarity. Seminars in Cell & Developmental Biology. 2006;17 doi: 10.1016/j.semcdb.2006.04.004. in press. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–33. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- Fedders H, Augustin R, Bosch TC. A Dickkopf- 3-related gene is expressed in differentiating nematocytes in the basal metazoan Hydra. Dev Genes Evol. 2004;214:72–80. doi: 10.1007/s00427-003-0378-9. [DOI] [PubMed] [Google Scholar]
- Ferkowicz MJ, Stander MC, Raff RA. Phylogenetic relationships and developmental expression of three sea urchin Wnt genes. Mol Biol Evol. 1998;15:809–19. doi: 10.1093/oxfordjournals.molbev.a025986. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–62. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Guder C, Pinho S, Nacak TG, Schmidt HA, Hobmayer B, Niehrs C, Holstein TW. An ancient Wnt-Dickkopf antagonism in Hydra. Development. 2006;133:901–11. doi: 10.1242/dev.02265. [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, Stemple DL, Smith JC, Wilson SW. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature. 2000;405:76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- Hino K, Satou Y, Yagi K, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. VI. Genes for Wnt, TGFbeta, Hedgehog and JAK/STAT signaling pathways. Dev Genes Evol. 2003;213:264–72. doi: 10.1007/s00427-003-0318-8. [DOI] [PubMed] [Google Scholar]
- Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407:186–9. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- Holland LZ. Heads or tails? Amphioxus and the evolution of anterior-posterior patterning in deuterostomes. Dev Biol. 2002;241:209–28. doi: 10.1006/dbio.2001.0503. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Ueno N, Gojobori T. A genome-wide survey of the genes for planar polarity signaling or convergent extension-related genes in Ciona intestinalis and phylogenetic comparisons of evolutionary conserved signaling components. Gene. 2003;317:165–85. doi: 10.1016/s0378-1119(03)00700-5. [DOI] [PubMed] [Google Scholar]
- Huang HC, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 2004;5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, Down T, Durbin R, Fernandez-Suarez XM, Gilbert J, Hammond M, Herrero J, Hotz H, Howe K, Iyer V, Jekosch K, Kahari A, Kasprzyk A, Keefe D, Keenan S, Kokocinsci F, London D, Longden I, McVicker G, Melsopp C, Meidl P, Potter S, Proctor G, Rae M, Rios D, Schuster M, Searle S, Severin J, Slater G, Smedley D, Smith J, Spooner W, Stabenau A, Stalker J, Storey R, Trevanion S, Ureta-Vidal A, Vogel J, White S, Woodwark C, Birney E. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–53. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–4. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Hwang SG, Yu SS, Lee SW, Chun JS. Wnt-3a regulates chondrocyte differentiation via c-Jun/AP-1 pathway. FEBS Lett. 2005;579:4837–42. doi: 10.1016/j.febslet.2005.07.067. [DOI] [PubMed] [Google Scholar]
- Illies MR, Peeler MT, Dechtiaruk A, Ettensohn CA. Cloning and developmental expression of a novel, secreted frizzled-related protein from the sea urchin, Strongylocentrotus purpuratus. Mech Dev. 2002;113:61–4. doi: 10.1016/s0925-4773(01)00657-8. [DOI] [PubMed] [Google Scholar]
- Imai K, Takada N, Satoh N, Satou Y. (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127:3009–20. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- Jockusch EL, Ober KA. Phylogenetic analysis of the Wnt gene family and discovery of an arthropod wnt-10 orthologue. J Exp Zool. 2000;288:105–19. [PubMed] [Google Scholar]
- Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–20. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- Katoh M. WNT/PCP signaling pathway and human cancer (review) Oncol Rep. 2005;14:1583–8. [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Comparative genomics on Wnt16 orthologs. Oncol Rep. 2005;13:771–5. [PubMed] [Google Scholar]
- Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- Kilian B, Mansukoski H, Barbosa FC, Ulrich F, Tada M, Heisenberg CP. The role of Ppt/Wnt5 in regulating cell shape and movement during zebrafish gastrulation. Mech Dev. 2003;120:467–76. doi: 10.1016/s0925-4773(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Klein TJ, Mlodzik M. Planar cell polarization: an emerging model points in the right direction. Annu Rev Cell Dev Biol. 2005;21:155–76. doi: 10.1146/annurev.cellbio.21.012704.132806. [DOI] [PubMed] [Google Scholar]
- Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005;38:439–46. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–60. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol. 2006;17:157–67. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Leveugle M, Prat K, Popovici C, Birnbaum D, Coulier F. Phylogenetic analysis of Ciona intestinalis gene superfamilies supports the hypothesis of successive gene expansions. J Mol Evol. 2004;58:168–81. doi: 10.1007/s00239-003-2538-y. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–7. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–5. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Marsal M, Pineda D, Salo E. Gtwnt-5 a member of the wnt family expressed in a subpopulation of the nervous system of the planarian Girardia tigrina. Gene Expr Patterns. 2003;3:489–95. doi: 10.1016/s1567-133x(03)00058-9. [DOI] [PubMed] [Google Scholar]
- Miller JR. The Wnts. Genome Biol. 2002;3:REVIEWS3001. doi: 10.1186/gb-2001-3-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M. Planar cell polarization: do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation? Trends Genet. 2002;18:564–71. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- Nusse R. WNT targets. Repression and activation. Trends Genet. 1999;15:1–3. doi: 10.1016/s0168-9525(98)01634-5. [DOI] [PubMed] [Google Scholar]
- Nusse R. An ancient cluster of Wnt paralogues. Trends Genet. 2001;17:443. doi: 10.1016/s0168-9525(01)02349-6. [DOI] [PubMed] [Google Scholar]
- Pongracz JE, Stockley RA. Wnt signalling in lung development and diseases. Respir Res. 2006;7:15. doi: 10.1186/1465-9921-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Lartillot N, Balavoine G, Adoutte A, Vervoort M. Phylogenetic analysis of the Wnt gene family. Insights from lophotrochozoan members. Curr Biol. 2002;12:1395. doi: 10.1016/s0960-9822(02)01068-0. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sasakura Y, Makabe KW. Ascidian Wnt-5 gene is involved in the morphogenetic movement of notochord cells. Dev Growth Differ. 2001;43:573–82. doi: 10.1046/j.1440-169x.2001.00602.x. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular Genetics of Axis Formation in Zebrafish. Annu Rev Genet. 2005 doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Schubert M, Holland LZ, Holland ND, Jacobs DK. A phylogenetic tree of the Wnt genes based on all available full-length sequences, including five from the cephalochordate amphioxus. Mol Biol Evol. 2000;17:1896–903. doi: 10.1093/oxfordjournals.molbev.a026291. [DOI] [PubMed] [Google Scholar]
- Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–61. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- Shulman JM, Perrimon N, Axelrod JD. Frizzled signaling and the developmental control of cell polarity. Trends Genet. 1998;14:452–8. doi: 10.1016/s0168-9525(98)01584-4. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–63. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Sullivan JC, Ryan JF, Watson JA, Webb J, Mullikin JC, Rokhsar D, Finnerty JR. StellaBase: the Nematostella vectensis Genomics Database. Nucleic Acids Res. 2006;34:D495–9. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony, Version 4 work. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–71. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooyen A, Kwee V, Nusse R. The nucleotide sequence of the human int-1 mammary oncogene; evolutionary conservation of coding and non-coding sequences. Embo J. 1985;4:2905–9. doi: 10.1002/j.1460-2075.1985.tb04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003a;5:367–77. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003b;13:680–5. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Fraser SE, Harland RM. Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Vogeli KM, Harland RM. Regulation of convergent extension in Xenopus by Wnt5a and Frizzled-8 is independent of the canonical Wnt pathway. Int J Dev Biol. 2001;45:225–7. [PubMed] [Google Scholar]
- Wang HY, Malbon CC. Wnt signaling, Ca2+, and cyclic GMP: visualizing Frizzled functions. Science. 2003;300:1529–30. doi: 10.1126/science.1085259. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shu W, Lu MM, Morrisey EE. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol. 2005;25:5022–30. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker CA, Bergeron K-F, Whittle JW, Brandhorst BP, Burke RD, Hynes RO. The Echinoderm Adhesome. Dev Biol. doi: 10.1016/j.ydbio.2006.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Wu CH, Nusse R. Ligand receptor interactions in the Wnt signaling pathway in Drosophila. J Biol Chem. 2002;277:41762–9. doi: 10.1074/jbc.M207850200. [DOI] [PubMed] [Google Scholar]
- Wu W, Glinka A, Delius H, Niehrs C. Mutual antagonism between dickkopf1 and dickkopf2 regulates Wnt/beta-catenin signalling. Curr Biol. 2000;10:1611–4. doi: 10.1016/s0960-9822(00)00868-x. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Butler K, Gurdon JB. Anterior endomesoderm specification in Xenopus by Wnt/beta-catenin and TGF-beta signalling pathways. Dev Biol. 1999;209:282–97. doi: 10.1006/dbio.1999.9257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


