Abstract
SH2-B, a JAK2-interacting protein has recently been identified as a key regulator of leptin and insulin sensitivity, glucose homeostasis and body weight in mice. The aim of this study was to determine whether single-nucleotide polymorphisms (SNPs) in the human SH2B gene are associated with these variables. A tagging SNP, Ala484Thr (rs7498665) was selected to represent five common SNPs (minor allele frequency (MAF) >0.05) in perfect linkage disequilibrium (LD) in a 16 kb region encompassing the SH2B gene. The tSNP was genotyped in 2455 Caucasian female twins (mean age 47.4±12.6 years) from the St Thomas’ UK Adult Twin Registry (Twins UK). Ala484Thr (minor allele frequency 0.38) was associated with serum leptin, total fat, waist circumference and body weight (Ps=0.02–0.04). The coding SNP has no predicted effect on protein structure or function and is likely to be in LD with an as yet unidentified functional variant in the SH2B gene. Our results support a role for SH2-B in modulating the regulation of body weight and fat by leptin in this female population. If SH2-B signalling is attenuated in diet-induced obesity, it could become a target for drug-induced leptin sensitization.
Keywords: fat distribution, genetic susceptibility, leptin, signal transduction
Leptin, a product of the obese gene [1], signals nutritional status to key regulatory centres in the hypothalamus, thereby acting as an important signal regulating energy homeostasis [2]. On binding to hypothalamic receptors, leptin induces tyrosine phosphorylation of JAK2 and STAT3, paralleled by a comparable increase in phosphoinositol 3-kinase (PI3K) activity associated with insulin receptor substrate (IRS) -2 [3]. The Src-homology-2 (SH2) domain-containing putative adapter SH2-B is a ubiquitously expressed cytoplasmic protein which binds via its SH2 domain [4] simultaneously to both JAK2 and IRS-2, promoting leptin-stimulated activation of the PI3K pathway in cultured cells [5]. Recently, it has been shown that SH2-B deficient mice develop insulin resistance and type 2 diabetes [6] as well as severe leptin resistance, hyperphagia and obesity [7]. Thus, SH2-B is a key cytoplasmic signalling molecule that acts as a positive regulator of leptin and insulin signal transduction in mice.
The aim of this study was to determine whether single-nucleotide polymorphisms (SNPs) in the human SH2B gene are associated with measures of body fat and insulin sensitivity in 2455 women from the St Thomas’ UK Adult Twin Registry (Twins UK). Characteristics of the subjects are shown in Table 1. Pairwise LD between five relatively common SNPs (MAF >0.05) located in a 16kb region including the SH2B gene: rs4788102, rs8055982, rs7498665, rs7359397 and rs3888190 was quantified by D′/r2. The five variants were in perfect linkage disequilibrium (LD; all pairwise r2 =1.00), i.e. define just two common haplotypes (Figure 1). Hence one tagging SNP was required to represent all HapMap variants with MAF>0.05. The only coding SNP, rs7498665 (Ala484Thr) was selected and genotyped in the complete cohort. The MAF of this tSNP was 0.38 and genotypes did not deviate significantly from Hardy Weinberg equilibrium (P=0.92).
Table 1.
General characteristics of subjects
| Variable | n | Mean ± SD |
|---|---|---|
| Age, years | 2455* | 47.4±12.6 |
| Postmenopausal, % | 2161 | 48.1 |
| Obesity-related variables: | ||
| Leptin, ng/ml | 2455 | 16.4±11.9 |
| BMI, kg/m2 | 2199 | 24.8±4.4 |
| Weight, kg | 2200 | 65.4±11.9 |
| Waist, cm | 2106 | 78.1±10.3 |
| Total fat, kg | 2204 | 23.2±8.9 |
| Total fat, % | 2066 | 35.2±8.1 |
| Central fat, kg | 2090 | 1.31±0.74 |
| Central fat, % | 2090 | 30.4±11.6 |
| Insulin sensitivity:† | ||
| Fasting glucose, mmol/L | 998 | 4.40±0.46 |
| Fasting insulin, μU/mL | 893 | 6.29±4.51 |
| HOMA | 889 | 1.25±0.91 |
| 2h-glucose, mmol/L | 647 | 5.19±1.10 |
| 2h-insulin, μU/mL | 647 | 34.3±25.5 |
| SiM | 647 | 88.9±70.7 |
Number of subjects (754 MZ, 1701 DZ) with leptin and genotype data;
Non-fasting subjects (n=166), patients with either type 1 or type 2 diabetes, fasting glucose>7.8mmol/L or 2h-glucose>11.1mmol/L (n=11) and patients on any anti-diabetic drugs (n=1) were all excluded.
Figure 1. Structure of SH2B gene and position of SNPs relative to start site in exon 1.
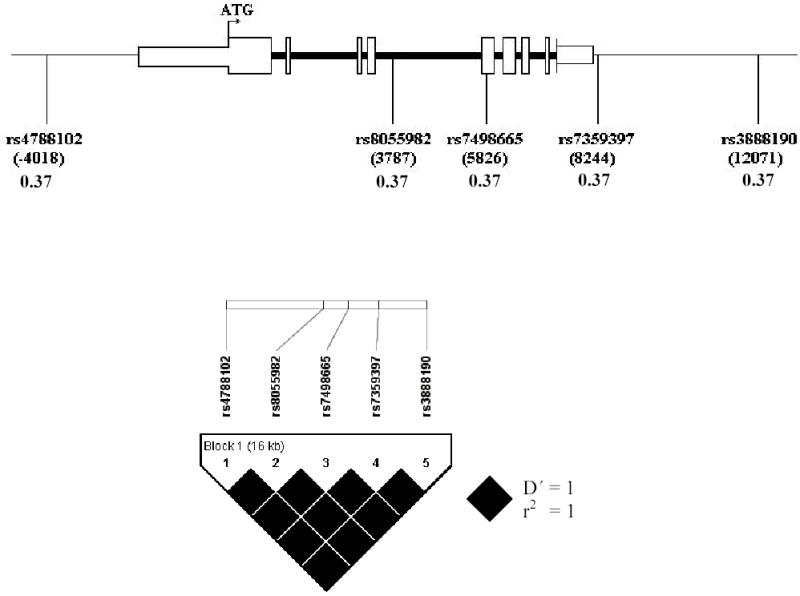
The SNPs span a distance of 16 kb including the 10.2 kb SH2B gene on chromosome 16. Minor allele frequencies and pairwise LD representation is from output of Tagger program, based on HapMap SNP genotype data in 30 Caucasian trios.
Carriers of the minor allele of SNP rs7498665 had a significantly higher general obesity factor score (P=0.02). Follow up analyses of individual obesity phenotypes revealed further significant associations with serum leptin (P=0.04), total fat mass (P=0.04), waist circumference (P=0.02) and weight (P=0.02) (Table 2). This tSNP explained between 0.21% and 0.34% of the variance for these traits. The higher level of serum leptin in allele 2 carriers infers an impaired sensitivity to leptin action (leptin resistance), which typically accompanies overweight. This is represented here by the parallel associations with fat mass, waist circumference and body weight. There were no significant associations with insulin resistance (HOMA) or sensitivity (SiM) measures. We tested association between SNP rs7498665 and obesity variables in the subgroups with HOMA (n= 889) and SiM (n=647) data and found outcomes similar to the full cohort. Significant associations were found with BMI, weight, total fat % and central fat % (Ps = 0.01–0.04) (data not shown).
Table 2.
Association of SH2B tSNP Ala484Thr (rs7498665) with obesity-related variables, HOMA and SiM
|
No.
|
Mean (SD)
|
P | ||||||
|---|---|---|---|---|---|---|---|---|
| Variables | 11/12/22 | 11 | 12 | 22 | 2 df Overall Test | Genetic model | P | Var.(%) |
| General obesity factor score | 899/1122/333 | −.06(.99) | .00(1.00) | .05(.96) | .04 | Additive | .02 | 0.27 |
| BMI, kg/m2 | 931/1164/344 | 24.56(4.40) | 24.76(4.38) | 24.85(4.24) | .12 | - | - | - |
| Weight, kg | 931/1165/344 | 64.61(11.76) | 65.39(11.87) | 66.06(11.48) | .03 | Additive | .02 | 0.34 |
| Total fat, kg | 909/1155/337 | 22.90(8.77) | 23.58(8.90) | 23.56(8.52) | .04 | Dominant | .04 | 0.21 |
| Total fat, % | 903/1131/315 | 35.12(8.05) | 35.66(8.03) | 35.59(8.00) | .18 | - | - | - |
| Leptin, ng/ml | 934/1175/347 | 15.86(11.58) | 16.67(12.11) | 17.02(11.64) | .04 | Additive | .04 | 0.23 |
| Central obesity factor score | 890/1115/326 | −.07(.99) | .00(1.00) | .00(.95) | .21 | - | - | - |
| Waist, cm | 919/1142/333 | 77.70(10.08) | 78.51(10.23) | 78.30(9.85) | .03 | Dominant | .02 | 0.26 |
| Central fat, kg | 902/1145/335 | 1.29(0.71) | 1.35(0.74) | 1.32(0.67) | .35 | - | - | - |
| Central fat, % | 902/1145/335 | 30.55(11.34) | 31.31(11.76) | 31.39(11.15) | .27 | - | - | - |
| HOMA | 352/411/126 | 1.18(0.68) | 1.28(1.11) | 1.32(0.75) | .39 | - | - | - |
| SiM | 254/305/88 | 82.86(56.44) | 92.93(73.83) | 92.44(92.69) | .89 | - | - | - |
1=Ala; 2=Thr.
The main strengths of our study lie in the large sample and the availability of detailed measurements of body fat by DEXA in all subjects. This is the first attempt to examine a possible role for SH2-B in predisposition to human obesity and insulin sensitivity and the first reported association of an SH2B variant with serum leptin, total fat mass, waist circumference and weight. The current study had 80% (a = 0.05) power to detect a biallelic quantitative trait locus, explaining as little as 0.51% of the variance in HOMA index (n=889) and 0.90% of the variance in SiM (n=647). However, sample sizes for these variables were not sufficient to detect an association accounting for a similar proportion of variance accounted for by the associations with leptin and fat in the full cohort.
It should be noted that we have established these associations only in a sample of female twins. We have previously found few differences between twins and singletons in the population generally, the only indication being that MZ twins had a slightly lower weight and a smaller variance for weight than DZ twins and singletons [8]. However, as measurements of leptin and fat were not made in that study, we are unable to generalise our present findings on general obesity, fat and leptin to age-matched females in the wider population.
The comprehensiveness of coverage of the SH2B gene by our selected tSNP was limited by the availability of SNPs on the HapMap database at the time of the study. Analysis of the genotypes of five SNPs with MAF >0.05 in a 16 kb region including the 10.2 kb SH2B gene, based on HapMap data from 30 Caucasian parent-offspring trios, showed all five to be in complete LD. The region containing the SH2B gene therefore appears to have undergone little or no recombination. Accordingly, we were able to choose only one tSNP to tag all the common SH2B variants available in HapMap. We are confident that other SNPs at this frequency within this region are unlikely to remain unmarked by our chosen tSNP. Although Caucasians are a heterogeneous population group in which haplotype composition, LD structure and LD decay with physical distance may vary within sub-groups, recent studies indicate that the HapMap data in Caucasians is representative of other Caucasian groups in terms of allele frequencies and LD structure, especially for the common SNPs [9]. HapMap genotype data further indicates that the five common SHP2 SNPs are also in perfect LD in Chinese (MAF=0.16) and Japanese (MAF=0.13) and in almost perfect LD in Africans (MAF=0.16). In Africans, Ala484Thr (rs7498665) has a slightly higher MAF (0.17) and a pairwise r2 of 0.94 with the other SNPs. Interestingly, one SNP, rs7359397, is not polymorphic in the African HapMap population.
Leptin regulates energy balance and body weight through activation of its receptor and several downstream signalling pathways. SH2-B has been identified as a key regulator of leptin sensitivity, energy balance, and body weight in mice on the basis of studies of null homozygotes [7]. The mice were severely hyperphagic and obese and developed a metabolic syndrome characterized by hyperleptinemia, hyperinsulinemia, hyperlipidemia, hepatic steatosis and hyperglycemia. Leptin-stimulated activation of hypothalamic JAK2 and phosphorylation of hypothalamic STAT3 and IRS2 were significantly impaired. This suggests that SH2-B is an endogenous enhancer of leptin and insulin sensitivity and is required for maintaining normal energy metabolism and body weight in mice. SH2-B is therefore a plausible candidate for involvement in human obesity, insulin resistance and glucose intolerance. The Ala484Thr polymorphism is a non-synonymous coding SNP (nsSNP) and therefore may be a functional variant. We investigated this possibility using two databases (SNPeffect and PolyPhen) [10,11], which predict the effects of nsSNPs on protein structure and function. Ala484Thr appears to be neutral, suggesting that it is in LD with an as yet unidentified functional variant in the SH2-B gene. Our results support a role for SH2B in modulating the regulation of body weight and fat by leptin in our female twin population, but we are as yet unable to draw any conclusions regarding an influence on insulin sensitivity.
The results of our study invite replication in a larger cohort to confirm these modest associations with obesity and detect any smaller effects on insulin sensitivity. In tagging only common SNPs, we excluded the possibility of discovering any substantial effect associated with rarer SNPs (MAFs<0.05). Also, potential LD markers of intergenic regulatory regions such as enhancers, which could lie 10–20 kb upstream of the gene, were not considered in a tagged region encompassing 16 kb. Future investigations could therefore extend the tagged region and include SNPs of lower frequency. If SH2-B signalling is eventually shown to be attenuated in diet-induced obesity, it could become a target for drug-induced leptin sensitization.
Methods
Study design
Twins UK comprises unselected, mostly female volunteers ascertained from the general population through national media campaigns in the UK [12]. The study cohort comprised 2455 subjects (754 MZ, 1701 DZ) with available leptin data. The number of individuals in the study with data on other phenotypic variables is shown in Table 1. Information on all twins was used in association analyses (see statistical analysis below).
Means and ranges of disease and lifestyle characteristics in Twins UK were similar to an age-matched sample of singleton women from the general population [8]. Informed consent was obtained from participants before they entered the study and approved by the local research ethics committee.
Zygosity, body composition and biochemical analyses
Zygosity confirmation and measurement of body composition by DEXA, serum leptin, insulin and glucose were performed as described previously [13]. A sub-sample of approximately 750 subjects, representing unselected female twins from the general population underwent an oral glucose tolerance test (OGTT) for which glucose and insulin levels were measured before and 2 hours after a 75-g oral-glucose load [13].
Selection of tSNPs
The SH2-B gene spans over 10kb and contains 9 exons. Five polymorphic SNPs with minor allele frequency (MAF) >0.05 in a 16 kb region including the SH2B gene are listed on the HapMap database (HapMap Data Release #20/Phase II Jan 2006; http://www.hapmap.org). Their positions with respect to the first coding base in exon 1 (shown in parentheses) are as follows: rs4788102 (−4018); rs8055982 (3787); rs7498665 (5826); rs7359397 (8244); rs3888190 (12071) (Figure 1). (Note that rs12599217 in the HapMap database lies between rs8055982 and rs7498665, but it is not polymorphic). Genotypes were downloaded from HapMap CEU 30 parent-offspring trio data into Haploview (http://www.broad.mit.edu/mpg/haploview/). The integral Tagger program (http://www.broad.mit.edu/tagger), was then used to collectively capture all SNPs at minimum r2 of 0.8, using the aggressive multi-marker tagging mode.
Genotyping in cohort
SNP rs7498665 was genotyped in the complete cohort by Pyrosequencing, (Biotage, Uppsala, Sweden). Genotyping accuracy as assessed by inclusion of duplicates (pairs of monozygotic twins) in the arrays was 98% and negative controls (water blanks) were included on each plate. Primers and PCR conditions for genotyping by Pyrosequencing can be obtained upon request.
Statistical analysis
For related individuals, conventional statistical analyses lead to inflated significance. Dependency of the observations within pairs was accounted for by use of the Generalized Estimating Equations (GEE) procedure [14] in which both monozygous (MZ) and dizygous (DZ) twins can be used in tests of association. The approach accounts for dependency of the observations within pairs and yields unbiased standard errors and p-values. Association analyses in the full cohort included both twin subjects from each pair. To limit the number of models tested, we first performed a 2-df overall test of genotypic association. Only in the presence of a significant association were additive, dominant and recessive models (all 1-df) further tested to find the best mode of inheritance. Factor analysis was used to combine strongly correlated indices of obesity into two measures: one for general obesity (serum leptin, BMI, weight, total fat mass and % total fat) and one for central obesity (waist, central fat mass and % central fat). We used two indices of insulin sensitivity, one based on fasting insulin and glucose data (HOMA) and the other based on both fasting and 2-h insulin and glucose data (SiM), as described previously [13]. To reduce the likelihood of generating false positive associations through multiple testing, single variables characterizing obesity were analysed only if initial tests with the general and central obesity scores yielded a positive association for at least one of these combined variables. This strategy was also used for the two indices of insulin sensitivity. Hardy-Weinberg equilibrium was tested by a χ2 test with 1 df in one twin of each pair chosen at random to prevent inflated significance. For all the phenotypes, age and menopausal status were included as covariates in the models. Phenotypes significantly (P<0.05) deviating from normal were log transformed to obtain normal distributions prior to analysis. A P-value of ≤0.05 was considered to be statistically significant.
Acknowledgments
This study was funded by the Wellcome Trust, Project grant No. 073142. The Twin Research and Genetic Epidemiology Unit received support from the Wellcome Trust, Arthritis Research Campaign, the Chronic Disease Research Foundation and the European Union 5th Framework Programme Genom EU twin no. QLG2-CT-2002-01254.
References
- 1.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 2.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 3.Niswender KD, Morton GJ, Stearns WH, Rhodes CJ, Myers MG., Jr Schwartz MW Intracellular signalling. Key enzyme in leptin-induced anorexia. Nature. 2001;413:794–795. doi: 10.1038/35101657. [DOI] [PubMed] [Google Scholar]
- 4.Rui L, Carter-Su C. Identification of SH2-beta as a potent cytoplasmic activator of the tyrosine kinase Janus kinase 2. Proc Natl Acad Sci USA. 1999;96:7172–7177. doi: 10.1073/pnas.96.13.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan C, Li M, Rui L. SH2-B promotes insulin receptor substrate (IRS)1- and IRS2-mediated activation of the phosphatidylinositol 3-kinase pathway in response to leptin. J Biol Chem. 2004;279:43684–43691. doi: 10.1074/jbc.M408495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan C, Yang H, White MF, Rui L. Disruption of the SH2-B gene causes age-dependent insulin resistance and glucose intolerance. Mol Cell Biol. 2004;24:7435–7443. doi: 10.1128/MCB.24.17.7435-7443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren D, Li M, Duan C, Rui L. Identification of SH2-B as a key regulator of leptin sensitivity, energy balance, and body weight in mice. Cell Metab. 2005;2:95–104. doi: 10.1016/j.cmet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–477. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 9.Montpetit A, Nelis M, Laflamme P, Magi R, Ke X, Remm M, Cardon L, Hudson TJ, Metspalu A. An evaluation of the performance of tag SNPs derived from HapMap in a Caucasian population. PLoS Genet. 2006;2:e27. doi: 10.1371/journal.pgen.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reumers J, Schymkowitz J, Ferkinghoff-Borg J, Stricher F, Serrano L, Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spector TD, MacGregor AJ. The St. Thomas’ UK Adult Twin Registry. Twin Res. 2002;5:440–443. doi: 10.1375/136905202320906246. [DOI] [PubMed] [Google Scholar]
- 13.Jamshidi Y, Snieder H, Wang X, Spector TD, Carter ND, O’Dell SD. Common polymorphisms in SOCS3 are not associated with body weight, insulin sensitivity or lipid profile in normal female twins. Diabetologia. 2006;49:306–310. doi: 10.1007/s00125-005-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trégouët D-A, Ducimetère P, Tiret L. Testing association between candidate-gene markers and phenotype in related individuals, by use of estimating equations. Am J Hum Genet. 1997;61:189–199. doi: 10.1086/513895. [DOI] [PMC free article] [PubMed] [Google Scholar]


