Abstract
Background
Variance component (VC) models are commonly used for Quantitative Trait Loci (QTL) mapping in outbred populations. Here, the QTL effect is given as a random effect and a critical part of the model is the relationship between the phenotypic values and the random effect. In the traditional VC model, each individual has a unique QTL effect and the relationship between these random effects is given as a covariance structure (known as the identity-by-descent (IBD) matrix).
Results
We present an alternative notation of the variance component model, where the elements of the random effect are independent base generation allele effects and sampling term effects. The relationship between the phenotypic vales and the random effect is given by an incidence matrix, which results in a novel, but statistically equivalent, version of the traditional VC model. A general algorithm to estimate this incidence matrix is presented. Since the model is given in terms of base generation allele effects and sampling term effects, these effects can be estimated separately using best linear unbiased prediction (BLUP). From simulated data, we showed that biallelic QTL effects could be accurately clustered using the BLUP obtained from our model notation when markers are fully informative, and that the accuracy increased with the size of the QTL effect. We also developed a measure indicating whether a base generation marker homozygote is a QTL heterozygote or not, by comparing the variances of the sampling term BLUP and the base generation allele BLUP. A ratio greater than one gives strong support for a QTL heterozygote.
Conclusion
We developed a simple presentation of the VC QTL model for identification of base generation allele effects in QTL linkage analysis. The base generation allele effects and sampling term effects were separated in our model notation. This clarifies the assumptions of the model and should also enhance the development of genome scan methods.
Background
Understanding the genetic architecture of complex traits controlled by many genes and environmental factors is currently one of the grand challenges in genetics. In this quest for the deciphering of the genetic code, Quantitative Trait Loci (QTL) mapping can be a powerful statistical tool. The basic idea of QTL analysis is to trace the inheritance of alleles from founders through a pedigree by using genetic markers. After estimating this gene flow through the pedigree, the allelic effects are estimated by relating the phenotypes with the different alleles. The position in the genome having the greatest statistical evidence for large allelic effects is the most likely position of a QTL.
In QTL studies of pedigrees in outbred populations, variance component (VC) models are commonly used to estimate the variance of the allelic effects [1], rather than the effect of each individual allele. The studied phenotype is the explanatory variable and the QTL effect is assumed to be a random part of the phenotype. It is random because the founders of the mapping population are assumed to have QTL alleles with effects drawn from a distribution of allelic effects in the entire population and also because the alleles are transmitted from ancestor to descendent by a random process. The model assumes that the random effect is sampled from a multivariate normal distribution with an infinite number of different alleles, and the model is therefore called the infinite alleles model. Simulations have shown that the model is capable of giving unbiased estimates also when the QTL is biallelic [2-5].
To be able to estimate the variance of the random QTL effect, a between-individual covariance structure has to be specified. For a non-inbred population this is equal to the proportion of genes that individuals share identical-by-descent at a specific position [6], and the matrix describing the covariance structure is therefore called the identity-by-descent (IBD) matrix. Since the IBD matrix is not known a priori, it has to be estimated from marker information. This matrix has applications beyond the VC QTL model [7,8], however, and a lot of effort has been put into developing IBD estimation algorithms. For small pedigrees and few markers, the most likely IBD matrix can be estimated [9], but for large pedigrees this is too computationally demanding and approximate algorithms have therefore also been developed [10-12]. Hence, the IBD matrix estimation algorithms have been described in detail, but explicit definitions of the random QTL effect in terms of independent levels are more difficult to find (see however [13,14]).
Several review articles have been published where the assumptions of the model are addressed [2,15-18]. The focus of these papers was on statistical testing of QTL effects and interpretation of genome scans. Several other papers have focused on the biological interpretation of the random QTL effects. Goddard [14] compares the assumptions of the biallelic and the infinite alleles model. He also explains how uncertainty is included in the infinite alleles model by adding a random sampling term to the expected allelic effects and that the variance of the sampling terms are proportional to the QTL variance under the infinite alleles model. Furthermore, Meuwissen and Goddard [19] developed a VC model where they related phenotypes with QTL allele effects by means of an incidence matrix. None of these papers show, however, how the random QTL effect can be given in terms of independent levels. Such a development would more clearly show the definitions of the levels in the random QTL effect and thereby help us to interpret the results from the VC QTL model.
The aim of this paper is to develop a simple presentation of the VC QTL model for identification of base generation allele effects in QTL linkage analysis. Although our main objective is to better understand the biological functions of the random QTL effect, we argue that our model formulation may also enhance the development genome scan methods.
Results
In this section we show how an alternative incidence matrix based VC QTL model is formulated, we give the prerequisites for the model to be equivalent to the IBD matrix based model, and we also present a general algorithm for constructing the incidence matrix. The choice of notation affects the results that can be obtained from the model in terms of best linear unbiased predictions (BLUP) [20]. This is shown with simulations and is also illustrated with an analysis of real data from a wild-domestic chicken cross. The theoretical details are given in the Methods section.
An incidence matrix based VC QTL model
The alternative incidence matrix notation breaks down the VC model to its most basic form where the levels of the random effect are independent. An advantage of this approach is that it is easy to identify the assumptions directly from the model. The present paper is restricted to VC models where there are additive and dominance effects but the models are easily extended to include polygenic, family specific effects, epistasis and genotype by environmental interactions following the models given in [1,21-23]. A VC model may consist of fixed and random effects (a mixed model) but the main parameter of interest is the variance of the random QTL effect, and the fixed effects are therefore ignored without loss of generality in the presentation below.
A restriction on the random QTL effect v can be given either by the covariance structure between individuals, i.e. the IBD matrix Π or alternatively by an incidence matrix Z relating individuals with the QTL alleles in the base generation. In the latter case, the elements in the vector of random effects v* are independent. The trait vector y is multivariate normal and the distribution of the random effects, i.e. the QTL allele effects, is given by Q ~ MVN(0, I) where I is the identity matrix and is the QTL genotypic effect. The genotypic value vi of individual i in the base generation is the sum of the pair of QTL allele effects at a specific position vi = Qk + Qk+1, where the QTL alleles are arbitrarily numbered k = 2i-1 in the base. Hence, by defining the variance of the random QTL genotypic effects as , the variance of the QTL allele effects is . The QTL alleles are all assumed to be independent in the base generation, i.e. Cov(Qi, Qj) = 0 where i and j are different indices for the m base alleles. The VC QTL model assumes that all alleles are different in the base generation so that m equals twice the number of base generation individuals.
The incidence presentation of the VC QTL model is:
y = μ + Zv* + e (1)
Here μ is the overall mean, v* is the vector of m independently normally distributed base generation alleles with v* ~ MVN(0,). The matrix Z is of size n × m, e is the vector of residuals with e ~ MVN(0, I) where is the residual variance. The variance of y is therefore
The traditional IBD matrix version [1] is given by:
y = μ + v + e (2)
where v is the vector of QTL genotype effects (length n) and v ~ MVN(0, Π) and e ~ MVN(0, I). Thus the variance of y is V = Π + I
The two models are equivalent if Π = ZZ' because they then result in the same log-likelihood function L(μ, V|y) = -ln(2π) - ln|V| - (y - μ)' V-1(y - μ) and the equivalence follows directly from the weak likelihood principle (see e.g. page 194 in [24]).
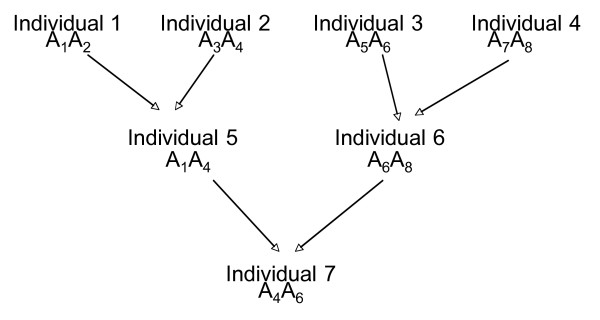
The following numerical example shows how Z is constructed at a marker position when the marker is fully informative (i.e. all marker alleles are unique), and also that Π = ZZ' Consider the example pedigree in Figure 1 with four individuals in the base generation, two in the second generation and one in the third generation, and a fully informative marker.
Figure 1.
An example pedigree with a fully informative marker A. The Z matrix, and corresponding IBD matrix, for this pedigree is given in the text.
The incidence matrix Z is obtained by letting the elements of v* be the random effects of the eight independent QTL alleles in the base generation. Let each row in Z be indexed according to the labels of the individuals in Figure 1 then:
For the pedigree in Figure 1 the corresponding IBD matrix (indexed according to the labels in the figure) is:
Note that the models (1) and (2) are equivalent since ZZ' = Π. In this example it was assumed that the QTL was completely linked to a fully informative marker, but a QTL can also be modelled at not fully informative markers, as well as non-marker positions, by adding additional levels to v* that account for the unknown sampling of QTL alleles. The theoretical details are given in Methods. A direct consequence of the presented theory is that Π will be positive definite at non-marker positions and that the number of non-zero eigenvalues at a fully informative marker is equal to twice the number of base individuals (i.e. when the number of marker allele types is equal to the number of base generation alleles). Hence, a VC estimation algorithm that assumes that Π can be inverted (which is the case in the computer packages DMU [25] and ASReml [26] for instance) cannot be used to identify QTL at marker positions where the marker is highly informative.
Including dominance effects
Model (1) can be extended to include dominance effects by adding an additional variance component d* for the dominance effects
y = μ + Zv* + Zdd* + e
where d* ~ MVN(0, ) and is the variance of the dominance effects. In this model there is a random effect for each allele combination Ai and Aj i.e. each allele combination has an additional dominance effect δij. For the pedigree in Figure 1, the matrix Zd (with rows indexed according to the labels in the figure) is given by:
where the number of columns in Zd and the length of d* is 36 since there are (9·8)/2 different allele combinations, and the ordering of columns is such that the element corresponding to δij is given in column number (i-1)*(8-i/2) + j. Note also that ZdZ'd is equal to the dominance IBD matrix defined by Xu [23].
A general algorithm for estimating Z
The matrix Z estimated with the algorithm outlined here gives 0.5ZZ' equal to the IBD matrix obtained from the single point algorithm developed by Wang et al. [27]. A fully detailed description of the algorithm is given in Methods.
The Z matrix is obtained in two steps. In the first step, the first m columns of the gametic IBD matrix is constructed, where m is twice the number of base generation individuals. Let this matrix be denoted W. Additional columns with the factors for the sampling terms due to uncertainty are added if the marker is not fully informative. In the second step, Z is obtained by adding the rows of W pairwise, so that a row in Z relates an element in y with a pair of QTL alleles from the base generation.
This is a single point algorithm, but the same principle could be used for multi-point estimation if a multi-point estimated gametic IBD is available.
Identification of base generation allelic effects
QTL mapping methods are in general based on strict assumptions regarding the genetic architecture of the base generation individuals for the mapping pedigree. Methods using fixed effect models generally assume bi-allelic QTL and that the founder lines are assumed to be fixed for alternative QTL alleles (e.g. [28]). In traditional variance component methods, all founders are assumed unrelated and contribute two alleles each with effects drawn from an allelic effect distribution [1]. QTL mapping experiments are often designed to be a first step in a process to fully dissect the genetic architecture of the trait. The ultimate aim could e.g. be to understand the molecular mechanisms underlying different allelic effects or to identify appropriate genetic markers for efficient marker assisted breeding. It is therefore desirable that one, already during the statistical QTL analysis, obtain as thorough understanding of the genetic architecture of the base generation individuals as possible by e.g. estimating their QTL allele effects before the genetic dissection process is continued. The primary parameter of estimation in the VC QTL model is the variance of the QTL effect, but it is also possible to estimate the effects of each element in the random effect vector using BLUP [20] (see also [6,29]).
The incidence matrix notation presented here provides new opportunities to estimate QTL allele effects of the base generation individuals and to separate these effects from the sampling term effects. The benefit of using the incidence matrix based parameterization of the VC model is shown in the results below.
Simulation setup
We simulated an F2 pedigree resulting from a cross between two phenotypically divergent outbred strains with a basic structure resembling the Jungle fowl × White leghorn chicken pedigree of Kerje et al. [30]. The base generation consisted of four individuals; one male and three females. In the two following generations there were 30 F1 and 800 F2 individuals. A biallelic QTL was simulated and the phenotypes were simulated as a QTL genotype effect plus a residual effect (i.e. with expected mean equal to 0). Each base generation allele was randomly assigned one of the biallelic types with a 0.5 probability for each type. Only the F2 individuals were assumed to be phenotyped. 100 replicates were simulated with a fully informative marker. The BLUP of the base generation QTL alleles were calculated from the mixed model equations (described in detail in the Methods section) with Z estimated using the algorithm outlined above. Variance component estimation was done using Fisher's scoring algorithm, and the simulation and estimation algorithms (see Appendix) were programmed in R [31].
Two different simulation experiments were performed to evaluate the possibility of identifying biallelic effects from the BLUP obtained from model (1) and to evaluate the possibility of identifying if a marker homozygote base individual is QTL heterozygous.
The parameters used in the first simulation experiment are given in Table 1. Three different additive biallelic effects were studied and one case with dominance was also included. For each replicate, the BLUP were partitioned into two clusters by minimizing the variance within clusters using the pam function in the cluster package of R [31,32]. The correspondence between the clustering and the generated biallelic QTL types in the base generation was subsequently analyzed by summing the number of alleles that had been incorrectly clustered.
Table 1.
Setup for the simulations with a fully informative marker. 800 F2 individuals were simulated with phenotype equal to the sum of the additive QTL effect, dominance QTL effect and residual effect
| Percentage of the phenotypic variance explained by the QTLa | Additive effect (a)b | Dominance effect (d) | Residual variance |
| 5% additive QTL | 3.162 | 0 | 95 |
| 10% additive QTL | 4.472 | 0 | 90 |
| 20% additive QTL | 6.324 | 0 | 80 |
| 20% add. QTL and 10% dominance | 6.324 | 3.162 | 70 |
a Expected additive QTL variance equal to 0.5a2
b The two alternative homozygous genotypes had additive effect a and -a in the simulated data
In the second set of simulation experiments a 5%, 10% and 20% additive QTL was simulated in the same way as described in Table 1, but Z was constructed with the base generation male being marker homozygous.
Simulation results
Our method based on the infinite alleles model is powerful in clustering biallelic effects even at moderate effect sizes and the power increases when the proportion of the total variance explained by the QTL increases (Table 2). In the case where dominance was included, only 13% of the dominance BLUP were correctly clustered. The clustering of these effects, however, can be deduced from the clustering of the main effects, because in the biallelic model the dominance effect is added only to the genotypes of the heterozygotes. Hence, it is possible to cluster the biallelic dominance effect accurately as well by identifying the QTL heterozygotes from the estimates of the main QTL effect.
Table 2.
Simulation results for clustering of biallelic effects using our presentation (eq. (1)) of the infinite alleles model when markers are fully informative. 100 replicates were simulated for each QTL effect
| Percentage of the phenotypic variance explained by the QTL | Proportion correctly clustered allele effects | Cluster differencea | Estimated allelic QTL varianceb () | Estimated dominance variance () | Estimated residual variance |
| 5% additive QTL | 0.79 | 2.63 (0.81) | 2.65 (1.16) | - | 95.11 (4.85) |
| 10% additive QTL | 0.93 | 3.93 (0.73) | 4.99 (1.58) | - | 89.85 (4.97) |
| 20% additive QTL | 0.97 | 5.91 (0.65) | 9.89 (2.44) | - | 78.98 (3.87) |
| 20% add. and 10% dom | 0.99 | 5.39 (1.85) | 9.61 (4.46) | 10.98 (3.1) | 70.43 (2.56) |
a Average differences between cluster means. Standard deviations within parentheses
b The allelic variance is estimated from model (1). The expected mean is half the proportion of the total variance explained by the QTL.
In the second set of simulations, the male in the base generation was marker homozygous. Depending on the genetic constitution of this male there will be a bias in the estimated QTL variance. If the male is a QTL heterozygote the QTL variance will be over estimated, and if it is a homozygote it will be underestimated (Table 3). The estimate is, however, consistent if there is a 50% chance of this male being a QTL heterozygote. The reason for the QTL variance being underestimated when the male is homozygous is that the true sampling terms are all zero and their variance will not be proportional to the QTL variance [14]. Since the estimated sampling terms for a QTL homozygote will be close to zero, and those for a heterozygote will differ from zero, it is possible to distinguish between base generation homozygotes and heterozygotes.
Table 3.
Variance component estimatesa from 100 replicates with a marker homozygous individual in the base generation. The marker homozygote had a 50% chance of being QTL heterozygous.
| Allelic QTL varianceb () | Residual variance () | |||||
| All | Homozygotes | Heterozygotes | All | Homozygotes | Heterozygotes | |
| 5% additive QTL | 2.90 (1.77) | 2.16 (1.03) | 3.76 (2.07) | 94.42 (4.80) | 94.17 (5.21) | 94.72 (4.30) |
| 10% additive QTL | 5.26 (2.81) | 3.63 (1.27) | 7.71 (2.71) | 89.59 (4.90) | 89.68 (4.48) | 89.45 (5.53) |
| 20% additive QTL | 10.27 (6.38) | 5.56 (1.30) | 17.32 (3.98) | 79.67 (4.19) | 79.59 (4.42) | 79.80 (3.85) |
The variance estimates are given as the mean of estimates from all simulations (All), and also as the mean of the estimates divided into two cases: marker homozygote simulated as QTL homozygous (Homozygotes) and marker homozygote simulated as QTL heterozygous (Heterozygotes)
a Standard deviations within parentheses.
b The allelic variance is estimated from model (1). The expected mean is half the proportion of the total variance explained by the QTL
In the simulated pedigree, there were four F0 and 40 F1 individuals and consequently there are 8 levels in the vector of random effects (v*) corresponding to the base alleles and 40 levels due to sampling terms. The ratio between the variance of the 40 sampling term BLUP and the variance of the 8 base allele BLUP is expected to be close to zero if the base male is a homozygote and it is expected to be greater than one if it is a heterozygote. This difference was clearly shown in the simulations (Table 4) and increased with the size of the QTL. A threshold of 0.5 for the variance ratio was empirically chosen to cluster the simulations into two groups so that all simulations below this threshold were classified into Group A (QTL homozygotes) and all simulations above the threshold were classified into Group B (QTL heterozygotes). Using this classification, all homozygotes and heterozygotes were correctly grouped into Group A and B, respectively, when a 20% QTL was simulated (Table 4). For the 10% QTL case, all homozygotes were correctly grouped and over 75% of the heterozygotes were correctly grouped. When a 5% QTL was simulated over 95% of the homozygotes were correctly grouped and nearly 50% of the heterozygotes were correctly grouped. Hence, this method gives good indication of whether the marker homozygote is QTL heterozygous or not and does not require knowledge of the true QTL variance. In the general case, there is strong support for a heterozygote if the QTL variance is high and the estimated variance ratio between allele effects and sampling terms is greater than one.
Table 4.
Quantiles for the variance ratios between sampling term BLUP and base allele BLUP split into the two cases where the marker homozygote was either simulated as QTL homozygous or QTL heterozygous
| Min. | 5% | 25% | 50% | 75% | 95% | Max. | ||
| 5% additive QTL | Homozygotes | 0.05 | 0.07 | 0.13 | 0.17 | 0.25 | 0.39 | 0.67 |
| Heterozygotes | 0.18 | 0.21 | 0.34 | 0.54 | 0.92 | 2.09 | 2.58 | |
| 10% additive QTL | Homozygotes | 0.08 | 0.10 | 0.12 | 0.15 | 0.21 | 0.37 | 0.40 |
| Heterozygotes | 0.26 | 0.30 | 0.81 | 1.19 | 1.83 | 2.74 | 7.06 | |
| 20% additive QTL | Homozygotes | 0.08 | 0.09 | 0.12 | 0.15 | 0.19 | 0.34 | 0.41 |
| Heterozygotes | 0.63 | 0.94 | 1.44 | 1.70 | 2.29 | 3.93 | 9.73 |
Analysis of a QTL in a wild – domestic chicken cross
In a previous standard QTL analysis [28], a QTL on Chicken chromosome 1 [30] was shown to explain 11% of the variance for body weight at 200 days of age, and subsequent analysis of the segregation patterns of the QTL within individual F1 sires indicated segregation of the QTL within the founder breeds. This QTL was close (12 cM) to a completely informative marker (LEI246). We calculated the BLUP for the base generation allele effects and subsequently clustered these into two groups assuming a bi-allelic QTL. The results showed that all Jungle fowl have a common allele and all Leghorn hens have common alleles except for one hen, which has one red Jungle fowl allele. Hence, we could identify which base individual that had an allele not fixed within lines. The difference between cluster means was 49.3 g. The estimated fixed effects and variance components were: mean = 1080.3 g, sex effect = 409.2 g, QTL genotype variance = 3251.8, and residual variance = 26028.9.
Discussion
We have developed a presentation of the VC QTL model where the random effect vector is given in terms of independent levels. Three main points can be concluded from the analytical description of the model and the results presented in the paper: more information can be extracted from QTL analysis by using our alternative model presentation, an improved understanding of VC estimation can be achieved, and the presentation of the model in terms of base allele effects should help to develop new models specifically designed for different structures of dependencies between base generation alleles.
Estimation of BLUP for base generation allelic effects and sampling term effects
Using our presentation of the infinite alleles model, we could accurately cluster a bi-allelic QTL when markers are fully informative. Furthermore, we were also able develop a measure of whether a marker homozygote is a QTL heterozygote or not, which can be used in both outbred populations and experimental crosses. To our knowledge these developments are novel, but the idea that the sampling variance should be different between the QTL heterozygote and homozygote individuals was mentioned already by Goddard [[14]; p.121]: "If the inheritance of an allele cannot be followed with certainty from parent to offspring [in the infinite alleles model], then a new allele is assumed with effect pg1 + (1-p)g2 + e where g1 and g2 are the effects of the parental alleles, p is the probability that allele 1 was inherited and e is a random effect with mean zero and variance equal to p(1-p) v(g). If there are only two alleles at this gene then the correct assumption would be that the offspring inherits g1 with probability p and g2 with probability 1-p. On average this implies the same segregation variance as the infinite alleles model but in a particular case, if g1 and g2 are similar, then the segregation variance is smaller in the two allele model than in the infinite allele model." Hence, the VC model is capable of giving unbiased estimates for a biallelic QTL [2-5] but this unbiased property of the estimator assumes that a marker homozygote in the base is equally probable of being QTL homozygous or heterozygous. More importantly, the estimate obtained from a single QTL analysis will be severely under- or overestimated depending on whether the marker homozygote is QTL homozygous or heterozygous, respectively.
The infinite alleles model has acquired its name not only because the base generation alleles are assumed to be drawn from an infinite number of different alleles, but also because the sampling terms have previously been included in the allelic effects (as explained in the citation above) and each new sampling term has been thought of as producing a new allelic effect. To us, this production of new QTL alleles through the pedigree due to uncertainty is contra-intuitive and our model notation separates the QTL allele effects from the sampling terms.
The clustering analysis of additive biallelic QTL effects at a fully informative marker presented in the results could have been performed based on the neat transformation of individual genotypic BLUP to individual allele BLUP developed by Nagamine et al. [33,34]. However, these allelic BLUP are calculated for each individual in the pedigree and does not give separate BLUP for the sampling terms, because each allelic effect in this model is a mixture of the base generation allele effects and the sampling terms. Our notation was also easy to extend so that dominance effects could be incorporated, which Nagamine et al. [33] did not attempt in their calculations of allelic BLUP. Furthermore, the transformation given by Nagamine et al. assumes that the IBD matrix is positive definite, which is not the case in marker positions.
Implications for VC estimation
Our decomposition of the IBD matrix into a low rank incidence matrix shows that IBD matrices will be singular in marker positions and the rank of the IBD matrix will depend on the size of the base generation and the informativeness of the marker. Furthermore, for positions close to markers the IBD matrix will be close to singular. The current VC estimation programmes that are used in human genetics (e.g. SOLAR [35] and MERLIN [36]) have not been developed to deal with large family pedigrees, because so far only small or moderately sized pedigrees have been used, but the need of obtaining fast algorithms for large family pedigrees will be a major task within the near future [37]. VC estimation programmes developed for animal breeding applications [25,26], however, have primarily been developed for estimating the variance of polygenic effects in large pedigrees. The covariance matrix of polygenic effects (i.e. the additive relationship matrix) is positive definite and the VC estimation algorithms that have been developed to estimate polygenic variances have therefore been optimized for situations with non-singular covariance structures, and are unable to handle those cases in VC QTL analysis where the covariance structure is singular (e.g. at marker locations). A way to get around this problem is to add a small positive value to the diagonal of the IBD matrix, which may lead to an extremely ill conditioned optimization problem since the IBD matrix will then be close to singular (i.e. with many eigenvalues close to zero). An alternative is to invert the variance of the response vector directly [38] but neither of these strategies take advantage of the fact that the IBD matrix is lower ranked or close to lower ranked when markers are dense (which was shown in the Methods section).
Modeling of dependencies between allelic effects in the base generation
We describe the VC QTL model in terms of independent random effects (base generation allele effects and sampling terms) instead of correlated random effects (as in the IBD version of the model). As a consequence it is easier to model correlation structures between base generation alleles, which has been an important issue in linkage-disequilibrium linkage mapping over the past decade [39] where the correlation structure is estimated from marker haplotypes. This method incorporates linkage disequilibrium in the base generation and gives greater power to position a QTL in fine mapping. It can also account for the possibility that marker homozygotes are more likely to be QTL homozygotes. Recently, we have shown [40] that VC modeling can be a powerful tool to identify within line variation in divergent intercrosses by including a correlation structure between the base generation allele effects that is estimated directly in the VC model. An important future development would be to combine these two models so that both the linkage disequilibrium information and the within line variation can be modeled simultaneously. This development, and other possible developments of the VC models where base generation correlation structures are included, should be enhanced by the model presentation given in this paper.
Conclusion
By defining VC QTL model in terms of independent levels of the random QTL effect, we have developed a simple presentation of the VC model for identification of base generation allele effects in QTL linkage analysis. This development clarifies the definition of the random QTL effect and will be helpful in applications where the aim is to trace allelic QTL effects through a pedigree. Our clarifications of the VC QTL model should also enhance the development of genome scan methods.
Methods
Adding uncertainty to the incidence matrix based VC QTL model
A QTL can be modelled at not fully informative markers, as well as non-marker positions, by adding additional levels to v* that account for the sampling of QTL alleles. These sampling terms can be shown to be independent of the QTL allele effects in the base [6,13,14], which motivates the inclusion of these as additional levels in v*. If, for instance, a QTL allele cannot be related to a base generation allele with absolute certainty but it is known that it is one of two alleles with equal probability, then the effect (Qx) of this unknown QTL allele is modelled as:
Qx = 0.5Q1 + 0.5Q2 + ·ε (3)
where Q1 and Q2 are the effects of the two alternative QTL alleles and ε is the random sampling term. The factor ensures that the variance of the sampling term ε is equal to the variance of random effects in v*.
In the following numerical example the QTL is linked to a marker that is not fully informative. Here, it is not possible to distinguish between the QTL alleles with absolute certainty since the marker is not fully informative. For example, if the marker alleles of individual 1 in Figure 1 were identical, it would not be possible to know which of the two that had been inherited by individual 5. The probability that individual 5 inherited the first of the two alleles is 0.5 and the effect Qx of the unknown allele is either Q1 or Q2. This uncertainty is incorporated into the VC model in two steps. In this example, the first step is to change the first and second elements on the fifth row in Z to 0.5. This would then be interpreted as Qx = 0.5Q1 + 0.5Q2, but since Qx is either Q1 or Q2 a sampling term must be added. This is achieved by applying equation (3): Qx = 0.5Q1 + 0.5Q2 + ·ε. The second step is therefore to extend v* to include a ninth element ε and add a ninth column to Z with element in the fifth row.
Thus, the sampling term is treated as an additional level in the normally distributed random effect.
Similarly v* and Z can also be extended to include further uncertainty in the inheritance of the QTL alleles caused by non-informative markers and QTL-scanning at non-marker positions. The formulation of Z above gives a VC model (1) that is equivalent to the IBD matrix based VC model (2) since Π = ZZ'.
Two important facts can be noted from our presentation. First of all, the rank of an IBD matrix at a marker position will depend on the informativeness of the marker and the size of the base generation. Secondly, if markers are dense then the IBD matrices at non-marker positions will be positive definite but many of the eigenvalues will be close to zero.
A general single-point algorithm of estimating Z
Here we describe a general algorithm for obtaining the incidence matrix in (1). The matrix Z estimated with this algorithm gives 0.5ZZ' equal to the IBD-matrix Π obtained from the single point algorithm developed by Wang et al [27].
The incidence matrix Z is obtained by first constructing a matrix W describing the haplotype specific inheritance of QTL alleles from the base generation. There are f base generation individuals and a total of n individuals in the pedigree.
W is a matrix of size 2n × 2n. An element l (for l = 1,...,2f) in row 2i-1 is the probability that the paternally inherited QTL allele is identical to the l:th allele in the base generation, and an element l (l = 1,...,2f) in row 2i is the corresponding probability for the maternally inherited QTL allele. Thus the first 2f columns in W are the same as the first 2f columns of the gametic IBD matrix (as defined in [27])
For simplicity of notation, let a row k of W be denoted Wk and an element in row k and column l be denoted Wk,l. The individuals are numbered i = 1,2,...,n (with ancestors preceding descendents) and the following subscripts are used:
h = 1 for paternal allele and 2 for maternal allele
jip = id-number of the father of individual i
jim = id-number of the mother of individual i
Each row 2f+1 to 2n is constructed in two steps. The first step is based on the concept of probability of descent of a gamete (PDQij) as defined in [27]. The PDQ matrix of size 2 × 4 gives the probabilities for the two alleles (row i) being inherited from the four possible parental alleles (column j).
Step 1: Construction of elements 1 to 2i-2 in row 2(i-1)+h
Step 2: Construction of the 2 × 2 block diagonal elements
W2(i-1)+h,2(i-1)+h = Eih
The matrix W will then contain columns with only zeros that correspond to the cases where transmission of alleles from parent to offspring is known without uncertainty. When the matrix W is completed the incidence matrix is obtained by summing up the rows in W pairwise for each individual:
Z = KW
where K = (1,1)⊗I, I is the identity matrix of size n × n and ⊗ is the Kronecker product.
Mixed model equations for the IBD and Z matrix based VC QTL models
The mixed model equations to solve in the incidence matrix notation (1) is [20]:
with G = I,
which gives BLUP for v*, i.e. the QTL allele effects of the base generation individuals and the additional sampling term effects.
The mixed model equations to solve in the IBD-matrix notation (2) is:
with G = Π,
which gives BLUP for v, i.e. the QTL genotype effects for all phenotyped individuals. Here X is the design matrix for the fixed effects.
Authors' contributions
OC initiated the study of VC QTL models. LR drafted the manuscript and initiated the idea of the incidence matrix notation. The paper was then developed through continuous discussions between LR and OC. All authors have read and approved the final version of this paper.
Appendix
Fisher scoring algorithm for REML estimates in R code
REML<-function(y,X,n_comp,conv_crit,n_maxiter,lambda_start,delta, Z1, Z2=0, print_results=FALSE, step=1) {
######################################################################################
## Responsible programmer: Lars Rönnegård (lars.ronnegard@lcb.uu.se)
## Date 20060925
#This is the Fisher Scoring algorithm for REML
#See e.g. Johnson & Thompson 1995 J. of Dairy Science 78:449–456.
#The function returns the estimated variance components and fixed effects, together with the log-likelihood
#and convergence information.
#INPUT PARAMTERS (Table 5)
Table 5.
| #y | Response vector |
| #X | Design matrix for fixed effects |
| #n_comp | Number of different random effects in the model (max.=2 in this version) |
| #conv_crit | Value that the change in loglikelihood should be less than |
| #n_maxiter | Maximum number of iterations |
| #lambda_start | Initial ratio of variance components |
| #delta | Minimum possible value of the variance component |
#WORKING VARIABLES AND OUTPUT PARAMETERS (Table 6)
Table 6.
| #A | A matrix which stores ZZ' for all variance components |
| #phi_start | Staring values for the variance components |
| #M_phi | Matrix with VC estimates at each iteration |
| #phi | VC estimate from the latest iteration |
| #DL | Gradient of the restricted log-likelihood |
| #FS | Fisher's Information matrix |
| #V | Variance matrix of y |
| #P | The projection matrix |
| #llh | Restricted log-likelihood |
| #beta_hat | Estimates of fixed effects |
| #conv_test | Binary variable equal to 1 if the algorithm converges within n_maxiter iterations |
######################################################################################
print("REML iteration number")
min.error<-10^-8
n_comp1<-n_comp+1
n_rows<-length(y)
A<-matrix(0,(n_comp1*n_rows),n_rows)
Aj<-matrix(0,n_rows,n_rows)
for (i_comp in 1:n_comp) {
if (i_comp==1) Aj<-Z1%*%t(Z1)
if (i_comp==2) Aj<-Z2%*%t(Z2)
A[((i_comp-1)*n_rows+1):(n_rows*i_comp),1:n_rows]<-Aj
}
A[(n_comp*n_rows+1):(n_comp1*n_rows),1:n_rows]<-diag(rep(1,n_rows))
res_var<-var(y-X%*%solve(t(X)%*%X)%*%t(X)%*%y)
phi_start<-numeric(n_comp1)
for (i in 1:(n_comp1-1)) {
phi_start[i]<-lambda_start*res_var
}
phi_start[n_comp1]<-res_var
dimIBD<-min(dim(A))
M_phi<-matrix(0,(n_maxiter+1),n_comp1)
M_phi[1,1:n_comp1]<-phi_start[1:n_comp1]
phi<-numeric(n_comp1)
DL<-numeric(n_comp1)
DL[1:n_comp1]<-conv_crit+1
FS<-matrix(0,n_comp1,n_comp1)
Aj<-matrix(0,dimIBD,dimIBD)
Ak<-matrix(0,dimIBD,dimIBD)
llh.prev<-1+conv_crit
llh<-0
for (i in 1:n_maxiter){
V<-matrix(0,dimIBD,dimIBD)
if (abs(llh-llh.prev)>conv_crit){
phi<-M_phi[i,]
for (j in 1:n_comp1){
Aj<-A[((j-1)*dimIBD+1):(j*dimIBD),1:dimIBD]
V<-V+phi[j]*Aj
}
invV<-solve(V)
temp<-solve(t(X)%*%invV%*%X)
P<-invV-invV%*%X%*%(temp)%*%t(X)%*%invV
for (j in 1:n_comp1){
Aj<-A[((j-1)*dimIBD+1):(j*dimIBD),1:dimIBD]
DL[j]<-sum(diag(Aj%*%P))-t(y)%*%P%*%Aj%*%P%*%y
for (k in j:n_comp1) {
Ak<-A[((k-1)*dimIBD+1):(k*dimIBD),1:dimIBD]
FS[j,k]<-sum(diag(Aj%*%P%*%Ak%*%P))
FS[k,j]<-FS[j,k]
}
}
FS.egen<-eigen(FS, only.values=TRUE)
FS.min<-min(FS.egen$values)
if (FS.min>min.error) M_phi[(i+1),]<-phi-step*solve(FS)%*%DL
if (FS.min<=min.error) {
print("Negative Hessian")
identitet<-diag(rep((0.5+abs(FS.min)),min(dim(FS))))
M_phi[(i+1),]<-phi-step*solve(FS+identitet)%*%DL
}
egen<-eigen(V, only.values=TRUE)
if (min(egen$values>0))ldV<-sum(log(egen$values))
egen2<-eigen(t(X)%*%invV%*%X, only.values=TRUE)
ldXVX<-sum(log(egen2$values))
llh.prev<-llh
if (min(egen$values)>0) llh<-(ldV+ldXVX+t(y)%*%P%*%y)*(-0.5)
if (min(egen$values)<=0) llh<-llh.prev-1
#Truncation at zero
M_phi[(i+1),]<-0.5*(M_phi[(i+1),]+delta+abs(M_phi[(i+1),]-delta))
conv_val<-abs(llh-llh.prev)
if (print_results==TRUE) {
print("--------------------------------")
print("Iteration:")
print(i)
print("Convergence criteria: Change in log-likelihood")
print(conv_val)
print("log-likelihood")
print(llh)
print("REML estimates of variance components")
print("Genotype variance [1] and residual variance [2]")
print(M_phi[(i+1),])
}
if (print_results==FALSE) print(paste(" ",i))
}
}
conv_test<-1
if (abs(llh-llh.prev)>conv_crit) conv_test<-0
beta_hat<-numeric(min(dim(X)))
beta_hat<-solve(t(X)%*%invV%*%X)%*%t(X)%*%invV%*%y
if (print_results==TRUE) {
print("Estimates of fixed effects")
print(beta_hat)
}
list(beta_hat=beta_hat,conv_test=conv_test,conv_val=conv_val,phi=phi,phi_iteration=M_phi,llh=llh)
}
Algorithm for construction of an F2 pedigree with a fully informative marker in R code
simple_F2<-function(n0.males,n0.females,n1.males,n1.females,n2.males,n2.females,QTLvar,RESvar) {
######################################################################################
## Responsible programmer: Lars Rönnegård (lars.ronnegard@lcb.uu.se)
## Date 20060925
## This function constructs an F2 pedigree for a fully informative marker.
## The function returns the simulated phenotypes (y) and the incidence matrix (Z)
## and also the true simulated QTL alleles of the base individuals.
## The phenotypes are simulated assuming a biallelic QTL at the marker position
#INPUT PARAMTERS (Table 9)
Table 7.
| #n0.males | No. of males in base generation |
| #n0.females | No. of females in base generation |
| #n1.males | No. of males in F1 generation |
| #n1.females | No. of females in F1 generation |
| #n2.males | No. of males in F1 generation |
| #n2.females | No. of females in F1 generation |
| #QTLvar | Genotypic QTL variance |
| #RESvar | Residual variance |
#WORKING VARIABLES AND OUTPUT PARAMETERS (Table 10)
Table 8.
| #n0 | Total no. of base generation individuals |
| #n1 | Total no. of F1 individuals |
| #n2 | Total no. of F2 individuals |
| #n | Total no. of individuals in pedigree |
| #index.mat | Stores indeces of alleles in the F1 generation |
| #Z | Incidence matrix for the random base allele effects |
| #v | Random base allele effects |
| #y | Vector of phenotypes |
| #e | Residual effects |
| #base.alleles | Simulated alleles of the base individuals |
######################################################################################
n0=n0.males+n0.females
n1=n1.males+n1.females
n2=n2.males+n2.females
n=n0+n1+n2
Z<-matrix(0,n2,(2*n0))
index.mat<-matrix(0,n1,2)
for (i in 1:n1) {
sire<-i-n0.males*floor(i/n0.males)+1
sire.allele<-sample(1:2,1)
dam<-i-n0.females*floor(i/n0.females)+1
dam=dam+n0.males
dam.allele<-sample(1:2,1)
index.mat[i,]<-c(((sire-1)*2+sire.allele),((dam-1)*2+dam.allele))
}
for (i in 1:n2) {
sire<-sample(1:n1.males,1)
sire.allele<-sample(1:2,1)
dam<-sample(1:n1.females,1)
dam=dam+n1.males
dam.allele<-sample(1:2,1)
Z[i,index.mat[sire,sire.allele]]=Z[i,index.mat[sire,sire.allele]]+1
Z[i,index.mat[dam,dam.allele]]=Z[i,index.mat[dam,dam.allele]]+1
}
base.alleles <-sample(0:1,(2*n0),replace=TRUE)
v<-base.alleles *sqrt(2*QTLvar)
e<-rnorm(n2,0,sqrt(RESvar))
y<-Z%*%v+e
list(y=y,Z=Z,base.alleles= base.alleles)
}
Acknowledgments
Acknowledgements
The study was funded by the Swedish Research Council FORMAS. We thank Ricardo Pong-Wong at Roslin Institute, UK, for many fruitful discussions on the contents of this paper.
Contributor Information
Lars Rönnegård, Email: lars.ronnegard@lcb.uu.se.
Örjan Carlborg, Email: orjan.carlborg@lcb.uu.se.
References
- Goldgar DE. Multipoint analysis of human quantitative genetic variation. American Journal of Human Genetics. 1990;47:957–967. [PMC free article] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Variance component methods for detecting complex trait loci. Advances in Genetics. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- Blangero J, Almasy L. Multipoint oligogenic linkage analysis of quantitative traits. Genetic Epidemiology. 1997;14:959–964. doi: 10.1002/(SICI)1098-2272(1997)14:6<959::AID-GEPI66>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grignola FE, Hoeschele I, Zhang Q, Tier B. Mapping quantitative trait loci via residual maximum likelihood: II. A simulation study. Genet Sel Evol. 1996;28:491–505. [Google Scholar]
- Xu S, Atchley WR. A random model approach to interval mapping of quantitative trait loci. Genetics. 1995;141:1189–1197. doi: 10.1093/genetics/141.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RL, Grossman M. Marker-assisted selection using best linear unbiased prediction. Genet Sel Evol. 1989;21:467–477. [Google Scholar]
- Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972;2:3–9. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sunderland, UK, Sinauer; 1998. [Google Scholar]
- Lander ES, Green P. Construction of multilocus maps in humans. Proc Natl Acad Sci. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath S. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. American Journal of Human Genetics. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pong-Wong R, George AW, Woolliams JA, Haley CS. A simple and rapid method for calculating identity-by-descent matrices using multiple markers. Genet Sel Evol. 2001;33:453–471. doi: 10.1051/gse:2001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AC, Pong-Wong R, Winding JJ, Woolliams JA. Precision of methods for calculating identity-by-descent matrices using multiple markers. Genet Sel Evol. 2002;34:557–579. doi: 10.1051/gse:2002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard ME. A mixed model for analyses of data on multiple genetic markers. Theoretical and Applied Genetics. 1992;83:878–886. doi: 10.1007/BF00226711. [DOI] [PubMed] [Google Scholar]
- Goddard ME. The validity of genetic models underlying quantitative traits. Livestock Production Science. 2001;72:117–127. doi: 10.1016/S0301-6226(01)00272-X. [DOI] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Quantitative trait locus mapping using human pedigrees. Human Biology. 2000;72:35–62. [PubMed] [Google Scholar]
- Feingold E. Methods for Linkage Analysis of Quantitative Trait Loci in Humans. Theoretical Population Biology. 2001;60:167–180. doi: 10.1006/tpbi.2001.1545. [DOI] [PubMed] [Google Scholar]
- Cherny SS, Sham PC, Cardon LR. Introduction to the special issue on variance components methods for mapping quantitative trait loci. Behavior Genetics. 2004;34:125–126. doi: 10.1023/B:BEGE.0000013938.54772.42. [DOI] [Google Scholar]
- Majumder PP, Ghosh S. Mapping quantitative trait loci in humans: achievements and limitations. Journal of Clinical Investigation. 2005;115:1419–1424. doi: 10.1172/JCI24757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Goddard ME. The use of marker haplotypes in animal breeding schemes. Gen Sel Evol. 1996;28:161–176. [Google Scholar]
- Henderson CR. Best linear unbiased estimation and prediction under a selection model. Biometrics. 1975;31:423–447. doi: 10.2307/2529430. [DOI] [PubMed] [Google Scholar]
- Stern MP, Duggirala R, Mitchell BD, Reinhart LJ, Sivakumar S, Shipman PA, Uresandi OC, Benavides E, Blangero J, O'Connell P. Evidence for linkage of regions on chromosome 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Research. 1996;6:724–734. doi: 10.1101/gr.6.8.724. [DOI] [PubMed] [Google Scholar]
- Blangero J. Statistical genetic approaches to human adaptability. Human Biology. 1993;65:941–966. [PubMed] [Google Scholar]
- Xu S. Computation of the full likelihood function for estimating variance at a quantitative trait locus. Genetics. 1996;144:1951–1960. doi: 10.1093/genetics/144.4.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan Y. In all likelihood: statistical modelling and inference using likelihood. Oxford, UK, Oxford University Press; 2001. [Google Scholar]
- Madsen P, Jensen J. A user's guide to DMU. Danish Institute of Agricultural Science, Research Centre Foulum, Denmark; 2000. [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R. ASReml user guide release 10. Hemel Hempstead, UK, VSN International Ltd; 2002. [Google Scholar]
- Wang T, Fernando RL, van der Beek S, Grossman M, van Arendonk JAM. Covariance between relatives for a marked quantitative trait locus. Genet Sel Evol. 1995;27:251–274. [Google Scholar]
- Haley CS, Knott SA, Elsen JM. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics. 1994;136:1195–1207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GK. That BLUP is a good thing: the estimation of random effects. Statistical Science. 1991;6:15–51. [Google Scholar]
- Kerje S, Carlborg Ö, Jacobsson L, Schütz K, Hartmann C, Jensen P, Andersson L. The twofold difference in adult size between the red junglefowl and White Leghorn chickens is largely explained by a limited number of QTLs. Animal Genetics. 2003;34:264–274. doi: 10.1046/j.1365-2052.2003.01000.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. Vienna, Austria, R Foundation for Statistical Computing; 2004. http://www.R-project.org [Google Scholar]
- Struyf A, Hubert M, Rousseeuw PJ. Integrating Robust Clustering Techniques in S-PLUS. Computational Statistics and Data Analysis. 1997;26:17–37. doi: 10.1016/S0167-9473(97)00020-0. [DOI] [Google Scholar]
- Nagamine Y, Knott SA, Visscher PM, Haley CS. Simple deterministic identity-by-descent coefficients and estimation of QTL allelic effects in full and half sibs. 2002;80:237–243. doi: 10.1017/s0016672302005918. [DOI] [PubMed] [Google Scholar]
- Nagamine Y. Transformation of QTL genotypic effects to allelic effects. Genet Sel Evol. 2005;37:579–584. doi: 10.1051/gse:2005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. American Journal of Human Genetics. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGene. Genetic Epidemiology. 1988;5:471–473. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- Chen W-M, Abecasis GR. Estimating the power of variance component linkage analysis in large pedigrees. Genetic Epidemiology. 2006;30:471–484. doi: 10.1002/gepi.20160. [DOI] [PubMed] [Google Scholar]
- Lee SH, van der Werf JHJ. An efficient variance component approach implementing an average information REML suitable for combined LD and linkage mapping with a general complex pedigree. Genet Sel Evol. 2006;38:25–43. doi: 10.1051/gse:2005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Goddard ME. Fine mapping of quantitative trait loci using linkage disequilibria with closely linked marker loci. Genetics. 2000;155:421–430. doi: 10.1093/genetics/155.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L, Carlborg Ö. A new efficient method for QTL mapping in divergent intercrosses incorporating within line variation. Paper no. 20-07 at the 8 th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brazil; 2006. [Google Scholar]