Abstract
Transcriptional cis-regulatory control regions frequently are found within non-coding DNA segments conserved across multi-species gene orthologs. Adopting a systematic gene-centric pipeline approach, we report here the development of a web-accessible database resource—GenomeTraFac (http://genometrafac.cchmc.org)—that allows genome-wide detection and characterization of compositionally similar cis-clusters that occur in gene orthologs between any two genomes for both microRNA genes as well as conventional RNA-encoding genes. Each ortholog gene pair can be scanned to visualize overall conserved sequence regions, and within these, the relative density of conserved cis-element motif clusters form graph peak structures. The results of these analyses can be mined en masse to identify most frequently represented cis-motifs in a list of genes. The system also provides a method for rapid evaluation and visualization of gene model-consistency between orthologs, and facilitates consideration of the potential impact of sequence variation in conserved non-coding regions to impact complex cis-element structures. Using the mouse and human genomes via the NCBI Reference Sequence database and the Sanger Institute miRBase, the system demonstrated the ability to identify validated transcription factor targets within promoter and distal genomic regulatory regions of both conventional and microRNA genes.
INTRODUCTION
Comparing evolutionarily conserved non-coding genomic sequences of divergent yet evolutionarily related species has proven to be an effective method for deciphering gene regulatory information because functional sequences tend to evolve at a slower rate than non-functional sequences (1–3). Prior efforts [see Ureta-Vidal et al. (1) and recent survey by GuhaThakurta (4)] at genome-wide human–mouse comparison have been successful in identifying potential gene regulatory regions (5), but a global human miRNA-centric approach with mineable pre-computed cis-regulatory region information for all known human–mouse ortholog miRNAs, to our knowledge, has not been accomplished.
Although microRNAs are implicated in both tissue differentiation and maintenance of tissue identity, the mechanisms underlying their regulation are not known fully. The precise mechanisms of miRNA-mediated gene regulation, the tissue-specific expression of specific miRNAs and the cascade of molecular events that lead to the biogenesis of miRNAs are beginning to emerge. Most miRNA genes are located far away from any annotated genes, implying independent transcription from their own promoters (6). For instance, the promoters of the miRNAs miR-1, miR-133, miR-124, miR-223 and miR-17 are reported to be regulated by specific transcription factors (TFs) (7–11). In this context a database of readily query-able promoter regions of miRNAs for conserved putative binding sites will accelerate our understanding of the microRNA functions and decipher the mini-circuitries comprised of miRNAs and TFs.
GenomeTrafac is built by serializing a version of TraFaC (12). At present, we have curated and processed over 260 miRNA genes obtained from miRBase (13) that have clear orthologs, along with >12 000 conventional genes obtained from the NCBI's Reference Sequence database (14) with human–mouse orthologs. GenomeTrafac also allows for the creation of gene groups based on GO functional associations, pathways, diseases or mammalian phenotypes. GenomeTrafac thus serves as both an analysis tool for any individual gene or miRNA for which there is an ortholog, as well as to serve as a pre-processed data source for the analysis of a list of genes or miRNAs using a tool such as CisMols (15) capable of evaluating shared cis-elements across a list of genes derived from a microarray transcriptional profiling experiment.
Computational methods capable of identifying authentic cis-regulatory sequences and predicting their cell-type, developmental stage, or signal transduction-specific activation functions has long been sought as a means to accelerate or overcome the intensive laboratory procedures required to identify and characterize genomic cis-regulatory regions (1,4,16,17). In order to facilitate a variety of search functions, we reasoned that as an initial step, access to pre-computed conserved cis-regulatory regions for most well-annotated conventional and miRNA genes would not only facilitate the exploration of novel gene cis-regulatory regions, but alsohelp guide strategies for the creation of transgenics and the construction of altered genes with minimal impact on potential regulatory regions. For example the design of target constructs necessary to create knock-in mice requires that the locP insertion sites do not disrupt cis-elements necessary for proper regulation, alteration of which could lead to unintended consequences.
Beyond the massive increase in curated content, search and management additions, we have also added a variety of cis-element feature tracks, ability to export data directly into the UCSC Genome browser, and a powerful new search capability called ConCisE (Conserved cis-Element) Scanner that allows all ortholog gene pairs in GenomeTrafac to be queried for potential targets of known TF(s) with defined cis-regulatory target specificity. Cis-regulatory modules that contain various combinations of cis-elements can be composed and used to query the database for potential additional targets.
DATA SOURCES AND DATA PROCESSING
Over 12 000 conventional ortholog genes and 260 miRNA genes were obtained from Homologene, NCBI and MGI (Mouse Genome Informatics)'s curated table of human–mouse orthologs (18) (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/) and miRBase (13), respectively. Only genes with a Reference Sequence (RefSeq) entry, specifically those with accession numbers starting with ‘NM’ were used. We selected the NCBI's RefSeq data set since it provides a comprehensive, integrated, non-redundant set of sequences and also provides a stable reference for gene identification and characterization (14).
For microRNA genes, we extracted the human–mouse ortholog pairs using human miRNA symbols as the common identity (241 of a total 462 human miRNAs were referenced by a mouse miRNA). An additional 28 mouse orthologs were identified based on sequence similarity by matching human miRNAs against the mouse genome using the BLAT algorithm (19). Two of the 241 symbol-based orthologous miRNAs (MIRN500 and MIRN448) and three of the sequence similarity-based human–mouse miRNA pairs (MIRN432, MIRN515-1 and MIRN593) did not show flanking region sequence conservation. These were marked in red font in the GenomeTrafac system as problematic (Supplementary Data 2). Corresponding genomic sequences for both the conventional genes and microRNA genes were extracted from the UCSC Golden path database (19) with 40 kb of flanking (upstream and downstream) nucleotide base pairs (see Supplementary Data 1 for an explanation of selection procedure for flanking region). For genes with multiple isoforms, we considered the longest isoform that encompassed the largest interval on the genome. In case of pseudogenes or genes with more than one chromosome mapping, we compared alignments of the transcript against the genome and distinguished ‘true’ genes from pseudogenes based on either their lack of introns, exons, or conserved intronic or flanking sequence. The exon annotations from the Table Browser of UCSC Golden path were used. However, exonic coordinates were mapped in context with downloaded genomic sequences so as to be able to subsequently indicate exon positions. The outstanding BlastZ algorithm (20), was used to align each of the orthologous genomic sequence pairs. The command-line version of MatInspector (version 7.4) (21) was used to identify the potential binding sites in each of the genomic sequences. The analysis parameters for identification of TF-binding sites were set as 0.7 for the core similarity and ‘optimal’ for the matrix similarity using the MatInspector vertebrate binding sites (Position Weight Matrix) library [Genomatix Matrix Family Library Version 5.0 (January 2005)]. TraFaC server [http://trafac.cchmc.org; (12)] was then used as previously described to find the conserved cis-elements within the BlastZ aligned regions of the ortholog genes by integrating the sequence, alignment and binding sites data for each gene/miRNA pair.
The ConCisE scanner functions are written in JAVA using JAVA Servlet Pages, standard taglibs and JAVA Servlets (for the GUI front end), and Oracle text and stored procedures—against an Oracle 9i database).
NEW FEATURES
Improved querying
The GenomeTrafac database can be now queried using Entrez Gene IDs or probeset IDs (Affymetrix, Illumina) also apart from accession nos or gene/miRNA symbols. Additionally, we have added additional annotation tables to the database and it is now possible to retrieve a list of genes using a GO term, disease term [OMIM (22) or GAD (23)], pathway term (Kegg, Biocarta, Biocyc or Reactome), mammalian phenotype (24), or gene family http://www.gene.ucl.ac.uk/nomenclature/genefamily.html. The GO-term-gene associations and gene-pathway associations were derived from NCBI's Gene Entrez (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/).
Enhanced visualization
The original TraFaC server, an integrated web-based software tool for comparing a pair of orthologous genomic sequences and generating graphical outputs, the Regulogram and Trafacgram, was described earlier (12) (Figure 1A and D). The new features added include the ability to plot the frequency and density of binding sites for each species separately, the display of repeat elements (as green blocks in parallel to the two sequences compared) (Figure 1A). Another new function is to be able to filter the ‘Hits’ (shared binding sites within a 200 bp window) in order to identify and display only those for which the order of occurrence is also conserved between both orthologs. Conservation of order provides an additional stringent filter for which cis-elements that pass have a higher likelihood of being functional. This filter can be invoked from both the Regulogram and Trafacgram images. A separate graph of the parallel elements is able to be requested in the Regulogram view, and their peak is thus able to be used as a decision basis for exploring a particular cluster. To do this, the Trafacgram is requested from the high-scoring peak, overall cis-elements are shown, from which a request for the parallel cis-elements can then be made. An important concern is to evaluate binding site definitions and the confidence in its potential to be functional. For most of the sites, the consensus sequence can give a rough idea whether it can be functional or not. To allow for this, we have also now shown for each of the shared conserved predicted cis-elements the actual sequence and its position in the genomic sequence as a table below each of the Trafacgrams. Links are also provided to the UCSC genomic browser. Another feature allows the sequences of any segments of interest to be downloaded.
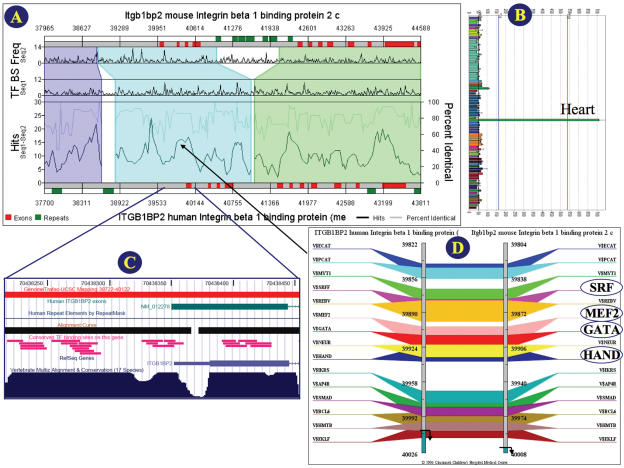
Figure 1.
(A) Regulogram depiction of shared cis-elements between two sequences in the context of their sequence similarity: The two sequences, mouse and human ITGB1BP2 (melusin), are represented as horizontal bars. The red colored segments on these bars are exons. The green colored bars shown parallel to the genomic sequences represent the repeat regions. The regions of sequence alignment are represented as different colored quadrilaterals that relate one sequence to another. Within each shaded block, the percent sequence similarity and the number of TF-binding sites are represented as two separate line graphs in the lower half. The frequencies of individual binding sites occurring in each of the sequences separately are shown as two running graphs in the top half of the pane. The percent similarity is the average sequence conservation as determined by the BlastZ algorithm and the shared cis-element hits are determined by an algorithm that uses a 200 bp moving window that looks through the cis-elements that are present within the conserved sequence block. Numbers are nucleotide positions. Regulogram can be clicked to zoom in or view the TF-binding sites that are in common between the two sequences at the click-point coordinate. (B) Example of GNF expression pattern for the human gene ITGBIBP2 showing very specific heart expression. The bar-graph is based on that from http://expression.gnf.org/ (C) Visualizing GenomeTrafac tracks on the UCSC Genome browser: The pink horizontal bars are the conserved TFBSs from GenomeTrafac. Image shown here corresponds to the flanking regions of the first exon of human gene ITGB1BP2. (D) TraFaC image of a cis-element dense upstream region of ITGB1BP2: The two gray vertical bars are the two genes (human and mouse ITGB1BP2) that are compared. The numbers represent the nucleotide positions with respect to the sequences used. The TF-binding sites occurring in both the genes are highlighted as various colored bars drawn across the two genes. The positional and extent of consecutive individual elements in potential regulatory clusters tend to be highly conserved. Putative binding sites for cardiogenic TFs (SRF, MEF2, GATA and HAND) are highlighted. This cardiogenic TFBSs cluster could be responsible for the cardiac overexpression (see panel B) of the human gene ITGB1BP2.
Non-conserved cis-elements within conserved sequence regions
When finding the conserved transcription factor binding sites (TFBSs) in orthologous human and mouse sequence conserved regions, most of the phylogenetic approaches return only sites which are aligned between the two orthologous sequences [e.g. ConSite (25)]. The drawback of such approach is the possibility of losing a fraction of TFBSs which although functional are not conserved. However, the GenomeTraFac system considers all sites in an overall conserved segment, rather than insisting on direct positional conservation at the cis-element site itself. This is important because it is possible for a predicted TFBS in human to be in a segment that aligns with mouse, and a predicted TFBS also be in the orthologous mouse sequence, but the two TFBSs may not align. For instance, in ENO1, one of the human canonical E-boxes in the promoter region is conserved in intronic mouse canonical E-box that lies in 1 kb downstream of the transcriptional start site. The two canonical E-boxes found in human promoter correspond to a mouse non-canonical E-box. The increased sensitivity of TraFaC in this case is due to less stringent criteria of including non-canonical E-boxes that occur in the larger region of at least 50% identity. TraFaC does not require local sequence alignment of the E-boxes for inclusion as predicted TF-binding sites (26).
Even though conserved non-coding regions are valuable for identification of regulatory switches, sequence similarity does not always decipher to cis-element conservation. For instance, the gene CALB3 (Calbindin), is transcriptionally regulated by estrogen in the uterus mediated by an estrogen-responsive element identified in the gene (27). A comparison of the human and mouse promoter regions reveals that this site is not conserved in the orthologs. To overcome this we have added a new feature that displays the individual TF-binding site frequency peaks of the orthologs on the regulograms (Figure 1A). This would help in identifying those cis-elements also which are not evolutionarily conserved in spite of sequence conservation. It also helps in understanding the subtle mechanism at the transcriptional level that might cause profound differences in the regulation of certain genes in humans and mice. Finally, the human or mouse specific cis-elements (cis-elements that are not shared) occurring within specific human–mouse sequence conserved blocks warrant a careful and in-depth analysis.
Order and extent of binding sites
The regulatory regions frequently manifest as clusters of cis-elements with the positions highly conserved. In other words, the elements are almost parallel to another. In the trafacgrams, a number of times, these parallel occurring clusters of cis-elements are camouflaged in the background noise of cis-elements crisscrossing all over. To filter the image in all such cases, we provide the option to display only those clusters which are uniformly spread. The user has the option to set the distance between two successive cis-elements, default being 10 bp. A genome-wide systematic study of the order and extent versus ortholog shuffling of cis-elements within a sequence conserved region is required.
Export features for plug-in to genome browsers
In addition to being able to view and browse potential regulatory cluster rich regions, it is also important that the data be readily connected to additional annotation sources, such that a user can easily determine the identity and attributes of a potential regulatory region, which for instance might be conserved in other species too. A common user request since the availability of TraFaC server is to add multispecies and not limit to a pairwise comparison. Of the mammalian genomes, human and mouse are the most complete so we decided to continue to use them. Additionally, the MultiPipMaker, ECR browser [http://ecrbrowser.dcode.org/; (28)] and the UCSC or Ensembl browsers to some extent can be used for this purpose. However, to cater to this specific request we have included the export feature of each gene as a general feature format (GFF) text files. It is a record (or feature) based tab-separated nine-column table with each row representing one feature. The definition of feature is very flexible, which can be exon, variation, repeat element, TFBS, etc. Our main concern was the compatibility with UCSC Genome browser, so we included browser and track lines for display purposes; a wiggle track was also included for displaying cross-species conservation. The detailed descriptions can be found at http://genome.ucsc.edu/goldenPath/help/customTrack.html. These UCSC browser-compatible GFF files can be uploaded to UCSC's golden path. This facilitates viewing a predicted region in the context of other features together with dozens of aligned annotation tracks (known genes, predicted genes, expressed sequence tags (ESTs), mRNAs, CpG islands, assembly gaps and coverage, chromosomal bands, gene expression, other species homologies and more) (Figure 1B and C), adding valuable additional insights to the predicted regulatory regions. Another frequent request is to add the experimentally validated enhancers or silencers info to the regulograms and trafacgrams. We intend to add these features as part of our future development. The biggest bottleneck however is the gross under representation of the enhancer/silencer information in the GenBank.
ConCisE scanner—identification of potential gene targets using cis-clusters as probes
Though servers and programs like TraFaC (12), rVISTA (5), ConSite (25), help in identification of potential regulatory regions through comparative sequence analysis, none of these will reveal or help in the identification of target genes through which the TFs exert their function. Even taking into account the >300 TFBSs, it would be impossible to search for targets of all combinations of these factors—there are too many possibilities. Even if we tried all possible pairs of factors, it is likely that every region of genome would have a high binding-site density for some collection of factors. And even though it's unlikely that all aspects of regulation can be inferred from comparative analyses, limiting the target regions to relatively highly conserved sequence blocks with ortholog-shared target sites could be fruitful. Providing a complement to the above listed phylogenetic approaches, the newly included ConCisE (Conserved cis Element) Scanner feature undertakes a more targeted search, finding phylogenetically conserved regulatory targets of defined TFs whose DNA binding site specificity is known. There have been reports of this approach for the genomes of Caenorhabditis elegans and Caenorhabditis briggsae (29) and Drosophila species (30). While the former, CisOrtho (29), is based on phylogenetic approach, the latter, Target Explorer (30), is not. The SynoR (31) also uses a similar approach for higher eukaryotes. The principal advantages SynoR has over our approach are the multi-species comparison and the annotations of the potential target genes. However, current version of SynoR doesn't support the Boolean ‘OR’ when searching for putative gene targets using TFBSs. The main advantage of our approach over SynoR is that the target genes are not only identified but also apart from the queried sites, other elements occurring in the cluster are also displayed. This helps in identifying potential novel interacting TFs.
Even though context-dependent methods such as combined promoter sequence analysis with TF perturbation experiment (32) data, or chromatin immunoprecipitation ‘ChIP-chip’ experiments (33) can greatly improve likelihood of functional validation of target gene regulatory regions and cis-element clusters, context-independent approaches identifying all TF-binding sites and combinations of sites in the genome on the basis of sequence analysis only [e.g. ConCisE Scanner and SynoR (31)] should continue to be highly valuable for extending and exploring knowledge gained in specific contexts (34).
CONCLUSION
Comparative genomics has greatly accelerated our ability to identify fundamental functional elements of genome structures. The development of new experimental and computational methods have also proven capable of enabling deep whole-genome annotation of sequences (35), including not only identification but also classification of functional regulatory elements. At the next stage of development, large-scale systematic comparative genomics approaches may be combined with extensive expression and other information to improve our ability to recognize transcriptional controls responsible for regulatory network behaviors. Towards this, our human–mouse miRNA and gene cis-regulatory database can be a particularly valuable resource for finding regulatory architecture providing experimental dissections specifically target factors within the regulatory networks. GenomeTrafac will be updated with new human–mouse gene/miRNA pairs regularly providing that corresponding reference sequences are available or the annotations are improved. Finally, even though experimental validation is the ultimate litmus test for any computationally predicted cis-regulatory modules, methods or resources that help to focus likely effects and defined mechanisms should provide vital direction.
AVAILABILITY
The GenomeTrafac database can be accessed freely at http://genometrafac.cchmc.org.
SUPPLEMENTARY DATA
Supplementary Data are available at http://genometrafac.cchmc.org/genome-trafac/supplementary.
Acknowledgments
This work was supported by grants NCI UO1 CA84291-07 (Mouse Models of Human Cancer Consortium), NIEHS ES-00-005 (Comparative Mouse Genome Centers Consortium) and NIEHS P30-ES06096 (Center for Environmental Genetics). Funding to pay the Open Access publication charges for this article was provided by CCHMC, Cincinnati, OH, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ureta-Vidal A., Ettwiller L., Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nature Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- 2.Cooper G.M., Sidow A. Genomic regulatory regions: insights from comparative sequence analysis. Curr. Opin. Genet. Dev. 2003;13:604–610. doi: 10.1016/j.gde.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Miller W., Makova K.D., Nekrutenko A., Hardison R.C. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- 4.GuhaThakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585–3598. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loots G.G., Ovcharenko I., Pachter L., Dubchak I., Rubin E.M. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohler U., Yekta S., Lim L.P., Bartel D.P., Burge C.B. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–1322. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 9.Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 10.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conaco C., Otto S., Han J.J., Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl Acad. Sci. USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jegga A.G., Sherwood S.P., Carman J.W., Pinski A.T., Phillips J.L., Pestian J.P., Aronow B.J. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 2002;12:1408–1417. doi: 10.1101/gr.255002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence project: update and current status. Nucleic Acids Res. 2003;31:34–37. doi: 10.1093/nar/gkg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jegga A.G., Gupta A., Gowrisankar S., Deshmukh M.A., Connolly S., Finley K., Aronow B.J. CisMols Analyzer: identification of compositionally similar cis-element clusters in ortholog conserved regions of coordinately expressed genes. Nucleic Acids Res. 2005;33:W408–W411. doi: 10.1093/nar/gki486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasserman W.W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 17.Michelson A.M. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc. Natl Acad. Sci. USA. 2002;99:546–548. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., et al. The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker K.G., Barnes K.C., Bright T.J., Wang S.A. The genetic association database. Nature Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 24.Smith C.L., Goldsmith C.A., Eppig J.T. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenhard B., Sandelin A., Mendoza L., Engstrom P., Jareborg N., Wasserman W.W. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J.W., Zeller K.I., Wang Y., Jegga A.G., Aronow B.J., O'Donnell K.A., Dang C.V. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darwish H., Krisinger J., Furlow J.D., Smith C., Murdoch F.E., DeLuca H.F. An estrogen-responsive element mediates the transcriptional regulation of calbindin D-9K gene in rat uterus. J. Biol. Chem. 1991;266:551–558. [PubMed] [Google Scholar]
- 28.Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigelow H.R., Wenick A.S., Wong A., Hobert O. CisOrtho: a program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics. 2004;5:27. doi: 10.1186/1471-2105-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sosinsky A., Bonin C.P., Mann R.S., Honig B. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 2003;31:3589–3592. doi: 10.1093/nar/gkg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovcharenko I., Nobrega M.A. Identifying synonymous regulatory elements in vertebrate genomes. Nucleic Acids Res. 2005;33:W403–W407. doi: 10.1093/nar/gki466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 33.Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 34.Li H., Wang W. Dissecting the transcription networks of a cell using computational genomics. Curr. Opin. Genet. Dev. 2003;13:611–616. doi: 10.1016/j.gde.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 35.ENCODE Project Consortium. The ENCODE (ENCyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]