Abstract
The global loss of B-cell-specific gene expression is a distinctive feature of the Hodgkin-Reed/Sternberg (HRS) cells of classical Hodgkin’s lymphoma (HL). The reasons for this loss remained largely unknown as transcription factors with pleiotropic effects on B-cell-specific gene expression, namely E2A, EBF, and PAX5, are present in primary HRS cells. We show here that ID2, which can inactivate E2A and perhaps PAX5, is not detectable in normal B cells but is strongly and uniformly expressed in HRS cells of all cases of classical HL. Recurrent chromosomal gains of the ID2 gene might contribute to this aberrant expression. Co-immunoprecipitation of E2A with ID2 from HRS-derived cell lines together with the high amount of ID2 relative to the B-cell transcription factors E2A and PAX5 in HRS-derived cell lines and primary HRS cells indicated that aberrant ID2 expression contributes significantly to the loss of the B-cell-specific gene expression in HRS cells. ID2 was also expressed in lymphocyte-predominance HL, mediastinal large B-cell, diffuse large B-cell, and Burkitt’s lymphoma, where lower amounts of ID2 relative to E2A and PAX5 compared with HRS cells might prevent a global down-regulation of B-cell-specific genes and ID2 may contribute to lymphomagenesis in other ways.
Hodgkin’s lymphoma (HL) is subdivided into the nodular lymphocyte-predominance (lp) and the classical (c) subtypes. A characteristic feature of all HL is the rarity of the tumor cells, the Hodgkin/Reed-Sternberg (HRS) cells in cHL and the lymphocytic and histiocytic (L&H) cells in lpHL, which represent only about 1% of the infiltrate.1 For the L&H cells of lpHL, the immunohistochemical detection of several B-cell markers indicated an origin from B cells.2 The HRS cells of cHL, however, coexpress markers of several lineages, and their origin remained enigmatic for a long time.3 Only with the demonstration of clonal V-gene rearrangements in single micromanipulated HRS cells was the B-cell origin of the vast majority of cases unequivocally clarified.4,5 The pattern of somatic mutations in the V-gene rearrangements indicated that L&H cells are derived from mutating germinal center (GC) B cells, which are still under selective pressure for expression of a functional B-cell receptor (BCR).6,7 HRS cells, however, are derived from preapoptotic GC-B cells, which frequently carry obviously crippling mutations in their V-gene rearrangements8 and are thus likely independent from BCR-generated survival signals that are essential for the survival of untransformed B cells.9
In most lymphomas derived from mature B lymphocytes, B-cell-specific differentiation is largely retained.10,11 For the HRS cells of cHL, however, global gene expression analysis using microarrays revealed that not only were a few B-cell genes not expressed, as previously recognized, but that with a few exceptions, nearly the complete B-cell-specific gene expression was lost.12
From early B-cell development, three transcription factors, namely E2A, EBF, and PAX5, are known to regulate the expression of several B-cell-specific genes in a pleiotropic fashion, among them CD19, CD79A, and the pre-B-cell receptor surrogate light chain constituents.13,14 E2A is the first one of the three to be expressed, and E2A together with EBF regulate the expression of PAX5.15,16 All three factors are also expressed in mature B cells, with the exception of plasma cells, where EBF and PAX5 are down-regulated.13,17 For PAX5, an essential role for maintenance of the B-cell-specific gene expression in mature B cells has been demonstrated by conditional inactivation of the PAX5 gene, and several B-cell genes directly regulated by PAX5, among them CD79A, CD19, and BLNK, have been identified.18,19 The functions of E2A and EBF in mature B cells are less clear. For E2A, an important role in regulation of class switch recombination and somatic hypermutation and in marginal zone B-cell differentiation has been shown.20,21
E2A is negatively regulated by direct interaction with the inhibitor of DNA binding ID2, and in vitro analysis showed that ID2 can also bind PAX5.16,22 All ID proteins dimerize with transcription factors, and, due to a lack of a DNA binding domain in the ID proteins, DNA binding of the heterodimers is prevented, thus inactivating transcription factors.23 ID2 expression in developing hematopoietic cells seems to repress B-cell development and B-cell-specific gene expression and to favor development of other lineages,24–28 whereas in mature B cells, ID2 is up-regulated on plasma cell differentiation with concomitant loss of expression of several B-cell genes.17 Furthermore, the balances between ID2 and E2A and ID2 and PAX5 seem to be important for B-cell differentiation in the spleen and the regulation of AID expression in GC-B cells, respectively.29,30
Given the loss of B-cell gene expression in HRS cells and the importance of E2A, EBF, and PAX5 for B-cell gene expression, the presence of these factors in HRS-derived cell lines and primary HRS cells has been analyzed by several groups. However, in most cell lines and in primary cases, all three factors are expressed, although mostly at reduced levels compared with normal B cells,29,31–33 and in an analysis of PAX5 transcripts in HRS-cell lines, no inactivating mutations were detected.12 We and others thus speculated that aberrant expression of negative regulators of these transcription factors could contribute to the loss of the B-cell-specific gene expression in HRS cells.12,33 The review of our global gene-expression data of HRS-cell lines indicated a strong ID2 expression in HRS-cell lines, and we present here our analysis of ID2 expression in HL and other lymphomas. Furthermore, we demonstrate the interaction of ID2 with E2A in HRS-cell lines and show that gains of the ID2 gene might contribute to the aberrant ID2 expression in primary HRS cells.
Materials and Methods
Cell Lines and Primary Cases
Cell lines used in this study were the HRS-cell lines L1236, L428, KMH2, and HDLM2, the Epstein-Barr virus-transformed lymphoblastoid cell line NCNC, the mantle cell lymphoma (MCL) lines Granta 519 and NCEB1, DEV (derived from a patient with lpHL), the diffuse large B-cell lymphoma (DLBCL) lines SuDHL6 and OCI-LY7, and the Burkitt’s lymphoma (BL) lines Raji and DG75. L1236, L428, KMH2, HDLM2, NCNC, and Granta 519 were grown according to the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany) recommendations (DSMZ), and NCEB1 and DEV were grown in RPMI 1640 with 10% fetal calf serum and antibiotics. All tissues used for immunohistochemistry (IHC) and immunofluorescence (IF) were fixed at room temperature overnight in 5% phosphate-buffered formalin and retrieved from the files of the Senckenberg Institute of Pathology of the University of Frankfurt. Cytogenetic suspensions of the 4 cHL cell lines quoted above and of 10 cHL used for fluorescence in situ hybridization (FISH) to determine ID2 gene copy numbers were retrieved from the Institute of Human Genetics, University Hospital Schleswig-Holstein Campus Kiel. All biopsies were originally submitted for diagnostic purposes and studied in accordance with national ethical principles.
Quantitative Real-Time PCR Analysis
RNA from the cell lines was isolated with an RNA isolation kit (Qiagen, Hilden, Germany) and quantified. cDNA was synthesized with the first strand cDNA synthesis kit (Roche, Mannheim, Germany) with 20 ng/μl RNA and random hexamer primers. One-microliter aliquots of the cDNAs were used as templates in real-time polymerase chain reactions (PCRs) using Assays-on-Demand (Hs00747379_m1 for ID2; Hs00413032_m1 for E2A; Hs00277134_m1 for PAX5; Hs00174333_m1 for CD19; Hs00233566_m1 for CD79A; and Hs00179459_m1 for BLNK; Applied Biosystems, Darmstadt, Germany) and the ABI7900HT Sequence Detection device. As a reference, the β-2-microglobulin (B2M; Applied Biosystems 4333766F) gene was used. For each gene-specific Assay-on-Demand experiment, a PCR product from one experiment was cloned into TOPO TA Cloning vector (Invitrogen, Karlsruhe, Germany) and sequenced, thus confirming specificities of the PCRs. Because of interference of the B2M assay with various other assays, PCRs were run as single and not as duplex PCRs. All PCRs were run as duplicates, and mean values were used for further calculations. Cycle threshold (Ct) values ranged from 15.9 to 20.8 for B2M, 19.1 to 28.8 for ID2, 16.4 to 36.8 for PAX5, 18.5 to 23.1 for E2A, 16.5 to 37.1 for CD79A, 19.3 to 40 for CD19, and 20.9 to 40 for BLNK. ΔCt values relative to B2M were calculated for all target genes and transformed into fold expression relative to B2M. For Figure 1, the reciprocal values of the fold expression values were used.
Figure 1-6920.
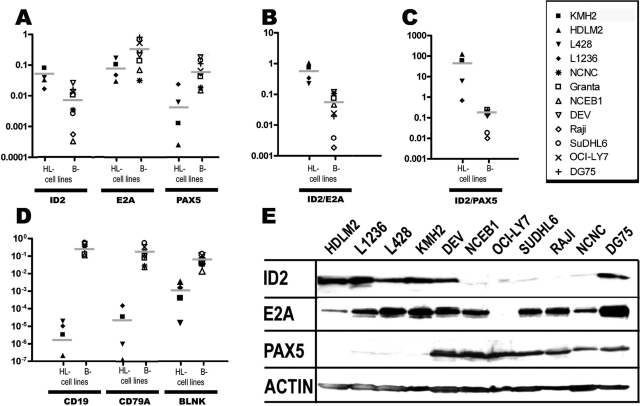
Quantitative real-time PCR and Western blot analysis of ID2, E2A, PAX5, CD19, CD79A, and BLNK expression in HRS-cell and other B-cell (lymphoma) cell lines. For all PCR, predesigned assays from ABI were used (Assays-on-demand). ΔCt values relative to B2M are shown as fold expression relative to B2M. For the PAX5 and CD79A ΔCts, to all values 1 and 3, respectively, were added to obtain positive numbers. A: Expression of ID2, E2A, and PAX5 in four cell lines derived from classical HL (L1236, L428, KMH2, and HDLM2), one cell line derived from lymphocyte-predominant HL (DEV), one Epstein-Barr virus-transformed B-cell line (NCNC), two MCL cell lines (NCEB1 and Granta 519), two DLBCL cell lines (SuDHL6 and OCI-LY7), and two BL cell lines (Raji and DG75) relative to B2M are shown. Bars represent average values. B and C: ID2 expression relative to PAX5 and E2A is shown. D: Expression values for the B-cell markers CD19, CD79A, and BLNK relative to B2M are shown. E: ID2, E2A, and PAX5 protein expression analyzed by Western blotting in the cell lines indicated is shown. Loading of equal amounts of protein was confirmed using an anti-actin antibody.
Western Blot Analysis
Cell lines or 10-μm sections of frozen tissues were lysed in Laemmli buffer by 10-minute incubation in boiling water. Lysates of approximately 1 × 105 cells per lane were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5 and 15%), blotted onto polyvinylidene difluoride (PVDF) membranes (Biorad, Munich, Germany), incubated overnight at 4°C with 1:1000 dilutions of anti-ID2 (ZMD.325, polyclonal rabbit; Zytomed, Berlin, Germany), anti-E2A (E12, polyclonal rabbit; Santa Cruz Biotechnology, Mannheim, Germany), anti-PAX5 (N19, polyclonal goat; Santa Cruz Biotechnology), or anti-actin (C11, polyclonal goat; Santa Cruz Biotechnology) antibodies and visualized using the ECL plus system (Amersham Biosciences, Freiburg, Germany).
Co-Immunoprecipitation of ID2 and E2A
Growing cells (1 × 107) were twice washed in PBS and then subjected to the ProFound Mamillian Co-Immunoprecipitation kit (Pierce, Rockford, IL) following the manufacturer’s instructions. A polyclonal rabbit anti-ID2 antibody (C20; Santa Cruz Biotechnology) was used for capturing the bait complexes. As a control, nonspecific rabbit serum IgG (Santa Cruz Biotechnology) was used. Precipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5 and 15%) and blotted onto PVDF membranes (Biorad). Precipitated proteins were detected by incubation of the membranes with 1:1000 dilutions of anti-E2A and anti-ID2 polyclonal antibodies (E12 and C20; Santa Cruz Biotechnology) overnight at 4°C and visualized with the ECL plus system (Amersham Biosciences).
IHC and IF
IHC and IF were performed on 5-μm sections of formalin-fixed, paraffin-embedded tissues. Antigen retrieval for both IHC and IF was performed by boiling the sections at 900 W for 15 minutes in a microwave oven in 20 mmol/L EDTA, pH 8. For ID2-IHC, sections were blocked by 15-minute incubation with a 1:100 dilution of normal goat serum IgG (Santa Cruz Biotechnology) in Tris-buffered saline (TBS) and then incubated with 1:100 dilution of anti-ID2 antibody (ZMD.325, polyclonal rabbit; Zytomed) in TBS overnight at room temperature. This antibody detected specifically a band of 15 kd in Western blots (in line with the molecular weight of ID2) (Figure 1; supplemental material at http://ajp.amjpathol.org) and is suitable for IHC with formalin-fixed tissues as shown in the supplemental Figure 1 (http://ajp.amjpathol.org) and by Russell et al.34 Bound primary antibody was visualized with the Envision system (Dako, Hamburg, Germany) with horseradish peroxidase and 3.3-diaminobenzidine as substrate. Negative controls without primary antibody were always performed in parallel.
For ID2/PAX5 and ID2/CD79A double IF, sections were incubated with a 1:100 dilution of anti-ID2 (ZMD.325) together with a 1:50 dilution of anti-PAX5 (sc-1974, polyclonal goat; Santa Cruz Biotechnology) or a 1:250 dilution of anti-CD79A (JCB117, monoclonal mouse; Dako) in TBS for 3 hours at room temperature. After washing, sections were incubated with fluorescent-labeled secondary antibodies (Alexa Fluor chicken-anti-rabbit 488 1:100 for ID2, Alexa Fluor chicken-anti-goat 594 1:100 for PAX5 and Alexa Fluor chicken-anti-mouse 594 1:200 for CD79A; Molecular Probes/Invitrogen, Karlsruhe, Germany) for 30 minutes, washed, counterstained with 4′,6-Diamidino-2-phenylindole, and evaluated using a Zeiss Axioscop-2 fluorescence microscope (Zeiss, Göttingen, Germany). For ID2/E2A double IF, sections were incubated with a 1:200 dilution of anti-E2A antibody (sc-349, polyclonal rabbit; Santa Cruz Biotechnology) for 3 hours at room temperature, washed, incubated with Alexa Fluor chicken-anti-rabbit 488 diluted 1:100 (Molecular Probes) for 30 minutes at room temperature, washed, and then incubated with directly labeled anti-ID2 diluted 1:100 for 2 hours at room temperature. Anti-ID2 was directly labeled with a fluorescence dye using the Zenon Alexa Fluor 594 Rabbit IgG Labeling kit (Molecular Probes) as recommended. After washing and 4′,6-Diamidino-2-phenylindole counterstaining, sections were evaluated as above. In Figure 4, the ID2/E2A double IF is shown in false colors (the red fluorescence for ID2 is shown in green and the green fluorescence for E2A in red) to maintain a uniform assignment of colors to antibodies throughout the figure.
Figure 4-6920.
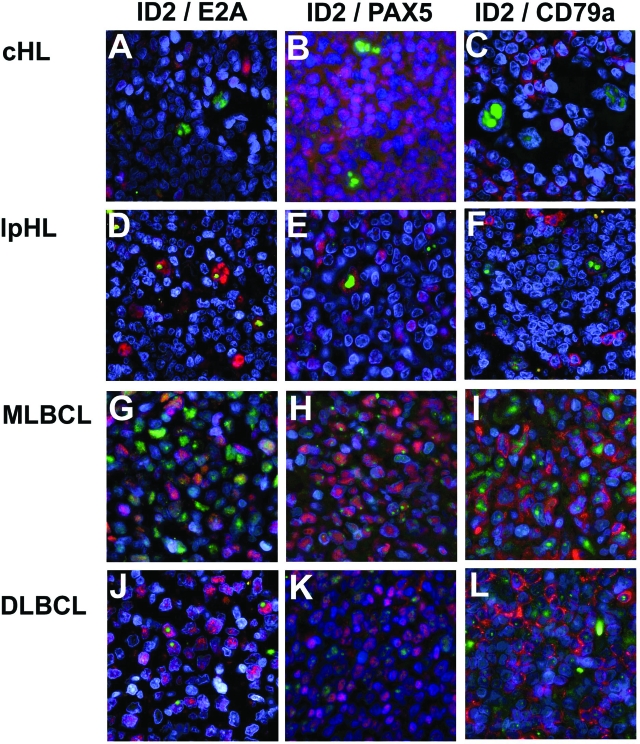
Double IF for ID2 with E2A, PAX5, or CD79A. ID2 staining is always shown in green, E2A, PAX5, and CD79A are always shown in red. A–C: In all three double IFs, strong nucleolar ID2 staining of HRS cells is shown. E2A, PAX5, and CD79A are detectable in several small bystanders but were below the detection level of our double IF in the HRS cells, also when only the red filter was used for examination (it should be noted that the large numbers of B cells seen in B are not typical for most cases of classical HL). D–F: Coexpression of ID2 with E2A, PAX5, and CD79A in L&H cells is shown. CD79A expression is weaker than in small bystanders. G–I: Coexpression of ID2 with E2A, PAX5, and CD79A in MLBCLs is shown. The expression levels of ID2 and E2A vary significantly between individual cells. Although in some cells, either ID2 (green) or E2A (red) expression was predominant, in other cells, similar staining intensities with both fluorescence dyes were observed (yellow). J–L: Strong E2A, PAX5, and CD79A expression relative to ID2 in DLBCL is shown.
FISH for ID2 Locus
FISH analyses for the detection of chromosomal rearrangements of the ID2 locus in 2p25 were performed on cytogenetic suspensions using differentially labeled BAC clones RP11-327F6 (centromeric to ID2) and RP11-434B12 (spanning ID2 and extending in the telomeric direction) as previously described.35 Slides were analyzed using a Zeiss Axioskop-2 fluorescence microscope (Zeiss) equipped with appropriate filter sets (AHF, Tübingen, Germany) and documented using an ISIS imaging system (MetaSystems, Altlussheim, Germany). Nuclei from HRS cells were identified by virtue of their larger size and frequent hyperploid genomic status. The ID2 genomic status was determined using the following criteria: signal numbers below 5 were defined as balanced (considering the characteristic tri- to tetraploidy of HRS cells), signal numbers between 5 and 8 were defined as gained, and signal numbers above 8 were defined as amplified.
Results
Expression Levels of ID2 and B-Cell-Specific Gene Transcripts in HRS- and B-Cell Lymphoma Cell Lines Are Inversely Correlated
Our global gene expression data of HRS-cell lines indicated strong, aberrant ID2 expression in HRS-cell lines compared with normal B cells and also to most other B-cell lymphomas and B-cell lymphoma-derived cell lines.36 To quantitatively analyze ID2 expression in relation to the expression of the B-cell transcription factors E2A and PAX5 and the B-cell markers CD19, CD79A, and BLNK in HRS- and other B-cell (lymphoma) lines, we used real-time PCR (Figure 1). Average ID2 expression in the HRS-cell lines was about 1 order of magnitude higher compared with the other B-cell (lymphoma) lines, and the relation of ID2 to E2A and PAX5 transcripts was on average about 1 and 2 orders of magnitude higher in HRS-cell lines compared with the B-cell (lymphoma) lines, respectively. For all three B-cell genes, CD79A, BLNK, and CD19, expression values were on average at least 1 order of magnitude higher in the B-cell (lymphoma) lines compared with the HRS-cell lines. Western blot analysis showed that differences in the expression of ID2, E2A, and PAX5 at the RNA level were (with the exception of E2A in OCI-LY7) also seen at the protein level (Figure 1E). These data indicate that the strong ID2 expression in HRS-cell lines relative to the B-cell transcription factors E2A and PAX5 is inversely correlated with low level expression of B-cell genes.
E2A Co-Immunoprecipitates with ID2 from HRS-Cell Lines
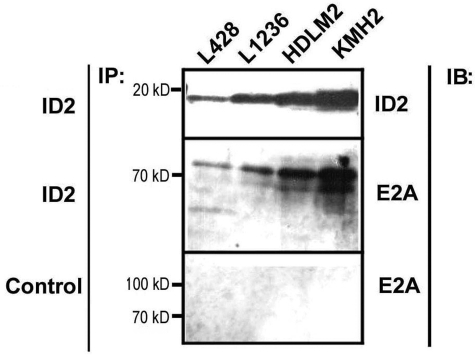
ID proteins and E2A can form ID2/E2A heterodimers in which E2A is inactivated,23,37 and we wished to analyze whether ID2/E2A heterodimers are detectable in HRS-cell lines. Therefore an anti-ID2 antibody was used for immunoprecipitation with lysates of all four HRS-cell lines. Subsequent analysis of the immunoprecipitates in Western blots revealed co-immunoprecipitation of E2A with ID2 from all four HRS-cell lines analyzed (Figure 2). This demonstrates that at least fractions of E2A are inactivated by the aberrantly expressed ID2 in HRS-cell lines.
Figure 2-6920.
Co-immunoprecipitation of E2A with ID2 from HRS-cell lines. A polyclonal anti-ID2 antibody and as negative control normal rabbit serum IgG were used for immunoprecipitation from lysates of 1 × 107 HRS cells using the ProFound Mammalian Co-Immunoprecipitation kit. Immunoprecipitates were separated on polyacrylamide gels, blotted onto PVDF membranes, and probed with anti-ID2 and anti-E2A antibodies. From all cell lines, E2A was co-immunoprecipitated with ID2, whereas no E2A was detected using normal rabbit serum IgG as a negative control for immunoprecipitation. IB, immunoblot; IP, immunoprecipitation.
ID2 Is Aberrantly Expressed in Primary HRS and L&H Cells of HL and in Large-Cell and Burkitt’s Lymphomas
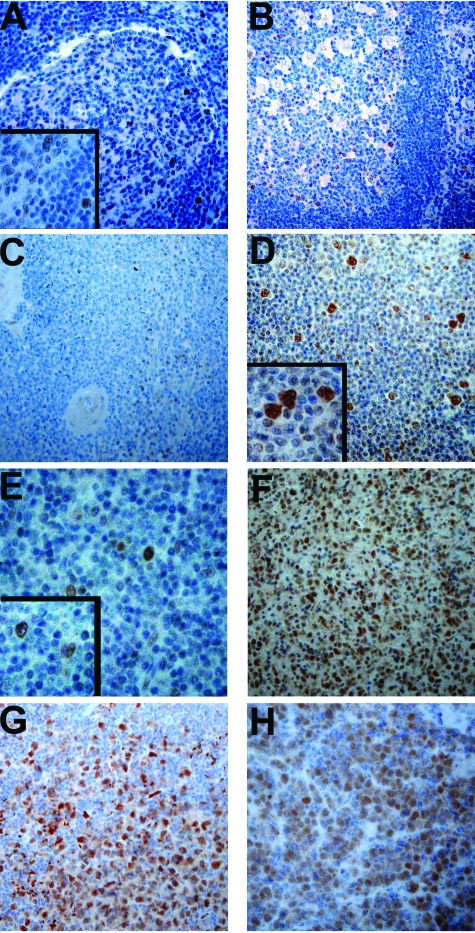
Given the above indications for an important role of ID2 in the loss of the B-cell-specific gene expression in HRS-cell lines, we used IHC to analyze the expression of ID2 in primary HL cases, in B-cell non-Hodgkin’s lymphomas (B-NHL), and also in normal lymphatic tissues (Figure 3). In normal lymphatic tissues, ID2 expression was largely restricted to a few GC and interfollicular cells. Based on morphology, the ID2-positive cells were dendritic cells and macrophages, in line with previous RNA-based expression analyses.24,25,27 Among the B-NHLs analyzed, no ID2 expression in the tumor cells of MCL and follicular lymphoma and chronic lymphocytic leukemia was observed (Table 1). In line with previous RNA analysis,38 ID2 expression was observed in the three BLs analyzed (Table 1; Figure 3). Also in DLBCL and mediastinal large B cell lymphoma (MLBCL), ID2 positivity was observed in the vast majority of cases. Among HL, all cHL showed strong positivity in the HRS cells, and also the vast majority of lpHL showed positivity for ID2 in the L&H cells. Thus, ID2 is aberrantly expressed in the tumor cells of all types of HL and in considerable fractions of large-cell lymphomas and BL.
Figure 3-6920.
IHC analysis of ID2 expression in normal lymphatic tissues, HL and B-NHL. For all stainings a polyclonal rabbit ID2-specific antibody was used and visualized with horseradish peroxidase and 3,3-diaminobenzidine as substrate. A–C: Staining of an unspecific lymphadenitis, a tonsil, and a spleen, respectively, are shown. Only a few GC and interfollicular cells with morphology of dendritic cells and macrophages were stained. D and E: Staining for a cHL (D) and for a lpHL (E) are shown. Tumor cells of both HL types were positive for ID2. F–H: Stainings for MLBCL, DLBCL, and BL, respectively, are shown.
Table 1.
IHC Detection of ID2 in HL and B-Cell Non-Hodgkin’s Lymphomas
| Lymphoma type | No. of positive/no. of cases analyzed |
|---|---|
| CLL | 0/10 |
| MCL | 0/10 |
| FL | 0/5 |
| BL | 3/3 |
| DLBCL | 12/16 |
| lpHL | 8/10 |
| MLBCL | 15/16 |
| cHL | 42/42 |
In cases scored as positive, the vast majority of tumor cells (>80% of cells) showed ID2 positivity. In negative cases, no or only a few cells were ID2 positive. CLL, chronic lymphocytic leukemia; FL, follicular lymphoma.
IF Analysis of ID2, E2A, PAX5, and CD79A Expression in HL and B-NHL
On the IHC analysis of ID2 expression, we recognized large variations in staining intensities among the different lymphoma types. To obtain a more quantitative impression of ID2 expression in relation to E2A, PAX5, and CD79A in the various lymphomas, we performed double IF stainings for ID2 together with E2A, PAX5, or CD79A (Figure 4). Each double IF staining for a specific antibody combination was performed for all cases analyzed in one experiment, and staining intensities were scored by visual inspection from 0 to 3 (Figures 4 and 5; Supplemental Figure 3 [available at http://ajp.amjpathol.org]). ID2 expression was strongest in the HRS cells of cHL, followed by MLBCL, lpHL, and DLBCL. In the IHC analysis, ID2 staining in several specimens seemed to be most prominent in nucleoli, and the higher spatial resolution of IF revealed that, especially in HRS and L&H cells, ID2 was predominantly localized to the nucleoli. On the other hand, the B-cell transcription factors E2A and PAX5 were hardly detectable in cHL and also had clearly weaker expression in lpHL compared with both types of large-cell lymphoma (Supplemental Figure 2 [available at http://ajp.amjpathol.org]). CD79A was not detectable in cHL but was clearly present in lpHL and both large-cell lymphomas. In several cases of MLBCL at the single cell level, a striking variation in ID2-to-E2A ratios was observed (Figure 4G).
Figure 5-6920.
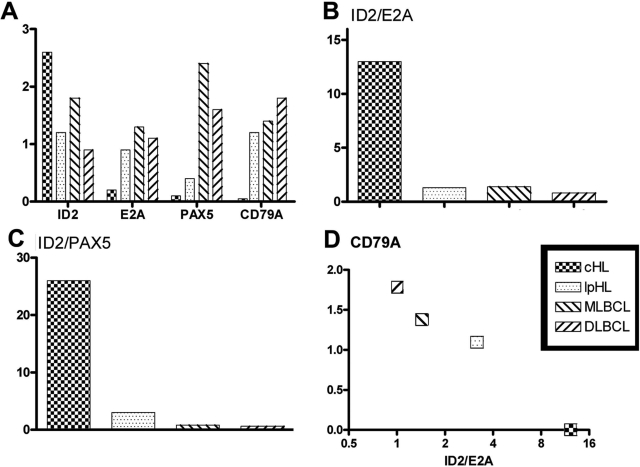
Quantitative analysis of ID2, E2A, PAX5, and CD79A expression in primary lymphomas. For quantitative evaluation of immunofluorescence double staining (ID2/E2A, ID2/PAX5, and ID2/CD79A), staining for each antibody combination was performed in one experiment for all cases analyzed, and expression levels were scored by visual inspection of two investigators from 0 to 3 (no detectable to very strong expression). Average values were calculated from 13 cHL, 9 lpHL, 8 MLBCL, and 10 DLBCL analyzed. A: The average expression values for ID2, E2A, PAX5, and CD79A are shown. B and C: The expression levels of ID2 relative to E2A and PAX5 levels, respectively, are shown. They were calculated using the average expression scores shown in A, which ranged from 0.1 for PAX5 in cHL to 2.6 for ID2 in cHL. D: The CD79A expression values are plotted against the ID2-to-E2A ratios for the various lymphoma types analyzed.
Because the balance between ID2 and E2A or ID2 and PAX5 seems to be important for the regulation of B-cell differentiation or expression of B-cell-specific genes,30,39 we calculated the ID2-to-E2A and ID2-to-PAX5 ratios for all lymphomas (Figure 5), using the average expression scores (from 0.1 to 2.6; Figure 5A) for each protein for each lymphoma. These ratios clearly showed the very high ID2 levels in HRS cells relative to the two transcription factors negatively regulated by ID2.
Thus, in line with the analysis of transcript levels in HRS- and other B-cell lymphoma-derived cell lines, the analysis of protein expression in primary lymphomas also revealed an inverse correlation between high ID2-to-E2A or ID2-to-PAX5 ratios and expression of the B-cell marker CD79A (Figure 5).
Recurrent Genomic Gains of the ID2 Locus in HRS Cells
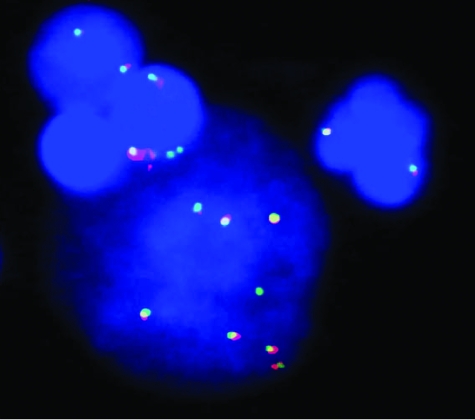
The ID2 gene is located on the short arm of chromosome 2, and gains of chromosome 2p are the most recurrent genomic alterations in HRS cells.40,41 To analyze whether genomic gains of ID2 could contribute to its aberrant expression in HRS cells, we performed FISH analyses with an assay for the ID2 locus on 4 cHL cell lines and 10 primary cHL cases (Figure 6). Copy number changes affecting the ID2 locus were observed in the HRS cells of 5 of the 10 primary cHL (50%), including four cases with genomic gain and one case with amplification. By FISH and array-based comparative genomic hybridization (data not shown), gains of ID2 were also observed in two (L1236 and KMH2) of four cHL cell lines, which is in agreement with findings recently obtained by conventional comparative genomic hybridization.42
Figure 6-6920.
Fluorescence in situ hybridization in a case of cHL using two differently colored probes for the ID2 locus. The large nucleus below shows multiple red and green hybridization, indicating the presence of a genomic gain of the ID2 locus. The small nucleus on the right shows the unaltered signal constellation, ie, two colocalized signals.
Discussion
The nearly complete loss of expression of B-cell-specific genes of the B-cell-derived HRS cells of cHL is outstanding among all other B-cell lymphomas and could be an important step in HL pathogenesis. Moreover, the loss of the B-cell-specific gene-expression program may allow and contribute to the aberrant expression of markers of several lineages,5 as indicated by the aberrant expression of genes of several lineages after down-regulation of PAX5 expression in mature B cells.18 For several of the aberrantly expressed genes, important roles for HL pathogenesis, eg, the attraction of T cells by thymus and activation regulated chemokine, have been demonstrated.43 In addition, HRS cells are the only B-cell lymphoma cells that are, due to disadvantageous somatic mutations in the BCR-encoding V-gene rearrangements, derived from pre-apoptotic GC-B cells.8,10,44 Normal B cells, and especially GC-B cells and also most B-NHL cells, depend on survival signals generated by a functional BCR.9,45,46 This BCR dependence of survival may be lost on loss of the B-cell-specific gene-expression program and thus supports the survival of the BCR-less HRS cells.
The pathogenic changes in the HRS cells causing the loss of B-cell gene expression remained so far unknown because all known transcription factors that regulate B-cell genes in a pleiotropic fashion, ie, E2A, EBF, and PAX5, are still expressed in HRS cells. An explanation for this contradictory situation could be the aberrant expression of negative regulators of these transcription factors. Here, we show that ID2, an inhibitor of E2A and PAX5, is aberrantly expressed in HRS cells. ID2, which was not detectable in normal B cells, was strongly expressed in nearly all HRS cells of all cases analyzed. Such a uniform expression in HL is rarely seen for other markers and may stress the importance of the aberrant ID2 expression for HRS-cell pathogenesis. Furthermore, we demonstrate a direct interaction of ID2 with E2A in HRS-cell lines, showing that at least E2A is inactivated by ID2 in HRS-cell lines. These results are in line with a study performed in parallel,33 where direct interactions of ID2 with E2A in HRS-cell lines and expression of ID2 transcripts in primary HRS cells also were demonstrated. It is, however, likely that other negative regulators of B-cell transcription factors also play important roles in HL.47 This has already been shown for the ABF-1 protein,33 which is aberrantly expressed in HRS cells.36 Another candidate is NOTCH1, which is also aberrantly expressed in HRS cells and can inhibit B-cell transcription factors.48–50
ID proteins are key regulators in several developmental and cellular processes. In general, aberrant ID expression seems to favor proliferation, to inhibit differentiation, and to facilitate tumor neoangiogenesis.23 For ID2, which is aberrantly expressed in several types of tumors,51 the situation is more complicated. Whereas ID2 can bind and inactivate the anti-proliferative effect of retinoblastoma (RB) and is likely an important effector of N-MYC in neuroblastoma,52,53 it can also suppress tumor formation in the intestinal epithelium and is important for maintenance of a differentiated, noninvasive phenotype of breast cancer cells.34,54 In HRS cells, however, our results and those of Mathas et al,33 clearly indicate a role in the dedifferentiation of the tumor cells by suppressing expression of B-cell genes. In addition to suppression of B-cell genes, the strong ID2 expression and interaction with E-box proteins in HRS cells could have also other effects on HRS cells, eg, on the proliferation by regulating RB activity.53,55
Another issue raised by this study is the role of ID2 in lymphomas other than cHL. Of the B-NHL entities in which we observed significant ID2 expression, so far, a partial loss of B-cell-specific gene expression has been described only for MLBCL. Ig is undetectable by IHC, and several components of the BCR signaling cascade are expressed at lower levels than in DLBCLs.56,57 In the present study, we observed a recognizable down-regulation of CD79A in L&H cells of lpHL, which is usually detected by IHC on the L&H cells.58 Thus, although the aberrant ID2 expression might contribute to the reduced expression of some B-cell genes in these two lymphomas, the ID2-to-E2A and ID2-to-PAX5 ratios in the B-NHLs seem to be too low to cause a down-regulation of B-cell genes comparable with HRS cells. Because it is hardly conceivable how a slight reduction in the expression of some B-cell-specific genes should contribute to lymphomagenesis in these entities, it is more likely that other aspects of ID2 function may be relevant for pathogenesis of these lymphomas.
The causes for the aberrant ID2 expression in cHL may be manifold. Our own molecular cytogenetic findings suggest that recurrent genomic gains of the ID2 locus might contribute to the aberrant ID2 expression in the HRS cells through increased gene dosage. Remarkably, 2p gains are also a recurrent aberration in MLBCLs, the vast majority of which also express ID2. ID2 expression, however, can be activated by multiple signaling pathways.23 Among them is transforming growth factor (TGF)-β signaling,59 and because HRS cells express both TGF-β and at least TGFβ-RII, autocrine TGFβ signaling could contribute to the aberrant ID2 expression in HRS cells.60,61
Taken together, ID2 is strongly and uniformly expressed in the HRS cells of cHL and likely represses B-cell-specific gene expression by inactivation of E2A (and perhaps also PAX5). ID2 is also expressed in lpHL, MLBCL, DLBCL, and BL, although at amounts that are too low to cause a global down-regulation of B-cell genes, and likely contributes in other ways to lymphomagenesis.
Supplementary Material
Acknowledgments
We thank Sabine Albrecht, Yvonne Blum, and Dorit Schuster for excellent technical assistance.
Footnotes
Address reprint requests to Andreas Bräuninger, Department of Pathology, University of Frankfurt, Theodor-Stern-Kai 7, 60590 Frankfurt, Germany. E-mail: braeuninger@em.uni-frankfurt.de.
Supported by the Deutsche Krebshilfe Verbundprojekt 102362 (to R.K. and A.B.), by the Deutsche Krebshilfe Network (grant 70-3173-Tr3 to R.S.), and by the Deutsche Forschungsgeinschaft (grants RK 1315/2-1 and BR 1238/6-1).
Supplemental material for this article appears on http://ajp.amjpathol.org.
References
- Mauch PM, Armitage JO, Diehl V, Hoppe RT, Weiss LM. Hodgkin’s Disease. Lippincott Williams Wilkins,; Philadelphia: 1999 [Google Scholar]
- Hansmann ML, Weiss LM, Stein H, Harris NL, Jaffe ES, editors. Lippincott Williams Wilkins,; Philadelphia: Pathology of Lymphocyte Predominance Hodgkin’s Disease. 1999:pp 168–180. [Google Scholar]
- Weiss LM, Chan JKC, MacLennan K, Warnke RA, editors. Lippincott Williams Wilkins,; Philadelphia: Pathology of Classical Hodgkin’s Disease. 1999:pp 101–120. [Google Scholar]
- Küppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R, Hansmann ML. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci USA. 1994;91:10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers R. Molecular biology of Hodgkin’s lymphoma. Adv Cancer Res. 2002;84:277–312. doi: 10.1016/s0065-230x(02)84009-x. [DOI] [PubMed] [Google Scholar]
- Braeuninger A, Küppers R, Strickler JG, Wacker HH, Rajewsky K, Hansmann ML. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci USA. 1997;94:9337–9342. doi: 10.1073/pnas.94.17.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marafioti T, Hummel M, Anagnostopoulos I, Foss HD, Falini B, Delsol G, Isaacson PG, Pileri S, Stein H. Origin of nodular lymphocyte-predominant Hodgkin’s disease from a clonal expansion of highly mutated germinal-center B cells. N Engl J Med. 1997;337:453–458. doi: 10.1056/NEJM199708143370703. [DOI] [PubMed] [Google Scholar]
- Kanzler H, Küppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Vardiman JW. IARC Press,; Lyon, France: WHO Classification of Tumors: Pathology and Genetics of Tumors of Haematopoietic and Lymphoid Tissues. 2001 [Google Scholar]
- Schwering I, Bräuninger A, Klein U, Jungnickel B, Tinguely M, Diehl V, Hansmann ML, Dalla-Favera R, Rajewsky K, Küppers R. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;101:1505–1512. doi: 10.1182/blood-2002-03-0839. [DOI] [PubMed] [Google Scholar]
- Bartholdy B, Matthias P. Transcriptional control of B cell development and function. Gene. 2004;327:1–23. doi: 10.1016/j.gene.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nat Rev Immunol. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- Underhill GH, George D, Bremer EG, Kansas GS. Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood. 2003;101:4013–4021. doi: 10.1182/blood-2002-08-2673. [DOI] [PubMed] [Google Scholar]
- Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14:779–790. doi: 10.1016/s1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Morrison AM, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Quong MW, Martensson A, Langerak AW, Rivera RR, Nemazee D, Murre C. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J Exp Med. 2004;199:1101–1112. doi: 10.1084/jem.20031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EC, Deed RW, Inoue T, Norton JD, Sharrocks AD. Id helix-loop-helix proteins antagonize pax transcription factor activity by inhibiting DNA binding. Mol Cell Biol. 2001;21:524–533. doi: 10.1128/MCB.21.2.524-533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Ishiguro A, Spirin KS, Shiohara M, Tobler A, Gombart AF, Israel MA, Norton JD, Koeffler HP. Id2 expression increases with differentiation of human myeloid cells. Blood. 1996;87:5225–5231. [PubMed] [Google Scholar]
- Cooper CL, Brady G, Bilia F, Iscove NN, Quesenberry PJ. Expression of the Id family helix-loop-helix regulators during growth and development in the hematopoietic system. Blood. 1997;89:3155–3165. [PubMed] [Google Scholar]
- Spits H, Couwenberg F, Bakker AQ, Weijer K, Uittenbogaart CH. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre-DC)2 but not into pre-DC1: evidence for a lymphoid origin of pre-DC2. J Exp Med. 2000;192:1775–1784. doi: 10.1084/jem.192.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, Kuhl C, Jorgas T, Kurz SM, Rose-John S, Yokota Y, Zenke M. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- Buitenhuis M, van Deutekom HW, Verhagen LP, Castor A, Jacobsen SE, Lammers JW, Koenderman L, Coffer PJ. Differential regulation of granulopoiesis by the basic helix-loop-helix transcriptional inhibitors Id1 and Id2. Blood. 2005;105:4272–4281. doi: 10.1182/blood-2004-12-4883. [DOI] [PubMed] [Google Scholar]
- Hertel CB, Zhou XG, Hamilton-Dutoit SJ, Junker S. Loss of B cell identity correlates with loss of B cell-specific transcription factors in Hodgkin/Reed-Sternberg cells of classical Hodgkin lymphoma. Oncogene. 2002;21:4908–4920. doi: 10.1038/sj.onc.1205629. [DOI] [PubMed] [Google Scholar]
- Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, Mori KJ, Yokota Y, Shimizu A. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss HD, Reusch R, Demel G, Lenz G, Anagnostopoulos I, Hummel M, Stein H. Frequent expression of the B-cell-specific activator protein in Reed-Sternberg cells of classical Hodgkin’s disease provides further evidence for its B-cell origin. Blood. 1999;94:3108–3113. [PubMed] [Google Scholar]
- Steimle-Grauer SA, Tinguely M, Seada L, Fellbaum C, Hansmann ML. Expression patterns of transcription factors in progressively transformed germinal centers and Hodgkin lymphoma. Virchows Arch. 2003;442:284–293. doi: 10.1007/s00428-002-0735-5. [DOI] [PubMed] [Google Scholar]
- Mathas S, Janz M, Hummel F, Hummel M, Wollert-Wulf B, Lusatis S, Anagnostopoulos I, Lietz A, Sigvardsson M, Jundt F, Jöhrens K, Bommert K, Stein H, Dorken B. Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat Immunol. 2005;7:207–215. doi: 10.1038/ni1285. [DOI] [PubMed] [Google Scholar]
- Russell RG, Lasorella A, Dettin LE, Iavarone A. Id2 drives differentiation and suppresses tumor formation in the intestinal epithelium. Cancer Res. 2004;64:7220–7225. doi: 10.1158/0008-5472.CAN-04-2095. [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Harder L, Gesk S, Schlegelberger B, Grote W, Martinez-Climent JA, Dyer MJ, Novo FJ, Calasanz MJ, Siebert R. Interphase FISH assays for the detection of translocations with breakpoints in immunoglobulin light chain loci. Int J Cancer. 2002;98:470–474. doi: 10.1002/ijc.10169. [DOI] [PubMed] [Google Scholar]
- Küppers R, Klein U, Schwering I, Distler V, Bräuninger A, Cattoretti G, Tu Y, Stolovitzky GA, Califano A, Hansmann ML, Dalla-Favera R. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J Clin Invest. 2003;111:529–537. doi: 10.1172/JCI16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera R, Murre C. The regulation and function of the Id proteins in lymphocyte development. Oncogene. 2001;20:8308–8316. doi: 10.1038/sj.onc.1205091. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Nilsson LM, Keller U, Yokota Y, Boyd K, Cleveland JL. Id2 is dispensable for myc-induced lymphomagenesis. Cancer Res. 2004;64:7296–7301. doi: 10.1158/0008-5472.CAN-04-2133. [DOI] [PubMed] [Google Scholar]
- Becker-Herman S, Lantner F, Shachar I. Id2 negatively regulates B cell differentiation in the spleen. J Immunol. 2002;168:5507–5513. doi: 10.4049/jimmunol.168.11.5507. [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Gesk S, Harder L, Sonoki T, Tucker PW, Schlegelberger B, Grote W, Novo FJ, Calasanz MJ, Hansmann ML, Dyer MJ, Siebert R. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood. 2002;99:1474–1477. doi: 10.1182/blood.v99.4.1474. [DOI] [PubMed] [Google Scholar]
- Joos S, Menz CK, Wrobel G, Siebert R, Gesk S, Ohl S, Mechtersheimer G, Trümper L, Möller P, Lichter P, Barth TF. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood. 2002;99:1381–1387. doi: 10.1182/blood.v99.4.1381. [DOI] [PubMed] [Google Scholar]
- Joos S, Granzow M, Holtgreve-Grez H, Siebert R, Harder L, Martin-Subero JI, Wolf J, Adamowicz M, Barth TF, Lichter P, Jauch A. Hodgkin’s lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int J Cancer. 2003;103:489–495. doi: 10.1002/ijc.10845. [DOI] [PubMed] [Google Scholar]
- van den Berg A, Visser L, Poppema S. High expression of the CC chemokine TARC in Reed-Sternberg cells: a possible explanation for the characteristic T-cell infiltrate in Hodgkin’s lymphoma. Am J Pathol. 1999;154:1685–1691. doi: 10.1016/S0002-9440(10)65424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuninger A, Schmitz R, Bechtel D, Renné C, Hansmann ML, Küppers R. Molecular biology of Hodgkin’s and Reed/Sternberg cells in Hodgkin’s lymphoma. Int J Cancer. 2006;118:1853–1861. doi: 10.1002/ijc.21716. [DOI] [PubMed] [Google Scholar]
- Küppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Küppers R, Bräuninger A. Reprogramming of the tumour B-cell phenotype in Hodgkin lymphoma. Trends Immunol. 2006;27:203–205. doi: 10.1016/j.it.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Jundt F, Anagnostopoulos I, Förster R, Mathas S, Stein H, Dorken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99:3398–3403. doi: 10.1182/blood.v99.9.3398. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- Smith EM, Akerblad P, Kadesch T, Axelson H, Sigvardsson M. Inhibition of EBF function by active Notch signaling reveals a novel regulatory pathway in early B-cell development. Blood. 2005;106:1995–2001. doi: 10.1182/blood-2004-12-4744. [DOI] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Boldrini R, Dominici C, Donfrancesco A, Yokota Y, Inserra A, Iavarone A. Id2 is critical for cellular proliferation and is the oncogenic effector of N-myc in human neuroblastoma. Cancer Res. 2002;62:301–306. [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Itahana Y, Singh J, Sumida T, Coppe JP, Parrinello S, Bennington JL, Desprez PY. Role of Id-2 in the maintenance of a differentiated and noninvasive phenotype in breast cancer cells. Cancer Res. 2003;63:7098–7105. [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, Aguiar RC, Li S, Salles G, Berger F, Jing W, Pinkus GS, Habermann T, Dalla-Favera R, Harris NL, Aster JC, Golub TR, Shipp MA. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- Banks PM, Warnke RA, editors. IARC Press,; Lyon, France: Mediastinal (thymic) large B-cell lymphoma. 2001 [Google Scholar]
- Kuzu I, Delsol G, Jones M, Gatter KC, Mason DY. Expression of the Ig-associated heterodimer (mb-1 and B29) in Hodgkin’s disease. Histopathology. 1993;22:141–144. doi: 10.1111/j.1365-2559.1993.tb00092.x. [DOI] [PubMed] [Google Scholar]
- Kee BL, Rivera RR, Murre C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat Immunol. 2001;2:242–247. doi: 10.1038/85303. [DOI] [PubMed] [Google Scholar]
- Skinnider BF, Mak TW. The role of cytokines in classical Hodgkin lymphoma. Blood. 2002;99:4283–4297. doi: 10.1182/blood-2002-01-0099. [DOI] [PubMed] [Google Scholar]
- Brown RE, Nazmi RK. The Reed-Steinberg cell: molecular characterization by proteomic analysis with therapeutic implications. Ann Clin Lab Sci. 2002;32:339–351. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.