Abstract
Myosin VI is an actin motor that moves to the minus end of the polarized actin filament, a direction opposite to all other characterized myosins. Using expression microarrays, we identified myosin VI as one of the top genes that demonstrated cancer-specific overexpression in clinical prostate specimens. Protein expression of myosin VI was subsequently analyzed in arrayed prostate tissues from 240 patients. Notably, medium-grade prostate cancers demonstrated the most consistent cancer-specific myosin VI protein overexpression, whereas prostate cancers associated with more aggressive histological features continued to overexpress myosin VI but to a lesser extent. Myosin VI protein expression in cell lines positively correlated with the presence of androgen receptor. Small interference RNA-mediated myosin VI knockdown in the LNCaP human prostate cancer cell line resulted in impaired in vitro migration and soft-agar colony formation. Depletion of myosin VI expression was also accompanied by global gene expression changes reflective of attenuated tumorigenic potential, as marked by a nearly 10-fold induction of TXNIP (VDUP1), a tumor suppressor with decreased expression in prostate cancer specimens. These results support that myosin VI is critical in maintaining the malignant properties of the majority of human prostate cancers diagnosed today.
Myosins are defined as actin-dependent Mg2+ ATPases that use the energy derived from ATP hydrolysis to move along the actin filaments within the cell.1 Structurally, myosins have a common domain organization consisting of a conserved N-terminal actin binding and ATPase domain (motor or head domain), a neck region containing IQ motifs that bind to myosin light chains, and a C-terminal tail domain for specific cargo binding.2 In the human genome, there are ∼40 myosin genes, representing 12 classes of actin motors that mainly participate in actin-based cellular processes.1,2 Only the class II myosins are known to form bipolar filaments that are essential for well-characterized contractile functions. The remaining classes of myosins are so-named unconventional myosins1,2 that are generally thought to function in nonmuscle cells as actin-bound monomers or dimers. Although not well characterized in terms of the precise mechanism, unconventional myosins have been implicated in F-actin-mediated cellular functions such as cell motility, vesicular trafficking, intracellular transport of macromolecules, and possibly regulation of signal transduction.2,3
The class VI unconventional myosin was initially identified and partially characterized in Drosophila and pig.4,5 In most organisms including human, a single gene encodes the class VI unconventional myosin. Myosin VI is a unique member of the myosin superfamily.6,7 Primarily because of a 53-amino acid insertion between the motor and the neck domain, myosin VI moves to the pointed/minus end of the polarized actin filament, a direction opposite to all other myosins characterized to date.8,9 Because actin filaments are believed to orient their pointed/minus ends away from the plasma membrane and internal organelles,6 the unique motor direction of myosin VI is potentially linked mechanistically to its functional roles in endocytosis (transport of vesicles away from the plasma membrane),10 secretion (transport of vesicles away from the Golgi),11 and cell migration (pushing of the barbed/plus ends of F-actin against the cell mem- brane).12–14
Although the role of actin motors (myosins) in human cancer is generally poorly documented, an intriguing connection between myosin VI and human cancer was recently reported.14 Based on the initial observation that myosin VI is required in border cell migration during Drosophila ovary development,12 Yoshida and colleagues14 examined protein expression of myosin VI in human ovarian cancers and discovered a functional link between myosin VI expression and aggressive ovarian cancer. In the present study, we initially discovered an unusually consistent cancer-specific overexpression of myosin VI mRNA through global gene expression analysis that emphasized the comparison between normal prostate epithelium and cancerous acini. Further, the role of myosin VI in human prostate cancer was investigated through immunohistochemical analysis in a cohort of 240 patients, as well as functional studies in human prostate cancer cell lines.
Materials and Methods
Human Prostate Tissues for Expression Microarrays
Prostate tissue samples used for cDNA microarray analysis were fresh frozen specimens collected at the time of prostate surgery from 1993 to 2000 at the Johns Hopkins Hospital. Tissue specimens used in this study were from nine patients undergoing surgery for symptomatic benign prostatic hyperplasia (BPH) and 25 patients undergoing radical prostatectomy for prostate cancer. Established procedures15 were followed for sample selection and processing. A total of 59 specimens were processed because normal-tumor paired tissues were retrieved from each of the 25 radical prostatectomy cases. Cryosections were cut from trimmed blocks enriched for tissues of interest before downstream RNA extraction. The first and last section from each sample was reserved for pathological confirmation and visual estimation of the percentage of epithelium. This study was approved by the Institutional Review Board at Johns Hopkins Medical Institutions.
Human Prostate Tissues for Immunohistochemistry
All prostate specimens used for immunohistochemical analysis were radical prostatectomy samples selected from the surgical pathology files at the Johns Hopkins Department of Pathology with Institutional Review Board approval. Tissue microarrays (TMAs) were constructed as previously described.16 Six high-density TMAs, each containing surgical prostate tissues from 40 cases (240 cases in total), were used for immunohistochemical staining. Each case was represented by eight cores (0.6 mm in diameter) that were predominantly matched normal and cancer tissues but may also have been high-grade prostatic intraepithelial neoplasia (HGPIN) and proliferative inflammatory atrophy (PIA) lesions.16 Standard tissue sections were selected and processed also as previously described.16
Expression Microarrays
Printed glass cDNA microarrays were used throughout the study. For prostate tissue profiling, microarrays containing 11,904 human expression sequence tags were used. Expression sequence tags were selected from human IMAGE clone plate sets, based on relative enrichment of annotated genes within the plate, and supplemented by six plates (576 clones) of custom arrayed IMAGE clones selected based on relevance to prostate biology after an extensive literature search. For profiling in cell lines, a recent version of cDNA microarrays containing 20,344 human expression sequence tags was used, after integration of additional plates enriched for annotated genes.
Gene Expression Analysis
The experimental design, total RNA extraction, labeling, hybridization, image analysis, and data analysis were modified based on the protocols described previously.15 Total RNA samples extracted from tissues or cultured cells were amplified once using the MessageAmp aRNA kit (Ambion, Austin, TX) using an input of 500 ng of total RNA, and labeled by direct incorporation of Cy3-dUTP (Amersham Pharmacia, Piscataway, NJ) in a reverse transcription reaction using random primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, California). For prostate tissue profiling, expression profiles were generated by co-hybridization of each of the 59 Cy3-labeled probes with a Cy5-labeled common reference sample, prepared from a pool of two BPH specimens as described15 and its RNA similarly amplified. For expression profiling of cultured cells, a common reference of nontreated LNCaP cells was used. The expression profile for each sample was represented as normalized ratios of sample/reference for all genes represented on the array. For expression data from tissues, genes associated with unreliable data points, defined as a mean fluorescence intensity less than 1000, were excluded from further analysis. To select genes whose expression varied most across the 59 samples, we applied a stringent filtration procedure based on the criteria of at least twofold expression change relative to the median in at least 15 samples to yield a list of 275 genes. An agglomerative hierarchical two-way clustering algorithm based on Euclidean distance measures15 was used to cluster the samples and the 275 genes. Statistical analyses of the differentially expressed genes were performed on expression data derived from the 59 tissue specimens and downloaded prostate tissue expression data from Lapointe and colleagues,17 using weighted gene analysis as described.15 For expression data derived from cultured cells, we first excluded unreliable data points by the same cutoff at mean intensity of 1000. The weighted gene analysis based on a modified distance-based w metric15 was again used to determine the extent of differential expression between siRNA treated samples and nontreated cells.
Immunohistochemical Staining
Immunohistochemical staining was performed using the Envision+ kit (DAKO Corp., Carpinteria, CA) as described.18 For myosin VI staining, a 1:400 dilution of the primary antibody (a gift from Mark Mooseker, Yale University, New Haven, CT)5 was used. For TMAs, we also performed keratin 8 staining to assist in automated scoring analysis as described.18 Sections of TMAs adjacent to those stained for myosin VI were stained for keratin 8 using a 1:800 dilution of the anti-CK8 antibody (InnoGenex, San Ramon, CA). For immunohistochemical staining in standard tissue slides, double labeling of α-methylacyl-CoA racemase and p63 (AMACR/p63) were performed as described16 in sections adjacent to those used for myosin VI staining.
TMA Analysis
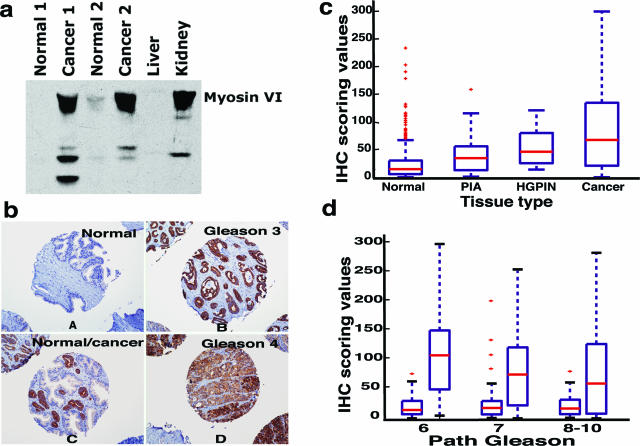
To avoid human bias during the assessment of immunohistochemical staining, we used the Chromavision ACIS II system (Clarient, Inc., San Juan Capistrano, CA), for semiautomated scoring.18 This approach uses two adjacent TMA slides in which one slide is stained with keratin 8 to determine epithelial content and the other is stained for myosin VI. For automated analysis we excluded TMA spots with a mixed diagnosis (mixed epithelial cells of normal/cancer/other lesions). Expression level of myosin VI within each individual TMA spot was evaluated by automatic and parallel calculation of pixel numbers in three staining categories (weak, moderate, and strong staining), yielding a composite score based on a previously described formula16 for each spot. The scores were normalized to the total brown pixel numbers for keratin 8 in the adjacent section to account for differential epithelial content across the TMA spots. Tissue histology in all TMA spots was re-examined by a pathologist (A.M.D.) in the adjacent hematoxylin and eosin (H&E)-stained TMA slides. Only spots annotated as containing a single diagnosis (no mixed normal/tumor/other lesions) were selected for further analysis. A nonparametric Wilcoxon’s rank-sum test was performed to test the statistical significance in expression levels between groups of interest. For Figure 2d, because multiple array spots are represented for each tissue type (normal or tumor) from each patient, the averaged values were used.
Figure 2.
TMA analysis of myosin VI. a: Confirmation of antibody specificity. Lanes 1 and 3: normal prostate tissues; lanes 2 and 4: prostate cancer tissues; lane 5: liver tissue as negative control; and lane 6: kidney tissue as positive control. b: Representative staining pattern of myosin VI in normal, Gleason grade 3 (medium grade), Gleason grade 4 (high grade), and prostate tissues with mixed normal and cancerous epithelial cells, as individually annotated. c: Box plots of myosin VI staining scores in normal (n = 665), PIA (n = 76), HGPIN (n = 18), and cancer tissues (n = 592). The score values were normalized to the epithelial content in each spot and displayed on the y axis. Each box is lined at lower quartile, median, and upper quartile score values for each group. The + symbols mark data values beyond the ends of the whiskers. d: Box plots of myosin VI staining scores. Patients were stratified by pathological Gleason scores (x axis), and myosin VI staining scores between normal and cancer samples were compared within each group. Number of patients represented in each category (normal versus cancer): Gleason 6 (80 versus 71), Gleason 7 (81 versus 61), Gleason 8 to 10 (34 versus 34).
Myosin VI Knockdown
The target sequences used to silence myosin VI expression were MYO6-siRNA-1, 5′-CCGCAAAAGTCCTGAGTAC-3′, and MYO6-siRNA-2, 5′-AGCTTGATCTCTTCCGGGT-3′ (Qiagen-Xeragon, Germantown, MD). The target sequence of nonsilencing control siRNA was 5′-TTCTCCGAACGTGTCACGT-3′ (Qiagen-Xeragon). LNCaP cells were transfected with siRNA duplexes by using Lipofectamine 2000 reagents (Invitrogen Corp.). Efficiency of myosin VI knockdown was tested by Western blot at different concentrations and various time points. Optimal gene knockdown conditions in LNCaP cells were achieved using 120 nmol/L siRNA at 96 hours after transfection.
Western Blot Analysis
Cultured cells and frozen human prostate tissues were subjected to standard Western blot analysis as described.16 For myosin VI detection, a polyclonal rabbit antibody (1:1000) raised against a C-terminal myosin VI peptide (Sigma, St. Louis, MO) was used. For VDUP1 detection, a monoclonal antibody (1:1000) was used (MBL International Cooperation, Woburn, MA). A monoclonal antibody (clone 36) for E-cadherin (BD Biosciences, San Jose, CA) was used at 1:3000 dilution. β-Actin was detected using a monoclonal antibody (AC-15) at 1:5000 dilution (Sigma).
Proliferation Assay
LNCaP cells that had been transfected 24 hours earlier with siRNA or without siRNA were seeded into a 96-well plate (8000 cells/well). The number of viable cells was determined daily with CellTiter 96 Aqueous nonradioactive cell proliferation assay (Promega, Madison, WI). In brief, 20 μl of the combined MTS/PMS solution was added to each well of the 96-well assay plate containing cells in 100 μl of culture medium. Optical density at 490 nm was recorded after 2 hours using an enzyme-linked immunosorbent assay plate reader.
Cell Migration Assay
For the in vitro migration assay, 24-well Costar transwell chambers (Corning Inc., Corning, NY) with 8-μm pore membrane were used. The under surface of the membrane was coated with fibronectin. LNCaP cells that had been transfected 96 hours earlier with and without siRNA were seeded (5 × 104/well) to the upper chambers and allowed to migrate for 16 hours at 37°C. At the end of the assay, after removal of nonmigratory cells on the upper surface, the migrated cells on the under surface were fixed and stained for 20 minutes with 0.5% crystal violet in 10% ethanol. Stained cells were eluted with 10% acetic acid, and the absorbance was determined. One-tailed Student’s t-test was used to assess the statistical significance (P < 0.05 considered to be significant).
Soft Agar Assay
The soft agar assay tests the anchorage-independent growth in vitro. In brief, 1 × 104 LNCaP cells that had been transfected 24 hours earlier with or without siRNA were resuspended with 3 ml of 0.3% agar (Invitrogen Corp.) in RPMI 1640 containing 10% fetal bovine serum. The cell-agar mixture was immediately seeded into six-well plates coated with 0.6% agar in RPMI 1640 with 10% fetal bovine serum. Culture media was replaced every 3 days. Colonies were stained with crystal violet as described above, at 2 weeks after seeding.
Results
Myosin VI mRNA Overexpression in Human Prostate Cancer
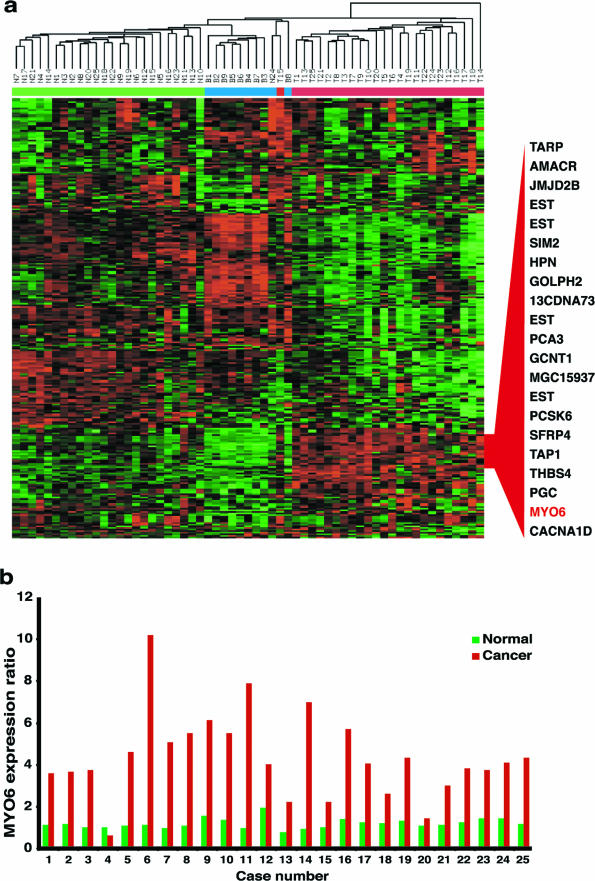
We generated gene expression profiles from 59 histologically characterized human prostate tissues (raw data available at http://www.oncomine.org). To highlight the expression differences across the samples, we applied an unbiased/unsupervised procedure (see Materials and Methods) to select 275 genes with expression that varied most across the 59 samples. A two-way clustering analysis was performed using this set of genes across the 59 samples, breaking down to 9 BPH (B1 to B9), 25 normal (N1 to N25), and 25 prostate cancer tissues (T1 to T25) that were matched with the normal prostate samples by number (Figure 1a). As shown, samples formed clusters based on their identities with few exceptions, and genes formed clusters based on differential expression patterns across the samples. We highlighted the identities of a cluster of 21 genes that demonstrated cancer-specific overexpression patterns (Figure 1a, fully annotated heatmap in Supplemental Figure 1 at http://ajp.amjpathol.org). Myosin VI clustered with many previously characterized prostate cancer markers, including prostate cancer antigen 3 (PCA3, DD3),19 AMACR,16 single-minded 2 (SIM2),20 hepsin,21 and TARP.22 Comparison of myosin VI expression ratios across the samples showed all but one of the 25 paired normal/cancer samples with higher expression in the cancer sample than in the paired normal sample (Figure 1b). On average, cancer samples (4.37 ± 2.05) showed a 3.7-fold higher expression of myosin VI mRNA when compared with the normal samples (1.20 ± 0.24), and a 4.6-fold increase when compared with the BPH samples (0.94 ± 0.10).
Figure 1.
Myosin VI is a novel prostate cancer marker identified in microarray analysis of surgical human prostate specimens. a: Heatmap representation of gene expression data for 59 histologically characterized samples. Columns represent samples, including 25 normal (N1 to N25), 9 BPH (B1 to B9), and 25 cancerous prostate tissues (T1 to T25). Rows represent genes. Normalized expression ratios for each gene are represented by red-green color scale, with red indicating overexpression relative to the median and green indicating underexpression relative to the median. The color bar above the color matrix denotes the sample identity, with blue marking BPH samples, green marking normal samples, and red marking cancer samples. A subcluster of genes representing those specifically overexpressed in cancer samples was shown in relation to their relative position in the color matrix. b: Comparison of myosin VI mRNA expression in 25 normal-tumor pairs. Normalized expression ratios of sample/reference were extracted from the microarray data and displayed for each individual pair of normal-tumor samples from each of the 25 cases. Green bars, normal; red bars, cancer; x axis, cases; y axis, expression ratios normalized to the common BPH reference denominator.
To further demonstrate the consistency and extent of myosin VI overexpression in human prostate cancers, we performed weighted gene analysis15 using two independent data sets, one from this study (raw data available at http://www.oncomine.org) and the other from Lapointe el al.17 As shown in Table 1, myosin VI was consistently identified as one of the top genes based on the test scores (w scores) comparing normal and cancerous human prostate tissues, with a P value less than 10−10 in both data sets (data not shown).
Table 1.
Top Ranked Genes Overexpressed in Human Prostate Cancer
| This study
|
Laponte et al17
|
||||
|---|---|---|---|---|---|
| wrank | IMAGE ID | Gene name | wrank | IMAGE ID | Gene name |
| 1 | 744944 | MYO6* | 1 | 318393 | TACSTD1 |
| 2 | 2119355 | CRISP3 | 2 | 120277 | AMACR |
| 3 | 344243 | UCK2 | 3 | 315796 | KIAA0872 |
| 4 | 1637504 | EST | 4 | 221880 | EST |
| 5 | 273546 | EST | 5 | 108810 | TACSTD1 |
| 6 | 809828 | E2F5 | 6 | 116644 | AMACR |
| 7 | 1034473 | AMACR | 7 | 226381 | AMACR |
| 8 | 855406 | EST | 8 | 226071 | NET-6 |
| 9 | 243159 | OCLN | 9 | 99827 | GJB1 |
| 10 | 502151 | SLC16A3 | 10 | 313671 | OCLN |
| 11 | 49630 | CACNA1D | 11 | 103681 | AMACR |
| 12 | 1571106 | JMJD2B | 12 | 223890 | KIAA1272 |
| 13 | 238821 | PLA2G7 | 13 | 312021 | EST |
| 14 | 22895 | INSM1 | 14 | 315526 | EST |
| 15 | 882506 | LOXL2 | 15 | 221393 | EST |
| 16 | 755239 | METTL1 | 16 | 112850 | MYO6* |
| 17 | 769945 | MGC13170 | 17 | 220886 | GPCR1 |
| 18 | 415962 | PACE4 | 18 | 117945 | GPCR1 |
| 19 | 81357 | BTEB1 | 19 | 100544 | NME1 |
| 20 | 160485 | GUCY1A3 | 20 | 107550 | SOX4 |
| 21 | 41569 | FLJ12650 | 21 | 239150 | PACE4 |
| 22 | 724615 | CHC1 | 22 | 107148 | UMPK |
| 23 | 241821 | GLYATL1 | 23 | 308201 | TNFRSF21 |
| 24 | 133130 | AMACR | 24 | 116796 | FBP1 |
| 25 | 486035 | UAP1 | 25 | 110119 | DNAH5 |
| 26 | 345034 | SCYB14 | 26 | 314538 | RNU17D |
| 27 | 810459 | FZD8 | 27 | 110896 | SPRY4 |
| 28 | 810284 | REXO2 | 28 | 308819 | SLC25A6 |
| 29 | 208413 | HPN | 29 | 109247 | TARP |
| 30 | 953262 | PCA3 | 30 | 100493 | MYO6* |
Two independent cDNA microarray datasets were analyzed using weighted gene analysis, and genes overexpressed in prostate cancer were ranked based on the wstatistic.15 Note that many genes are represented by multiple cDNA clones. Myosin VI clones were marked with asterisks.
TMA Analysis of Myosin VI Expression
An affinity purified polyclonal antibody against the tail domain of porcine myosin VI was used5 for immunohistochemical (IHC) analysis of myosin VI expression in human prostate cancer tissues. The antibody recognized a major band of ∼150-kd human myosin VI (Figure 2a) in prostate cancer tissues and a kidney tissue sample (positive control, 5) but not in normal prostate tissues or a liver sample (negative control, 5), thus confirming the binding specificity of the antibody and suitability for tissue staining. High-density TMAs were used for immunohistochemical analysis of myosin VI expression. A visual evaluation of stained TMAs confirmed the strongly positive myosin VI staining in the majority of cancerous epithelial cells but generally negative or weak staining in normal epithelium and negative staining in stromal components, as shown in representative array spots (Figure 2b).
Semiautomated scoring analysis18 was performed for IHC data from six TMAs. We first focused on the comparison of myosin VI protein expression among four histological lesions of interest: normal epithelium, PIA, HGPIN, and cancer epithelium. After histological evaluation of individual array spots by a pathologist (A.M.D.) and exclusion of array spots with poor quality and mixed diagnosis, IHC scores were obtained from 665 normal, 76 PIA, 18 HGPIN, and 592 cancer lesions. As shown in Figure 2c, cancer tissues had significantly higher myosin VI protein expression when compared with normal and PIA lesions (P < 10−10 and P < 10−5, respectively). Interestingly, when compared with the normal tissue, myosin VI protein expression is statistically higher in the two putative premalignant lesions, PIA and HGPIN (P < 10−6 and P < 10−5, respectively), suggesting that overexpression of myosin VI is an early event during prostate carcinogenesis.
Comparative analysis of myosin VI protein expression levels between normal and cancerous tissues was performed in three groups of patients stratified by pathological Gleason scores (Figure 2d). The majority of prostate cancers diagnosed today present Gleason scores of 6 or 7, typically containing a predominant component of grade 3 cancer that is characterized by infiltrative growth of well-formed acini (Figure 2b, B and C). As shown in Figure 2d, patients in these categories (Gleason scores 6 or 7) demonstrated the most consistent overexpression of myosin VI in the cancer tissues when compared with normal tissues (P < 10−10 and P < 10−6, respectively). Within the group of Gleason 6 patients in particular, the median score of the cancer samples was six times higher than the median score for the normal samples. High-grade prostate cancers (Gleason score 8 to 10) typically present back-to-back fused glands or loss of glandular differentiation (Figure 2b, D). These histologically more aggressive cancers (Gleason score 8 to 10) also showed marked overexpression of myosin VI when compared with the normal tissues (Figure 2d) (P < 0.02), although there was a decreased overall extent of cancer-specific myosin VI overexpression in comparison to medium-grade cancers (Gleason score 6 and 7) (P < 0.01). Consistent with its decreased cancer-specific expression in more aggressive cancer lesions, myosin VI levels were negatively correlated with the presence of seminal vesicle invasion and pelvic lymph node metastasis (P < 0.03) (data not shown).
IHC Analysis Using Standard Slides
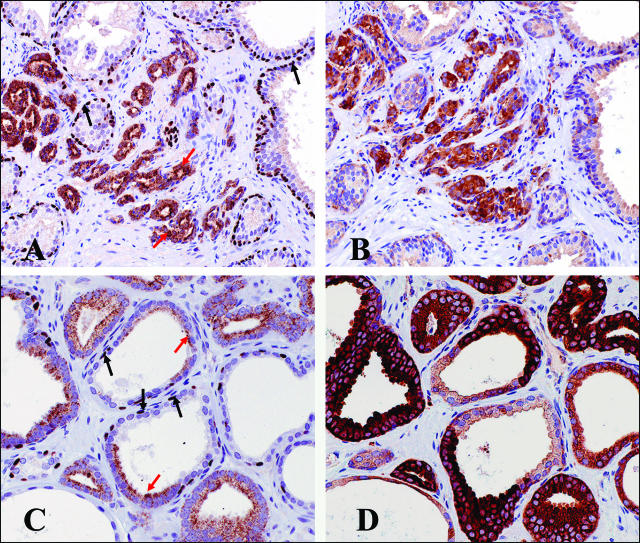
Histologically defined prostate cancer presents an invasive phenotype characterized by the absence of basal cells and local stromal invasion by the cancerous acini.23 Combined staining for cytoplasmic AMACR and basal cell-specific nuclear protein p63 can be used to reliably detect such cancer lesions.16 To illustrate the spatial pattern of myosin VI protein expression in relation to the cancerous histology as well as histological details surrounding the lesions of interest, we performed AMACR/p63 and myosin VI staining in adjacent cuts of standard sections (as opposed to arrayed tissues) from cases that were myosin VI-positive. As shown in Figure 3, myosin VI staining patterns were highly correlated with a readily discernible cancerous morphology, in tissues where normal and cancerous histology are both present (Figure 3, A and B) and even adjoined within the same acini (Figure 3, C and D). Intense myosin VI staining (Figure 3, B and D) was invariably seen in cancer lesions, as marked by positive cytoplasmic AMACR and negative nuclear p63 staining in adjacent sections (Figure 3, A and C), whereas normal epithelial cells with intact basal cell layer and negative AMACR staining (Figure 3, A and C) were weakly positive or negative for myosin VI (Figure 3, B and D).
Figure 3.
Myosin VI staining correlates with cancer morphology and stromal invasion in standard prostate tissue sections. Sections were double stained for AMACR/p63 (A and C), and adjacent cuts from the same paraffin blocks were stained for myosin VI (B and D). Black arrows: positive p63 nuclear staining that is specific for normal basal cells and absent in cancerous lesions; red arrows: positive cytoplasmic AMACR staining that is highly specific for prostate cancer cells.
Western Blot Analysis of Myosin VI in Cell Lines
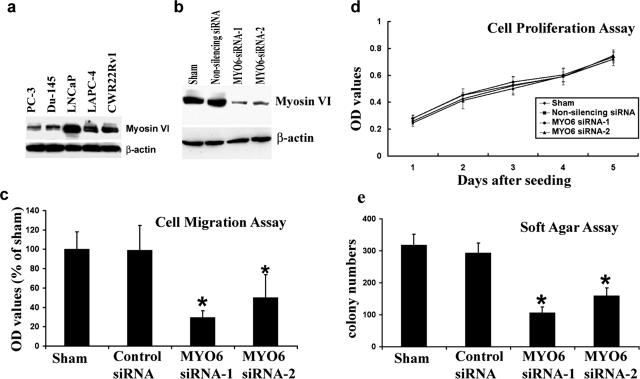
To establish an in vitro cell line model for functional studies, we examined protein expression of myosin VI in a panel of five human prostate cancer cell lines (Figure 4a). LNCaP cells were originally isolated from pelvic lymph node metastases of human prostate cancer. These cells retain many biological features of human prostate cancer including relatively slow growth and androgen sensitivity. As shown in Figure 4a, the LNCaP cell line expressed the most abundant myosin VI protein expression, followed by two other androgen receptor-positive lines (LAPC-4 and CWR22Rv1) that were derived from xenographs of locally advanced human prostate cancer. PC-3 and Du145 lines were established from androgen-refractory distant metastasis of human prostate cancer and expressed less myosin VI than the androgen-sensitive cancer cell lines. The expression pattern of myosin VI in cultured human prostate cells is again in line with the expression changes observed in clinical tissue specimens, in which there was a general trend of decreased cancer-specific myosin VI expression in more aggressive cancers.
Figure 4.
Myosin VI expression and function in prostate cancer cell lines. a: Protein expression of myosin VI in five commonly used human prostate cancer cell lines. Protein levels of β-actin were examined in the same blot and serve as loading controls. b: Myosin VI expression was inhibited by two siRNA duplexes (lanes 3 and 4), but not affected by control siRNA treatment (lane 2). Protein levels of β-actin were examined in the same blot and serve as loading controls. c: Bar graph showing impaired LNCaP cell migration after myosin VI knockdown (siRNA-MYO6-1, siRNA-MYO6-2). Data were compiled from three replicates for each treatment conditions. *Significantly lower number of migratory cells when compared with the sham-transfected cells. d: Cell proliferation curve in a span of 5 days. Data were complied from five replicates for each treatment and time point. e: Decreased soft agar colony formation after inhibition of myosin VI expression. Data were complied from four replicates for each treatment condition. *Significantly less colonies when compared with the sham-transfected cells.
Functional Roles of Myosin VI
Because LNCaP human prostate cancer cells demonstrated the most abundant expression of myosin VI, we performed in vitro functional assays after inhibition of myosin VI expression in these cells. As shown in Figure 4b, myosin VI protein expression was dramatically decreased by both siRNA duplexes designed to target the specific degradation of myosin VI RNA (target sequences are myosin VI-specific sequences in the motor domain) but was not affected by control nonsilencing siRNA under identical conditions. No gross morphological changes were observed in cultured cells after siRNA treatment. Consistent with previous findings,14 the inhibition of myosin VI expression resulted in impaired cell migration (Figure 4c) but did not affect the proliferation rate of cells in the culture medium (Figure 4d). However, experimental knockdown of myosin VI significantly reduced the number of soft agar colonies 14 days after inoculation (Figure 4e), suggesting a role of myosin VI in anchorage-independent growth, a hallmark of transformed phenotype.
Global Expression Changes after Inhibition of Myosin VI Expression
Additional clues regarding the biological impact of myosin VI expression was examined by cDNA microarray analysis after myosin VI knockdown in LNCaP cells. We compared expression differences between two siRNA-transfected samples and the two control cell samples (including cells treated with nonsilencing control siRNA). Genes were ranked based on a w metric15 that measures the extent of gene expression change as a function of myosin VI knockdown (Supplemental Figure 2 at http://ajp.amjpathol.org). After myosin VI inhibition, the majority (13 of 15) of the genes (Supplemental Figure 2 at http://ajp.amjpathol.org) showed expression suppression by approximately twofold. The list of suppressed genes included myosin VI (ranked no. 5) (Supplemental Figure 2 at http://ajp.amjpathol.org), the intended target of siRNA-mediated knockdown. Exceptionally, myosin VI knockdown resulted in a nearly 10-fold increased expression for TXNIP,24 whereas no other genes in the whole dataset consistently demonstrated more than threefold expression changes in either direction.
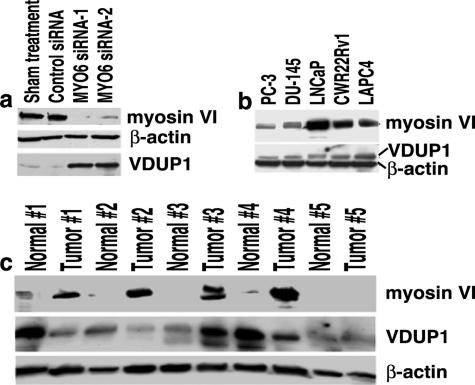
Validation of TXNIP/VDUP1 Expression
TXNIP (thioredoxin-interacting protein 1), also named VDUP1 (vitamin D3 up-regulated protein 1), is a tumor suppressor that also participates in transcriptional repression to inactivate oncogenic signals.24 The protein expression of TXNIP was dramatically increased after inhibition of myosin VI expression in both LNCaP (Figure 5a) and CWR22Rv1 cells (data not shown), as validated by Western blot analysis. In addition, protein expression of myosin VI and TXNIP (Figure 5b) appeared to be inversely correlated in unperturbed androgen receptor-positive cell lines (LNCaP, CWR22Rv1, LAPC-4), whereas the AR-negative PC-3 and DU-145 cells did not express higher levels of TXNIP despite lower expression of myosin VI. Protein expression of TXNIP was subsequently examined in five paired normal and tumor samples from radical prostatectomy specimens. Despite the heterogeneity of the overall expression pattern, TXNIP was generally decreased in the cancer specimens when compared with their matched normal counterparts (Figure 5c).
Figure 5.
Inverse correlation between myosin VI and VDUP1 protein expression. a: Western blot analysis of TXNIP after inhibition of myosin VI expression in LNCaP cells. b: Western blot analysis of TXNIP and myosin VI in the cell lines. c: Western blot analysis of TXNIP and myosin VI in five normal-tumor pairs of human prostate specimens. Expression levels of β-actin were used as loading controls in all analysis.
Discussion
In the human prostate, normal ducts and acini are lined by a double cell layer: a flat basal cell layer oriented parallel to the basement membrane and a secretory tall columnar luminal cell layer. A defining histological feature of human prostate cancer is the complete absence of basal cells and local stromal invasion/infiltration by the cancerous acini.23 Gain of invasive potential, therefore, is required for the establishment of histologically defined human prostate cancer. Global expression analysis emphasizing the comparison between normal prostate epithelium and cancerous acini15,17,25–31 may reveal molecular alterations accompanying this critical gain of function. In this follow-up study on a top gene identified in global transcriptome analysis of surgical prostate specimens, we uncovered a novel connection between an actin motor, myosin VI, and human prostate cancer. Although myosin VI may participate in diverse cellular functions,6,7 the observed association of myosin VI expression with histological characteristics of human prostate cancer may be linked to an established role of myosin VI in cell migration.12–14 When combined with pericellular proteolysis and proliferative force, an enhanced migratory potential may facilitate cancer cell/acini invasion in the tissue environment.32
Stromal invasion is a hallmark of virtually all human cancers of epithelial origin.33 Transcriptional up-regulation of myosin VI, however, does not appear to be a universal phenomenon for all carcinomas. Based on mRNA expression data in the public database, cancer-specific myosin VI overexpression is primarily restricted in human prostate and breast cancers (http://genome-www5.stanford.edu/cgi-bin/source/sourceSearch and http://www.oncomine.org). It is unclear whether steroid hormone receptors play a role in regulating myosin VI mRNA expression. Although expression levels of myosin VI appear to correlate with androgen receptor status (Figure 4a), it is not regulated by synthetic DHT analog R1881 (unpublished observation) nor is it affected by complete knockdown of androgen receptor in human prostate cancer cells (unpublished observation). The regulatory mechanism accounting for myosin VI overexpression in human prostate cancer is currently unknown.
A hypothesis-driven approach has found elevated myosin VI protein levels in ovarian cancers as well as a positive correlation with ovarian cancer aggressiveness both in vitro and in vivo.14 We did not observe a similar correlation between myosin VI expression and any of the clinical and pathological indicators of prostate cancer aggressiveness. There was instead a general trend of slightly decreased cancer-specific myosin VI expression in prostate cancer cases with aggressive histological and clinical features. Therefore, human cancers of different tissue origin may display different modes of regulation and different patterns of alteration in myosin VI expression. It is worth noting that advanced human prostate cancers may have acquired other properties, such as enhanced pericellular proteolysis,34 for invasion-associated functions and may have thus become less reliant on the participation of myosin VI. Moreover, more dedifferentiated prostate cancers often express reduced levels of E-cadherin,35 which may lead to decreased myosin VI expression as previously suggested.12 Therefore, it is reasonable to speculate that myosin VI may regulate coordinated movement of a cluster of cells as seen in well-differentiated, E-cadherin-positive prostate cancer lesions but may not be as critical in advanced cancers in which E-cadherin-mediated cell adhesion is perturbed.
The aforementioned emphasis on cell migration should not prelude a role of myosin VI in other cellular processes that may also contribute to the development of human prostate cancer. In studies unrelated to human cancer, myosin VI was found to play critical roles in spermatogenesis,36 inner ear hair cell differentiation,37 asymmetric stem cell division,38 endocytosis,10 and secretion.13 Although these seemingly diverse functions may have a common underlying mechanism linked to the unique myosin VI motor direction, they appear to be species-, organ-, and tissue-specific and possibly depend on specific isoforms of myosin VI as well as the presence of critical myosin VI binding partners.6 In this study, inhibition of myosin VI expression in human prostate cancer cells resulted in reduced anchorage-independent growth (Figure 4), as well as a nearly 10-fold induction of the tumor suppressor TXNIP (VDUP1) (Figure 5b), suggesting a key role for myosin VI in maintaining the malignant phenotype of human prostate cancer cells. TXNIP may play a key role in regulating oncogenic signaling because expression analysis also identified dramatically reduced expression of TXNIP after transfection of an oncogenic ETS transcription factor.39 Despite the fact that the TXNIP expression may be regulated by the ischemic conditions encountered during the surgical tissue collection process and that the protein product is very labile,40,41 examination in clinical specimens indeed revealed a generally decreased pattern of TXNIP expression in cancer samples (Figure 5c). This novel observation should be followed up pending the availability of an antibody suitable for immunohistochemical analysis.
Our functional studies primarily relied on siRNA technology because of lack of expression constructs for human myosin VI. Off-target effects, which lead to changes in expression in genes other than the target gene, cannot be efficiently controlled unless a rescue construct is available.42 It remains to be definitively determined whether the pattern of gene expression alterations (Supplemental Figure 2 at http://ajp.amjpathol.org) was a direct response to loss of myosin VI, or a result of off-target gene regulation by synthetic siRNA duplexes. A recent study,43 however, revealed distinctive, nonoverlapping patterns of off-target gene suppression among experiments targeting seven different locations of the same MAPK14 transcript, suggesting that off-target effects are specific to the target sequence but not to the target gene. Therefore our observation that the two different myosin VI siRNA sequences led to almost identical gene suppression patterns (Supplemental Figure 2 at http://ajp.amjpathol.org) argued against an off-target effect. In addition, the observed gene expression alterations were specific to myosin VI knockdown because these alterations were not observed in our expression analysis after knockdown of other genes (J.L., unpublished observation). Therefore, in this study, it is unlikely that the off-target effects played a dominant role in regulating gene expression and mediating the biological effect after myosin VI gene knockdown.
Because ∼80 to 90% of human prostate cancers diagnosed today present a pathology (Gleason score 6 and 7) that highly correlates with cancer-specific myosin VI overexpression (Figure 2), relevant studies may have an impact in clinical management of human prostate cancer. However, unlike AMACR,16 we do not expect myosin VI to be useful as a tissue marker for prostate cancer diagnosis by IHC given that many of the normal and atrophy lesions were also positive for staining and a subset of the cancers were negative or weak (Figure 2, c and d). Inherited inactivating deletions and mutations of myosin VI gene in both mice and humans result in hearing loss but do not affect viability,6 suggesting that myosin VI may be amenable to therapeutic intervention. Myosin VI function may be mediated by its interaction with multiple binding partners6 through its tail domain. Detailed structural and functional studies in the context of molecular interactions may help to identify specific therapeutic targets.
In summary, we discovered a novel connection between myosin VI and human prostate cancer. Myosin VI is one of the top genes and also the only myosin gene that has demonstrated cancer-specific overexpression in our expression data, shedding light on the nature and scale of dysregulated myosin VI expression in human prostate cancer. Previously characterized as a backward motor, myosin VI moves toward the minus end of the actin track, a direction opposite to all other known myosin members. Myosin VI may have unique properties and functions that are yet to be fully characterized, particularly in the context of human cancer. This novel connection should stimulate a thorough investigation of the unique structural and functional properties of myosin VI in a broader context.
Supplementary Material
Acknowledgments
We thank Dr. Mark Mooseker (Yale University, New Haven, CT) for providing the myosin VI antibody for immunohistochemical analysis, and Dr. Denise Montell (The Johns Hopkins University, Baltimore, MD) for critical reading and discussion of the manuscript.
Footnotes
Address reprint requests to Jun Luo, Department of Urology, 411 Marburg Building, 600 N. Wolfe St., Baltimore, MD 21287, or Angelo M. De Marzo, Department of Pathology, CRB153, 1650 Orleans St., Baltimore, MD 21231. E-mail: jluo1@jhmi.edu and ademarz@jhmi.edu.
Supported by the US Department of Defense (grant W81XWH-04-1-0873 to J.L.), the National Institutes of Health (National Cancer Institute Specialized Program in Research Excellence in Prostate Cancer no. P50CA58236 to Johns Hopkins), The Donald and Susan Sturm Foundation (to A.M.D.), and The Prostate Cancer Foundation (to J.L. and A.M.D.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology. 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- Wu X, Jung G, Hammer JA., III Functions of unconventional myosins. Curr Opin Cell Biol. 2000;12:42–51. doi: 10.1016/s0955-0674(99)00055-1. [DOI] [PubMed] [Google Scholar]
- Kellerman KA, Miller KG. An unconventional myosin heavy chain gene from Drosophila melanogaster. J Cell Biol. 1992;119:823–834. doi: 10.1083/jcb.119.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Mooseker MS. Porcine myosin-VI: characterization of a new mammalian unconventional myosin. J Cell Biol. 1994;127:425–440. doi: 10.1083/jcb.127.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Noguchi T, Miller KG. Myosin VI: a structural role in actin organization important for protein and organelle localization and trafficking. Curr Opin Cell Biol. 2004;16:189–194. doi: 10.1016/j.ceb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- Menetrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T. Myosin VI: two distinct roles in endocytosis. J Cell Sci. 2003;116:3453–3461. doi: 10.1242/jcs.00669. [DOI] [PubMed] [Google Scholar]
- Warner CL, Stewart A, Luzio JP, Steel KP, Libby RT, Kendrick-Jones J, Buss F. Loss of myosin VI reduces secretion and the size of the Golgi in fibroblasts from Snell’s waltzer mice. EMBO J. 2003;22:569–579. doi: 10.1093/emboj/cdg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht ER, Montell DJ. Myosin VI is required for E-cadherin-mediated border cell migration. Nat Cell Biol. 2002;4:616–620. doi: 10.1038/ncb830. [DOI] [PubMed] [Google Scholar]
- Buss F, Luzio JP, Kendrick-Jones J. Myosin VI, an actin motor for membrane traffic and cell migration. Traffic. 2002;3:851–858. doi: 10.1034/j.1600-0854.2002.31201.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Cheng W, Hung J, Montell D, Geisbrecht E, Rosen D, Liu J, Naora H. Lessons from border cell migration in the Drosophila ovary: a role for myosin VI in dissemination of human ovarian cancer. Proc Natl Acad Sci USA. 2004;101:8144–8149. doi: 10.1073/pnas.0400400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- Luo J, Zha S, Gage WR, Dunn TA, Hicks J, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DA, Isaacs WB, Morgan JD, Fedor HL, Hicks JL, Mangold LA, Walsh PC, Partin AW, Platz EA, Luo J, De Marzo AM. Trefoil factor 3 overexpression in prostate carcinoma: prognostic importance using tissue microarrays. Prostate. 2004;61:215–227. doi: 10.1002/pros.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- DeYoung MP, Tress M, Narayanan R. Identification of Down’s syndrome critical locus gene SIM2-s as a drug therapy target for solid tumors. Proc Natl Acad Sci USA. 2003;100:4760–4765. doi: 10.1073/pnas.0831000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Chevillet J, Mirosevich J, Roberts RL, Matusik RJ, Vasioukhin V. Hepsin promotes prostate cancer progression and metastasis. Cancer Cell. 2004;6:185–195. doi: 10.1016/j.ccr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Wolfgang CD, Essand M, Vincent JJ, Lee B, Pastan I. TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus. Proc Natl Acad Sci USA. 2000;17:9437–9442. doi: 10.1073/pnas.160270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- Jeon JH, Lee KN, Hwang CY, Kwon KS, You KH, Choi I. Tumor suppressor VDUP1 increases p27(kip1) stability by inhibiting JAB1. Cancer Res. 2005;65:4485–4489. doi: 10.1158/0008-5472.CAN-04-2271. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 2001;61:5692–5696. [PubMed] [Google Scholar]
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;6:5974–5978. [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- Ernst T, Hergenhahn M, Kenzelmann M, Cohen CD, Bonrouhi M, Weninger A, Klaren R, Grone EF, Wiesel M, Gudemann C, Kuster J, Schott W, Staehler G, Kretzler M, Hollstein M, Grone HJ. Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: a gene expression analysis on total and microdissected prostate tissue. Am J Pathol. 2002;160:2169–2180. doi: 10.1016/S0002-9440(10)61165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- Kohn EC, Liotta LA. Molecular insights into cancer invasion: strategies for prevention and intervention. Cancer Res. 1995;55:1856–1862. [PubMed] [Google Scholar]
- Clark WH. Tumour progression and the nature of cancer. Br J Cancer. 1991;64:631–644. doi: 10.1038/bjc.1991.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- De Marzo AM, Knudsen B, Chan-Tack K, Epstein JI. E-cadherin expression as a marker of tumor aggressiveness in routinely processed radical prostatectomy specimens. Urology. 1999;53:707–713. doi: 10.1016/s0090-4295(98)00577-9. [DOI] [PubMed] [Google Scholar]
- Hicks JL, Deng WM, Rogat AD, Miller KG, Bownes M. Class VI unconventional myosin is required for spermatogenesis in Drosophila. Mol Biol Cell. 1999;10:4341–4353. doi: 10.1091/mbc.10.12.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self T, Sobe T, Copeland NG, Jenkins NA, Avraham KB, Steel KP. Role of myosin VI in the differentiation of cochlear hair cells. Dev Biol. 1999;214:331–341. doi: 10.1006/dbio.1999.9424. [DOI] [PubMed] [Google Scholar]
- Petritsch C, Tavosanis G, Turck CW, Jan LY, Jan YN. The Drosophila myosin VI Jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Dev Cell. 2003;4:273–281. doi: 10.1016/s1534-5807(03)00020-0. [DOI] [PubMed] [Google Scholar]
- Myers-Irvin JM, Van Le TS, Getzenberg RH. Mechanistic analysis of the role of BLCA-4 in bladder cancer pathobiology. Cancer Res. 2005;65:7145–7150. doi: 10.1158/0008-5472.CAN-05-1142. [DOI] [PubMed] [Google Scholar]
- Xiang G, Seki T, Schuster MD, Witkowski P, Boyle AJ, See F, Martens TP, Kocher A, Sondermeijer H, Krum H, Itescu S. Catalytic degradation of vitamin D up-regulated protein 1 mRNA enhances cardiomyocyte survival and prevents left ventricular remodeling after myocardial ischemia. J Biol Chem. 2005;280:39394–39402. doi: 10.1074/jbc.M502966200. [DOI] [PubMed] [Google Scholar]
- Dutta KK, Nishinaka Y, Masutani H, Akatsuka S, Aung TT, Shirase T, Lee WH, Yamada Y, Hiai H, Yodoi J, Toyokuni S. Two distinct mechanisms for loss of thioredoxin-binding protein-2 in oxidative stress--induced renal carcinogenesis. Lab Invest. 2005;85:798–807. doi: 10.1038/labinvest.3700280. [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci USA. 2005;102:2396–2401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.